Introduction
Hepatocellular carcinoma (HCC) is one of the most
common types of cancer worldwide, and ranks second as a cause of
cancer-related mortality in China (1). The majority of the patients are
diagnosed at an intermediate or advance stage owing to lack of
surveillance. Currently, the Barcelona Clinic Liver Cancer (BCLC)
staging system is the most common staging system for HCC. BCLC
divides HCC into three stages, namely early, intermediate and
advanced (2,3). According to the BCLC recommendations,
intermediate and advance HCC are not candidates for curative
resection, indicating a relatively poor prognosis of these patients
(4). Therefore, it is crucial to
identify new biomarkers for early diagnosis and prediction of
prognosis of HCC.
MicroRNAs (miRs) are a class of non-coding RNAs
(18–25 nucleotides) that bind to the 3′-untranslated region and
regulate translation at the post-translational level (5). More than 3,700 miRs have been
registered in the miRBase to date (6). Over the past decades, miRs have been
found to play key roles in tumorigenesis, cancer progression and
metastasis. In particular, miR-21 expression has been found to be
upregulated in breast, lung and cervical cancer, as well as in HCC
(7), and numerous studies have
investigated the association between miR-21 expression and the
prognosis of HCC (8–10). miR-21 is considered as a promising
biomarker and treatment target in HCC, and several systematic
reviews have investigated the association between miR-21 expression
and the prognosis of HCC. Wang et al investigated the
diagnostic and prognostic value of miR-21 in cancer, and observed
that high expression of miR-21 was significantly correlated with
poor prognosis of cancer patients; however, only two studies in
their analysis reported survival data from HCC patients (11). Yan et al reported that miR-21
maybe complementary to α-fetoprotein (AFP) in the diagnosis of HCC,
and that high expression of miR-21 indicated poor prognosis of HCC
(12); however, only 4 studies were
included in their review, which limits the reliability of their
conclusions. Taking into consideration that different investigators
reported inconsistent information on the role of miR-21 in HCC, the
present systematic review was performed to investigate the value of
miR-21 expression in predicting the prognosis of HCC.
Data collection methods
Search strategy and study
selection
‘miR-21’ or ‘microRNA-21’ and ‘hepatocellular
carcinoma’ or ‘HCC’ were used as mesh terms to search the PubMed,
Science Citation Index, EMBASE and CNKI databases. There was no
limitation in terms of publication year and region, but the
language was limited to English. The cited references of the
obtained articles were also scanned to identify additional relevant
studies.
Studies were included if they met the following
criteria: Studies on the role of miR-21 expression in patients with
HCC; and studies with adequate data on survival outcome and
clinical characteristics. Studies were not eligible if they were
reviews, letters, or conference abstracts. Studies designed on
animals or cell lines, and studies without adequate data for
survival analysis, were also excluded. The study selection process
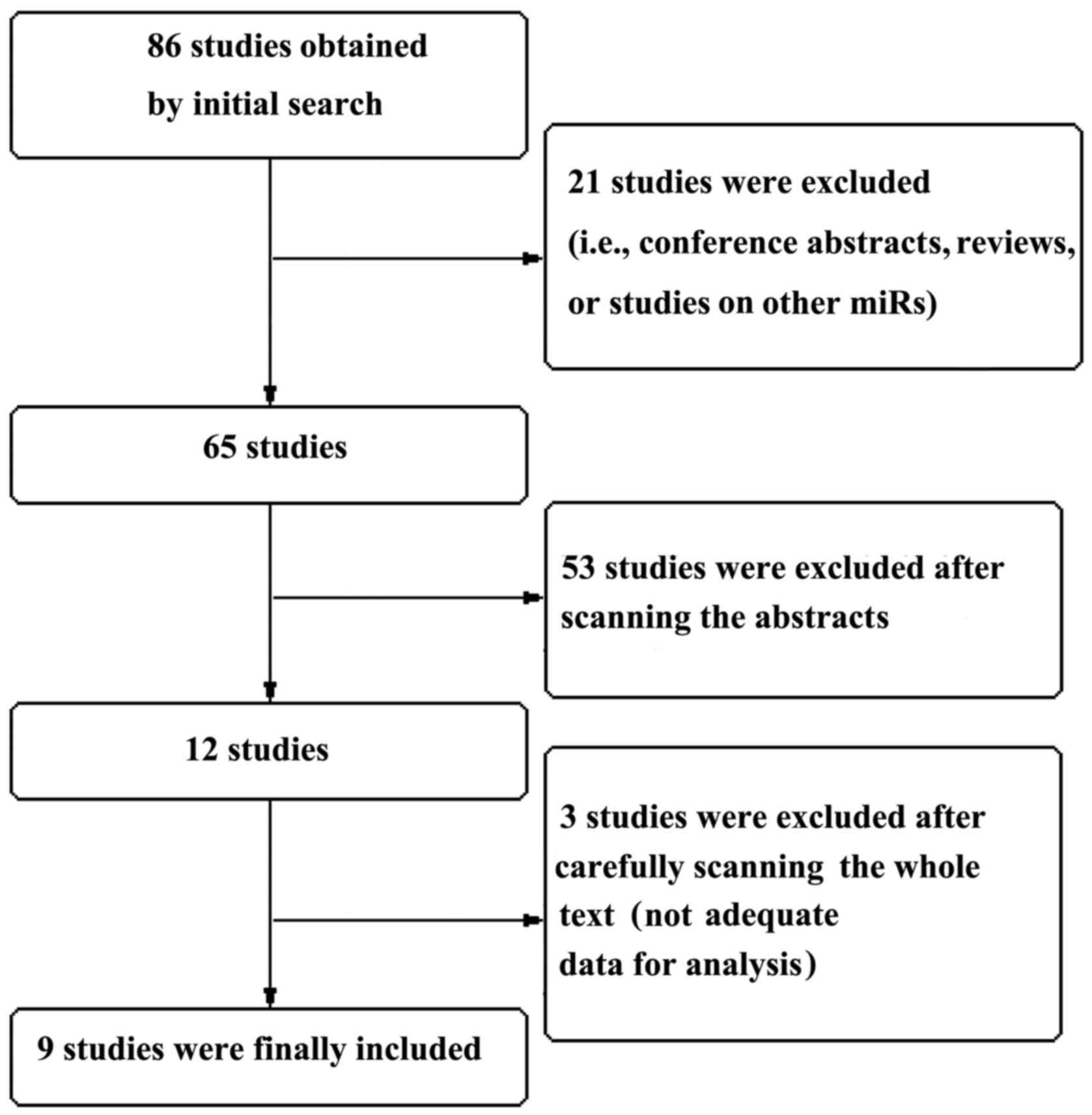
is summarized in Fig. 1.
Quality assessment
Two investigators independently assessed the quality
of the included studies; if a disagreement emerged, a third
investigator was contacted. The quality of the studies included in
the meta-analysis was evaluated based on the Newcastle-Ottawa scale
(13). Three aspects were generally
assessed: Selection of the cohort, comparability of the cohort and
ascertainment of the outcome (Table
I). A study with a score of >5 was considered as being of
high methodological quality (14).
 | Table I.Quality assessment of the included
studies. |
Table I.
Quality assessment of the included
studies.
| Study | Selection of
cohort | Comparability | Ascertainment of the
outcome | Total score | (Refs.) |
|---|
| Wang et
al | 3 | 2 | 3 | 8 | (25) |
| Huang et
al | 2 | 1 | 2 | 5 | (26) |
| Wang et
al | 2 | 1 | 3 | 6 | (27) |
| Wang et
al | 2 | 1 | 3 | 6 | (28) |
| Liu et al | 2 | 1 | 2 | 5 | (29) |
| Gyöngyösi et
al | 2 | 1 | 3 | 6 | (30) |
| Karakatsanis et
al | 3 | 1 | 3 | 7 | (10) |
| Tomimaru et
al | 3 | 2 | 3 | 8 | (9) |
| Tomimaru et
al | 2 | 1 | 3 | 6 | (8) |
Data extraction
Two investigators independently extracted baseline
characteristics from each study, including study name, publication
year, country, patient number, tumor stage, specimens, RNA
detection method and cut-off value of miR-21 expression. Survival
data and clinical characteristics were also extracted. The survival
outcome was measured by overall survival (OS) rate, and the
clinical characteristics included tumor size, tumor number,
differentiation, venous invasion, TNM stage and liver cirrhosis.
Differentiation was classified as high or low (moderate
differentiation was classified as low), and the TNM stage was
divided into stages I+II and III+IV.
Statistical analysis
Prognostic efficacy was measured by pooled hazard
ratio (HR) of OS with 95% confidence interval (95% CI). If the HR
of OS was not provided directly in the main text, it was calculated
from Kaplan-Meier curves, as described by Tierney et al
(15). The correlation between
miR-21 expression and clinical characteristics was measured by odds
ratio (OR) with 95% CI. All OR values were calculated by the
Chi-square test using data provided by the investigations.
Heterogeneity was assessed using Cochran's Q test and Higgins's
I2 statistics; P<0.1 was considered to indicate
significant heterogeneity. If heterogeneity was detected, the
random-effects model was used; otherwise, the fixed-effects model
was adopted. Publication bias was detected by funnel plots.
Statistical significance was set at P<0.05. All the statistics
were performed by RevMan software, version 5.3 (Cochrane, London,
UK).
Results
Characteristics of included
studies
A total of 86 studies were obtained following an
initial search through the PubMed, Science Citation Index, Embase
and CNKI databases. A total of 21 studies were initially excluded
(conference abstracts, reviews, systematic reviews, or studies
investigating other miRs). A further 53 studies were excluded after
scanning the abstracts, as they did not meet the inclusion
criteria, and another 3 studies were excluded following a full-text
review. Finally, 9 studies were included in the analysis. The
baseline characteristics of the included studies are summarized in
Table II.
 | Table II.Baseline characteristics of the
included studies. |
Table II.
Baseline characteristics of the
included studies.
| Study | Year | Country | Stage | Patient no. | Specimen | Method | Cut-off value | (Refs.) |
|---|
| Wang et
al | 2015 | China | TNM I–IV | 97 | Serum | RT-PCR | Median | (25) |
| Huang et
al | 2015 | China | TNM I–IV | 112 | Tissue | RT-PCR | Median | (26) |
| Wang et
al | 2014 | China | TNM I–IV | 119 | Tissue | RT-PCR | Median | (27) |
| Wang et
al | 2014 | China | TNM I–IV | 30 | Serum | RT-PCR | 5-fold of
control | (28) |
| Liu et
al | 2014 | China | BCLC A-C | 136 | Serum | RT-PCR | Median | (29) |
| Gyöngyösi et
al | 2014 | Italy | NR | 20 | Tissue | RT-PCR | Median | (30) |
| Karakatsanis et
al | 2013 | Greece | TNM I–IV | 60 | Tissue | RT-PCR | 3.07-fold of
control | (10) |
| Tomimaru et
al | 2012 | Japan | TNM I–IIIa | 126 | Plasma | RT-PCR | 0.754 | (9) |
| Tomimaru et
al | 2010 | Japan | BCLC C | 30 | Tissue | RT-PCR | Median | (8) |
Correlation between high miR-21
expression and prognosis of HCC
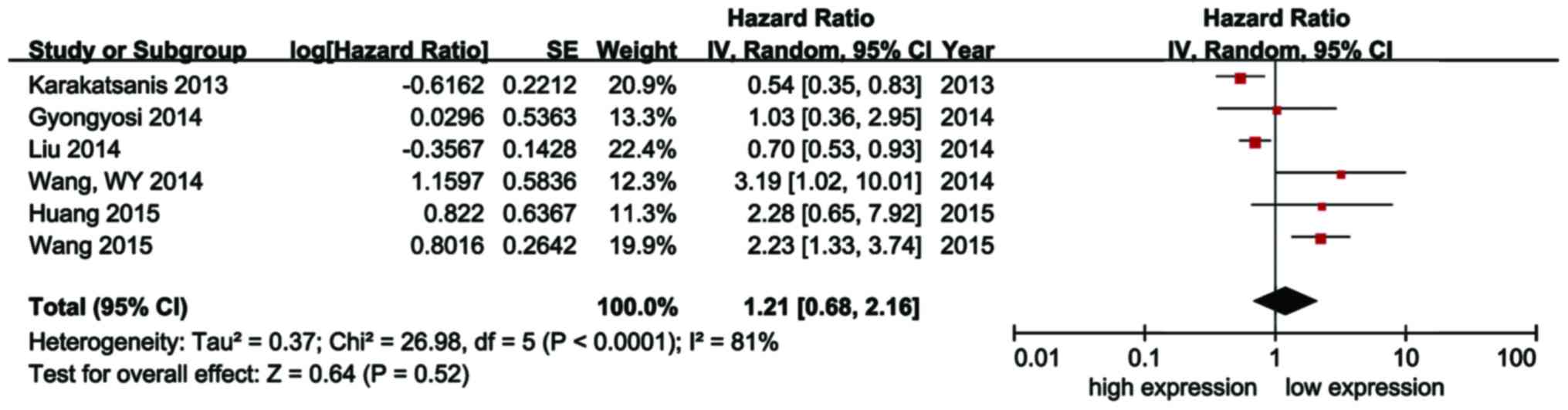
A total of 6 studies reported the HR of the OS of
patients with HCC. Significant heterogeneity was detected among
these 6 studies (I2=81%, P<0.1) and the random-effects model was
used to estimate the overall effect. It was observed that high
expression of miR-21 was not correlated with poor OS in HCC
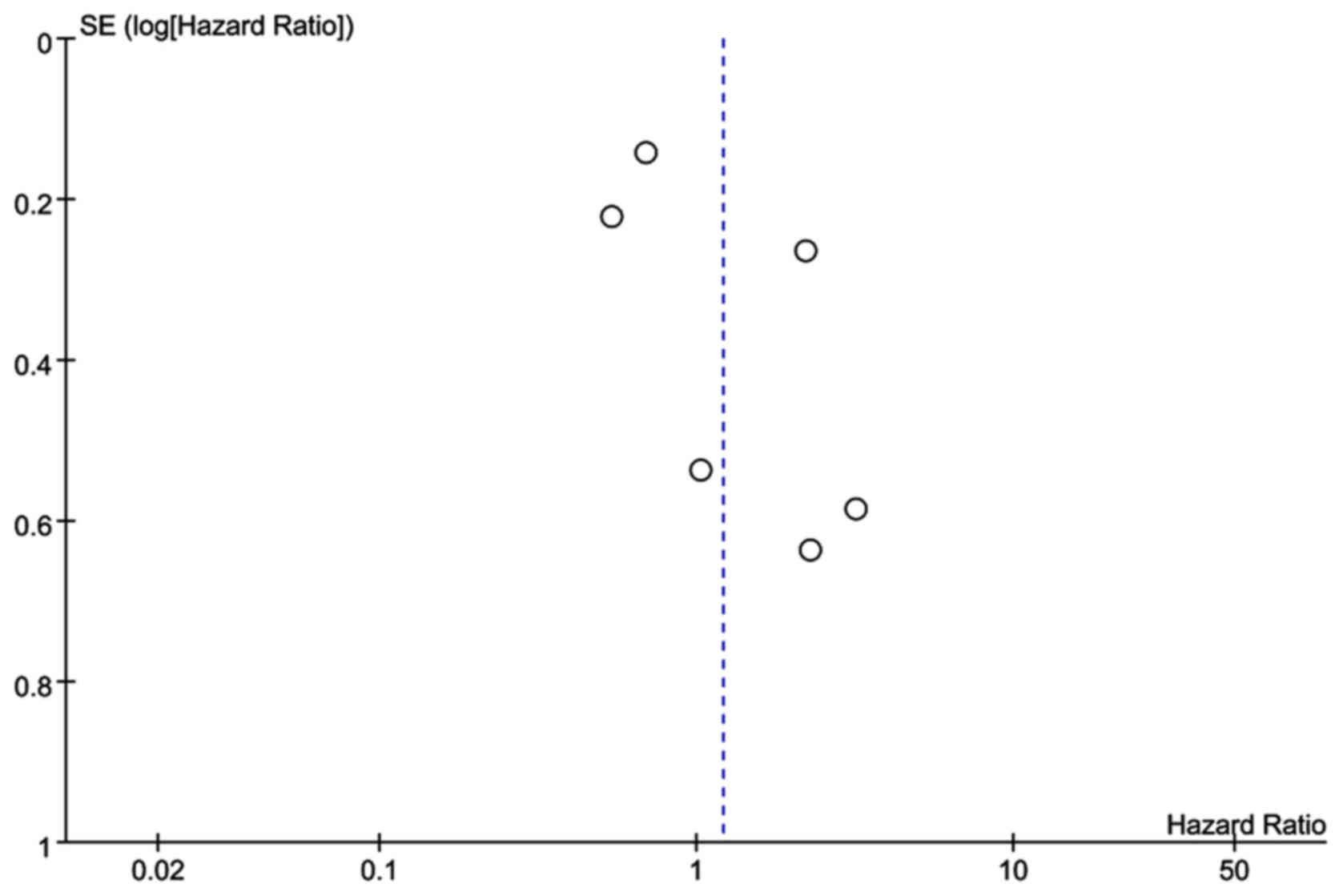
patients (HR=1.21; 95% CI: 0.68–2.16; P=0.52; Fig. 2). No publication bias was identified
by the Begg's test (P>0.05; Fig.
3).
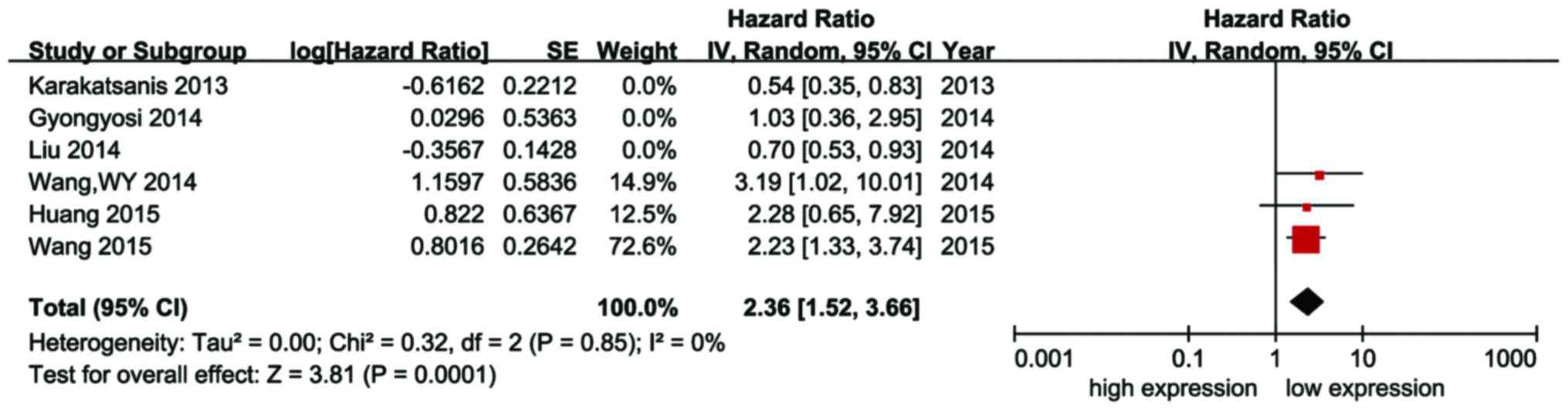
A total of 3 studies reported data on the
association between the expression of miR-21 and the survival of
HCC patients receiving curative resection. No significant
heterogeneity was observed among these 3 studies and the
fixed-effects model was used to estimate the overall effect.
Subgroup analysis revealed that high expression of miR-21 was
associated with poor OS (HR=2.36; 95% CI: 1.52–3.66; P<0.01) in
HCC patients receiving curative resection (Fig. 4).
Association of high expression of
miR-21 with clinical characteristics of HCC
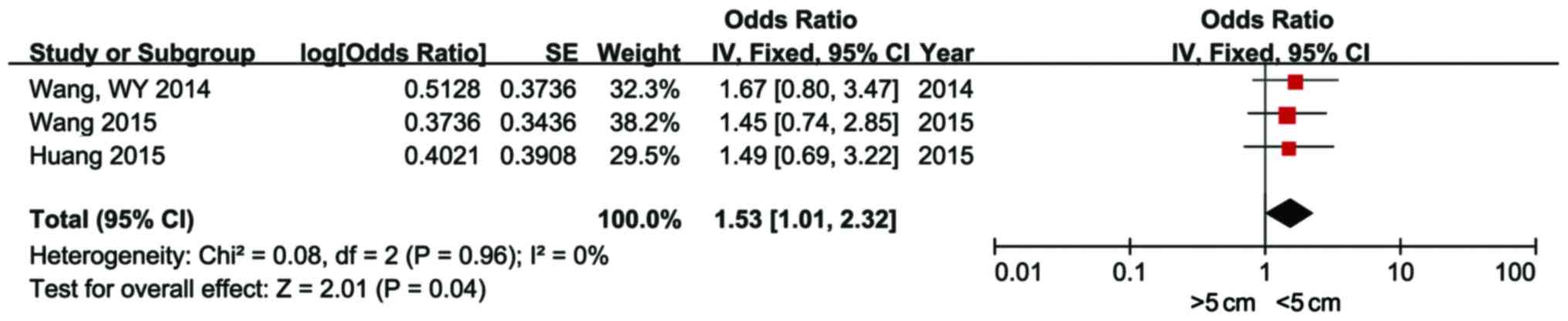
It was observed that high expression of miR-21 was
correlated with tumor size >5 cm (OR=1.53; 95% CI: 1.01–2.32;
P=0.04), venous invasion (OR=4.86; 95% CI: 1.47–16.08; P=0.01), TNM
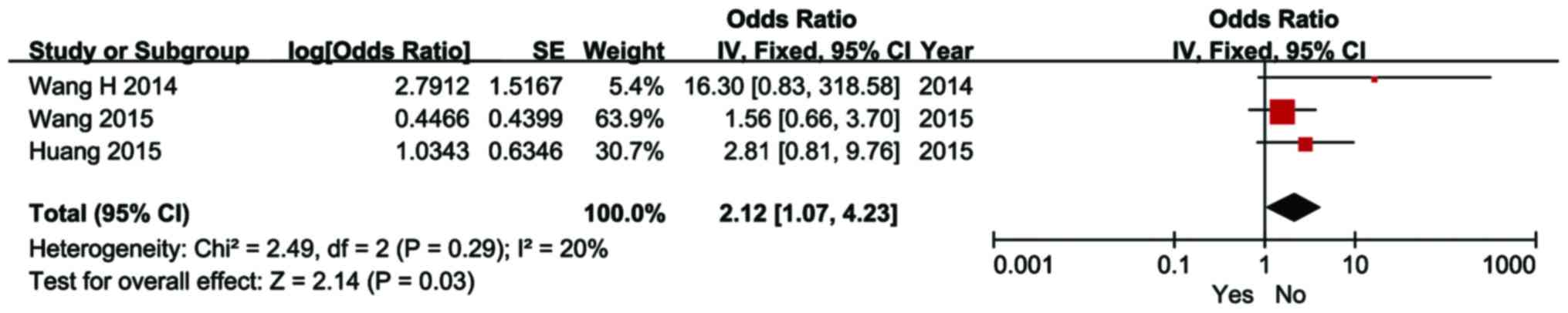
stage (OR=3.44; 95% CI: 1.56–7.59; P<0.01) and liver cirrhosis
(OR=2.12; 95% CI: 1.07–4.23; P=0.03). However, high expression of
miR-21 was not found to be significantly correlated with tumor
number (P=0.44) or differentiation (P=0.83). Details on the
association of high expression of miR-21 with the clinical
characteristics of HCC are presented in Figs. 5–8.
Discussion
In the present analysis, higher miR-21 expression
was not found to be significantly associated with poor prognosis of
all HCC patients; however, high expression of miR-21 was found to
be correlated with poor prognosis of HCC patients receiving
curative resection. Zhou et al reported that miR-21 may be a
potential prognostic biomarker for several carcinomas, including
breast, colon and lung cancer, with an HR of 1.91, and the subgroup
analysis revealed that elevated miR-21 expression was associated
with poor prognosis of liver cancer based on the results of 3
studies (16); however, those 3
studies differed in the pathological type of the liver cancer (2
studies enrolled patients diagnosed with cholangiocarcinoma,
whereas1 enrolled patients with HCC). Thus, it is difficult to
assess the prognostic value of miR-21 in HCC from that study.
Subsequently, Wang et al investigated the value of miR-21 in
cancer diagnosis and prognosis, and concluded that miR-21 may be a
prognostic marker in digestive tract cancers with an HR of 5.77
(11), whereas only 2 studies were
designed to explore the diagnostic and prognostic value of miR-21
in HCC, in which the small patient sample limited the reliability
of the conclusions. Yan et al investigated the prognostic
and diagnostic value of miR-21 in early HCC, based on the pooled
results of sensitivity, specificity and area under the curve, and
concluded that miR-21 may be a complementary factor in HCC
diagnosis (12); in addition, the
pooled results suggested that high expression of miR-21 was
associated with poor OS of early HCC. That study by Yan et
al only enrolled patients with early HCC; however, as our study
enrolled patients with early and intermediate HCC, our patient
sample was more representative of that patient population.
In our review, 6 studies with a total of 730
patients were eligible for analysis, covering all stages of HCC,
and the larger patient sample in our analysis made the findings
more reliable compared with the abovementioned studies. Subgroup
analysis revealed that high expression of miR-21 was correlated
with poor prognosis of HCC patients receiving curative resection.
Previous studies demonstrated that miR-21 may promote liver
regeneration following partial hepatectomy (17,18). It
may be hypothesized that miR-21 can simultaneously promote liver
regeneration in normal hepatocytes and tumor cells, resulting in
poor prognosis of HCC patients receiving liver resection; however,
the mechanism underlying the association of miR-21 expression and
poor prognosis of HCC following resection requires further
investigation.
On investigating the correlation between higher
miR-21 expression and the clinical characteristics of HCC, it was
observed that higher miR-21 expression was significantly correlated
with tumor size >5 cm, venous invasion, TNM stage and liver
cirrhosis. Yan et al also observed that higher expression of
miR-21 was associated with HCC TNM stage (12), which was consistent with our
findings. However, compared with the findings of Yan et al,
we found that more clinical characteristics were associated with
higher miR-21 expression.
Considering the mechanism underlying the role of
miR-21 in HCC, various studies have reported potential signaling of
miR-21 in tumorigenesis, progression and metastasis of HCC.
Verified targets of miR-21 include RASGRP1, BCL2, RPS7, PTEN, E2F1
and PDCD4 (6,19). Modulation of miR-21 has been shown to
regulate the translation of PTEN; hence, miR-21 may play a key role
in PTEN-dependent pathways involved in cancer cell growth,
migration and invasion (20,21). Recently, miR-21 has been demonstrated
to mediate sorafenib resistance of HCC by suppressing autophagy via
the PTEN/Akt pathway (22). PDCD4 is
a known tumor suppressor mediating the apoptosis of tumor cells and
repressing the development of HCC. miR-21 was demonstrated to
promote migration and invasion via the miR-21-PDCD4-AP-1 feedback
loop in HCC (23), and Qiu et
al reported that miR-21 may deregulate the expression of PDCD4
in patients with hepatitis B virus infection, and promote
tumorigenicity (24). Taken
together, these findings indicate that miR-21 may promote tumor
cell growth, migration and invasion, and inhibit tumor cell
apoptosis through various pathways, resulting in tumor lesions of a
larger size, higher incidence of venous invasion and more advance
stage.
The present study had certain limitations: First,
heterogeneity was observed between studies included in the survival
analysis and the test specimens of miR-21 differed among these
studies, which increased the selection bias and decreased the
reliability of our results. Second, the OR values were calculated
from data provided in the main text, and most investigators had not
provided the precise OR values in their studies. Third, the sample
size of the studies included in our analysis was still
comparatively small, and more well-designed studies are required to
elucidate the prognostic value of miR-21 in HCC.
In conclusion, miR-21 cannot be considered as a
factor complementary to AFP, microvascular invasion and advanced
tumor stage in predicting the prognosis of HCC. However, higher
expression of miR-21 may be a promising biomarker associated with
certain clinicopathological characteristics of HCC, such as tumor
size, venous invasion, TNM stage and liver cirrhosis.
Acknowledgements
The authors would like to acknowledge the support of
the staff of the Department of Pathology and Clinical Sample Bank
of West China Hospital.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 71673193), and the Key
Technology Research and Development Program of the Sichuan Province
(grant no. 2015SZ0131).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JSL designed this study, PSY wrote the paper and
performed data analysis. Both authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Dr PSY majors in liver surgery and basic research on
hepatology; Professor JSL is the Chief Director of Department of
Hepatobiliary Pancreatic Disease of the Affiliated Hospital of
North Sichuan Medical College (Sichuan, China).
References
|
1
|
Chen KW, Ou TM, Hsu CW, Horng CT, Lee CC,
Tsai YY, Tsai CC, Liou YS, Yang CC, Hsueh CW and Kuo WH: Current
systemic treatment of hepatocellular carcinoma: A review of the
literature. World J Hepatol. 7:1412–1420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cillo U, Vitale A, Grigoletto F, Farinati
F, Brolese A, Zanus G, Neri D, Boccagni P, Srsen N, D'Amico F, et
al: Prospective validation of the Barcelona Clinic Liver Cancer
staging system. J Hepatol. 44:723–731. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yi PS, Zhang M and Xu MQ: Management of
the middle hepatic vein in right lobe living donor liver
transplantation: A meta-analysis. J Huazhong Univ Sci Technolog Med
Sci. 35:600–605. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yi PS, Zhang M, Zhao JT and Xu MQ: Liver
resection for intermediate hepatocellular carcinoma. World J
Hepatol. 8:607–615. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Montani F and Bianchi F: Circulating
cancer biomarkers: The macro-revolution of the micro-rna.
EBioMedicine. 5:4–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pfeffer SR, Yang CH and Pfeffer LM: The
role of mir-21 in cancer. Drug Dev Res. 76:270–277. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomimaru Y, Eguchi H, Nagano H, Wada H,
Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M,
Umeshita K, et al: MicroRNA-21 induces resistance to the
anti-tumour effect of interferon-α/5-fluorouracil in hepatocellular
carcinoma cells. Br J Cancer. 103:1617–1626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomimaru Y, Eguchi H, Nagano H, Wada H,
Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I,
Umeshita K, et al: Circulating microRNA-21 as a novel biomarker for
hepatocellular carcinoma. J Hepatol. 56:167–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karakatsanis A, Papaconstantinou I,
Gazouli M, Lyberopoulou A, Polymeneas G and Voros D: Expression of
microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c,
miR-221, miR-222, and miR-223 in patients with hepatocellular
carcinoma or intrahepatic cholangiocarcinoma and its prognostic
significance. Mol Carcinog. 52:297–303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao
H, Sun Q, Yan F, Yan C, Li H and Ren X: Diagnostic and prognostic
value of circulating miR-21 for cancer: A systematic review and
meta-analysis. Gene. 533:389–397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan SR, Liu ZJ, Yu S and Bao YX:
Investigation of the value of miR-21 in the diagnosis of early
stage HCC and its prognosis: A meta-analysis. Genet Mol Res.
14:11573–11586. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in meta-analyses. Eur J Epidemiol.
25:603–605. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lo CK, Mertz D and Loeb M:
Newcastle-Ottawa Scale: Comparing reviewers' to authors'
assessments. BMC Med Res Methodol. 14:452014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tierney JF, Stewart LA, Ghersi D, Burdett
S and Sydes MR: Practical methods for incorporating summary
time-to-event data into meta-analysis. Trials. 8:162007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou X, Wang X, Huang Z, Wang J, Zhu W,
Shu Y and Liu P: Prognostic value of miR-21 in various cancers: An
updating meta-analysis. PLoS One. 9:e1024132014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Juskeviciute E, Dippold RP, Antony AN,
Swarup A, Vadigepalli R and Hoek JB: Inhibition of miR-21 rescues
liver regeneration after partial hepatectomy in ethanol-fed rats.
Am J Physiol Gastrointest Liver Physiol. 311:G794–G806. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen X, Song M, Chen W,
Dimitrova-Shumkovska J, Zhao Y, Cao Y, Song Y, Yang W, Wang F,
Xiang Y and Yang C: Microrna-21 contributes to liver regeneration
by targeting pten. Med Sci Monit. 22:83–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu S, Tao R, Wang S, Wang C, Zhao X, Zhao
H, Li L, Zhu S, He Y, Jiang X and Gao Y: MicroRNA-21 promotes cell
proliferation in human hepatocellular carcinoma partly by targeting
HEPN1. Tumour Biol. 36:5467–5472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bao L, Yan Y, Xu C, Ji W, Shen S, Xu G,
Zeng Y, Sun B, Qian H, Chen L, et al: MicroRNA-21 suppresses PTEN
and hSulf-1 expression and promotes hepatocellular carcinoma
progression through AKT/ERK pathways. Cancer Lett. 337:226–236.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He C, Dong X, Zhai B, Jiang X, Dong D, Li
B, Jiang H, Xu S and Sun X: MiR-21 mediates sorafenib resistance of
hepatocellular carcinoma cells by inhibiting autophagy via the
PTEN/Akt pathway. Oncotarget. 6:28867–28881. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu Q, Wang Z, Hu Y, Li J, Li X, Zhou L
and Huang Y: miR-21 promotes migration and invasion by the
miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma.
Oncol Rep. 27:1660–1668. 2012.PubMed/NCBI
|
|
24
|
Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao
Y, Li Y, Zeng T, Hu J, Zhang L, et al: HBx-mediated miR-21
upregulation represses tumor-suppressor function of PDCD4 in
hepatocellular carcinoma. Oncogene. 32:3296–3305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Zhang J, Zhou L, Lu P, Zheng ZG,
Sun W, Wang JL, Yang XS, Li XL, Xia N, et al: Significance of serum
microRNA-21 in diagnosis of hepatocellular carcinoma (HCC):
Clinical analyses of patients and an HCC rat model. Int J Clin Exp
Pathol. 8:1466–1478. 2015.PubMed/NCBI
|
|
26
|
Huang CS, Yu W, Cui H, Wang YJ, Zhang L,
Han F and Huang T: Increased expression of miR-21 predicts poor
prognosis in patients with hepatocellular carcinoma. Int J Clin Exp
Pathol. 8:7234–7238. 2015.PubMed/NCBI
|
|
27
|
Wang WY, Zhang HF, Wang L, Ma YP, Gao F,
Zhang SJ and Wang LC: miR-21 expression predicts prognosis in
hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
38:715–719. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Hou L, Li A, Duan Y, Gao H and
Song X: Expression of serum exosomal microRNA-21 in human
hepatocellular carcinoma. Biomed Res Int.
2014:8648942014.PubMed/NCBI
|
|
29
|
Liu M, Liu J, Wang L, Wu H, Zhou C, Zhu H,
Xu N and Xie Y: Association of serum microRNA expression in
hepatocellular carcinomas treated with transarterial
chemoembolization and patient survival. PLoS One. 9:e1093472014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gyöngyösi B, Végh É, Járay B, Székely E,
Fassan M, Bodoky G, Schaff Z and Kiss A: Pretreatment microrna
level and outcome in sorafenib-treated hepatocellular carcinoma. J
Histochem Cytochem. 62:547–555. 2014. View Article : Google Scholar : PubMed/NCBI
|