Introduction
Melanoma is the deadliest form of skin cancer and
continues to have an increasing incidence in the last decades
(1). About ~50% of melanomas harbor
a T to A substitution in codon 600 of the BRAF gene,
resulting in a substitution of Valine to Glutamic Acid
(BRAFV600E), causing tonic activation of the RAF/MEK/ERK
pathway, proliferation, and cell survival (2,3). While a
combination of selective RAF/MEK inhibitors results in brisk
responses in most patients with BRAFV600E, treatment with
these drugs in BRAF wild-type (WT) tumors leads to paradoxical
activation of the pathway with the potential to accelerate tumor
growth (4,5). Determining the BRAF mutation status is
therefore critical prior to initiation of RAF/MEK-inhibitors. In
contrast, immune checkpoint inhibitors, which are monoclonal
antibodies that target CTLA-4 (i.e., ipilimumab) or the PD-1/PD-L1
axis (e.g., nivolumab or pembrolizumab) exhibit activity
irrespective of the BRAF mutation status, and are therefore the
preferred first-line therapy for patients with BRAF WT melanoma
(6–8). Testing for the BRAF mutation is usually
performed on a tissue biopsy, such as a core needle biopsy.
However, there are instances, in which potentially significant
morbidity prohibits procedures for obtaining tissue. Primary
leptomeningeal melanoma (PLM) is a rare and very aggressive type of
melanoma with an estimated frequency of 1 in 10 million individuals
(9–12). Due to its localization involving the
letptomeninges, biopsies cannot be easily performed, which may
limit potential therapeutic benefits for these patients. Here, we
describe a case of a patient with PLM who underwent BRAF
mutation testing from cell-free DNA (cfDNA) isolated from
cerebrospinal fluid (CSF) to guide therapy choices.
Case report
A 62-year-old Caucasian man with a past medical
history of essential hypertension and obstructive sleep apnea was
admitted to Beth Israel Deaconess Medical Center with altered
mental status, headaches and gait difficulties in March of 2016.
Three months prior to presentation, the patient had noticed lower
back pain with radiation to the buttocks. Magnetic resonance
imaging (MRI) without contrast of the lumbar spine at that time was
unrevealing. Over the following 6 weeks, the patient developed
worsening gait difficulties and intermittent confusion and, 3 days
prior to presentation, he developed headaches and was persistently
confused.
On arrival to our emergency room, the patient was
somnolent and only oriented to name. The vital signs were notable
for a temperature of 102°F, heart rate 74 beats/min, blood pressure
162/98 mmHg, respiratory rate 20 breaths/min, and oxygen saturation
97% at ambient air. Physical examination revealed somnolence with
responses only to noxious stimuli, nuchal rigidity with a positive
Brudzinski sign, and bilateral papilledema. An immediate
non-contrast head computed tomography (CT) scan showed extensive
communicating hydrocephalus and transependymal flow. The patient
underwent a large-volume lumbar puncture where the opening pressure
was 34 cm H2O, with subsequent mental status
improvement. A complete cell count and chemistry of the CSF is
summarized in Table I. The patient
was admitted to the neurological intensive care unit for further
care.
 | Table I.CSF cell count and chemistry from two
LPs performed on hospital days 1 and 4. |
Table I.
CSF cell count and chemistry from two
LPs performed on hospital days 1 and 4.
| LP no. | WBC (/µl) | RBC (/µl) | PMN (%) | LYM (%) | MONO (%) | MACRO (%) | OTHER (%) | Glucose (mg/dl) | Protein (mg/dl) |
|---|
| 1 (HD1) | 28 | 16,750 | 42 | 48 | 7 | 1 | 4 | 60 | 516 |
| 2 (HD4) | 5 | 7,950 | 7 | 57 | 6 | 23 | 6 | 53 | 459 |
| Reference range | <5 | 0 | 0 | 0 | 0 | 0 | 0 | 45–80 | 15–45 |
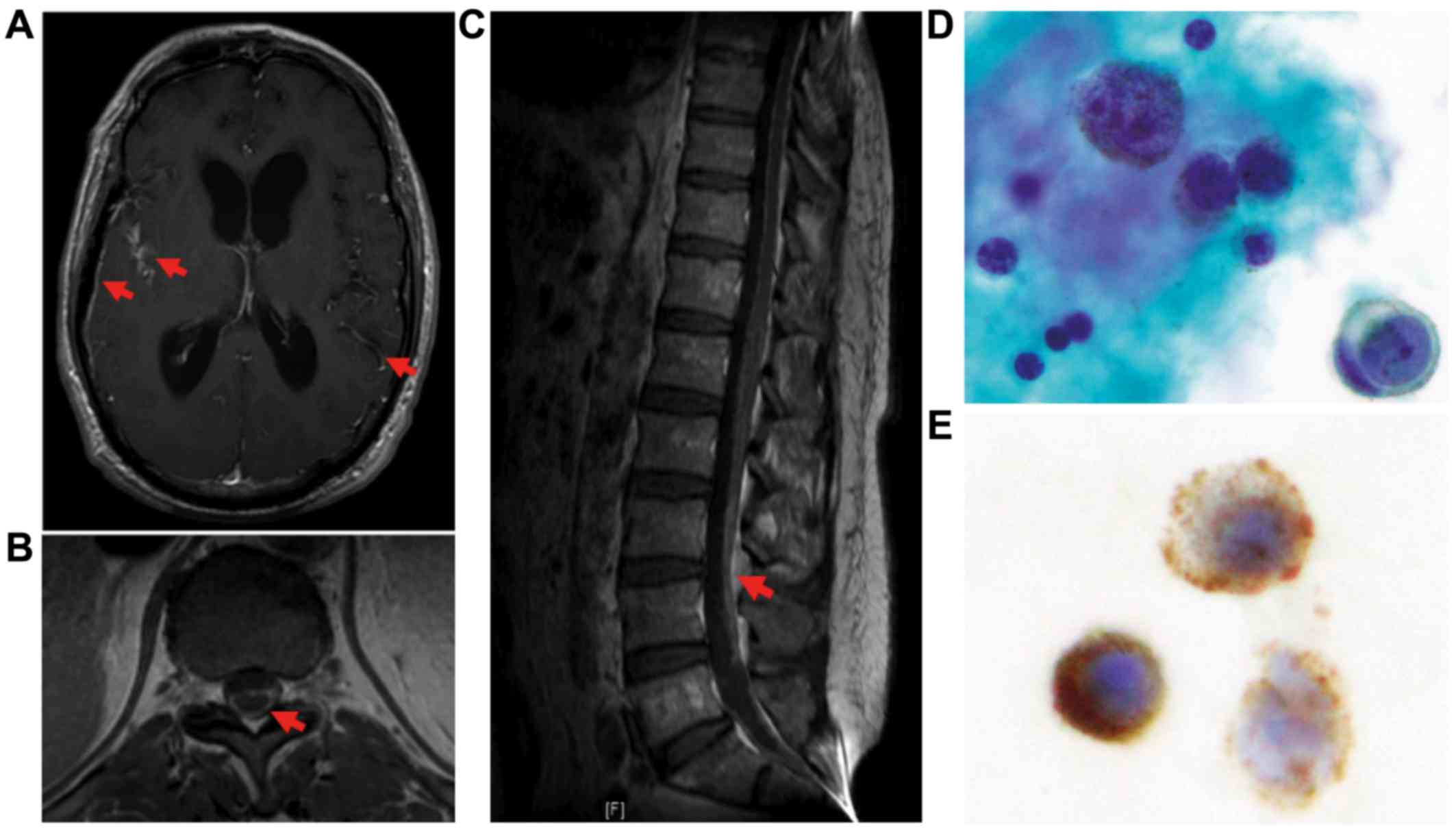
An MRI scan of the head and spine with and without
contrast revealed diffuse leptomeningeal enhancement involving
intracranial and spinal meninges on post-contrast T1-weighted
imaging, as well as communicating hydrocephalus. There was no
parenchymal central nervous system (CNS) involvement (Fig. 1A-C). A CT scan of the chest, abdomen
and pelvis did not reveal evidence of visceral metastatic disease.
A full skin examination by a dermatologist reported no evidence of
a primary cutaneous melanoma, and the ophthalmic examination did
not reveal any suspicious lesions. Cytology of the CSF revealed
malignant cells with strong staining for human melanoma black-45
(HMB-45), confirming a diagnosis of malignant melanoma (Fig. 1D and E).
Overall, this presentation was consistent with a
primary leptomeningeal melanoma (PLM). The patient was treated with
dexamethasone and required emergent placement of an external
ventricular drain (EVD) due to interval worsening mental status in
the setting of hydrocephalus. CSF was collected for isolation of
cfDNA and sequencing of the BRAF gene. Plasma was collected
at the same time for cfDNA sequencing. BRAF mutation testing
of CSF and plasma was performed using the Biocept Target
Selector™ assay.
The patient received five fractions of whole-brain
radiation therapy (2,000 cGy) and palliative radiation of the spine
from level T12 to S3. He had significant improvement of his
neurological symptoms, allowing for removal of the EVD on day 12 of
his hospitalization, and was discharged from the hospital on day
20.
Although previous efforts have used targeted
next-generation sequencing to evaluate small panels of genes
involved in melanoma biology, including BRAF and NRAS
(13), only mutant BRAF is a
currently actionable target and may help guide the choice of
targeted therapy vs. immunotherapy. Sequencing of cfDNA revealed
wild-type (WT) BRAF gene in both the CSF and plasma. Based
on this finding, treatment with either ipilimumab, a PD-1
checkpoint inhibitor, or temozolomide was discussed. Given the
patient's good clinical status, treatment with ipilimumab was
initiated, with a plan to administer four cycles, potentially
followed by PD-1 checkpoint blockade. Although the patient received
his first infusion without treatment-related complications, the
course was complicated by the development of a pulmonary embolism,
which delayed a planned second infusion. Five weeks after the first
ipilimumab infusion (~9 weeks after the initial diagnosis), the
patient developed rapidly progressing confusion and gait
instability with worsening hydrocephalus and succumbed to the
disease 3 days later.
Discussion
Primary leptomeningeal melanoma (PLM) is a very rare
type of cancer that is considered to arise from melanocytes in the
pia and arachnoid (10). Diagnostic
criteria for PLM have been suggested (9), including hyperintensity of the meninges
on T1-weighted MRI and cytology with positive immunostaining for
lineage-specific HMB-45 and S-100 (14–16).
In ~25% of patients, PLM is associated with giant
melanocytic nevi, which frequently carry treatment-sensitizing
oncogenic driver mutations (17,18)
including BRAFV600E and NRASQ61. Among patients with
metastatic cutaneous melanoma, ~50% harbor sensitizing BRAF
mutations, most commonly BRAFV600E. Treatment with RAF or
RAF/MEK-inhibitors in this subset of patients has resulted in
unprecedented response rates and improvement of progression-free
and overall survival (4,19). However, patients with wild-type
BRAF melanoma are not candidates for RAF/MEK inhibition, as
BRAF inhibitors may promote growth of BRAF-WT cells and
further exacerbate the disease (20), highlighting the importance of
targeted BRAF testing in this patient. In patients with
BRAF-WT melanoma, first-line immunotherapies are currently
under investigation as an alternative strategy. To this end, the
Food and Drug Administration has approved immunotherapies, such as
the CTLA-4 inhibitor ipilimumab and PD-1 checkpoint inhibitors,
including nivolumab and pembrolizumab, as first-line therapies for
patients with metastatic melanoma. Single-agent treatment with any
of these compounds or combinations of ipilimumab and PD-1
inhibitors produce deep and long-lasting responses in a subset of
patients (6–8,21),
including those with leptomeningeal disease (22).
The ideal choice of first-line therapy,
RAF/MEK-inhibitors or immunotherapies, in patients with
BRAF-mutant melanoma remains unclear and is mostly guided by
the clinical setting. For example, in patients with rapidly
progressing BRAF-mutant melanoma, treatment with BRAF/MEK
inhibitors may induce faster responses and is the preferred
treatment modality. In patients without sensitizing BRAF
mutations (BRAF-WT), as in the present case, immunotherapy
is the first-line treatment.
Understanding the molecular profile of these tumors
is crucial for employing treatments such as targeted therapies or
immune checkpoint inhibitors, which may induce dramatic responses
in leptomeningeal melanoma (22,23).
However, testing from malignant CSF with as few as 1 malignant cell
per µl, as in the case presented here, is challenging with current
approaches. We herein report the successful use of targeted
BRAF sequencing of cfDNA isolated from the CSF as well as
the plasma in a patient with PLM. The results from BRAF
mutation testing were instrumental in selecting the treatment for
this patient, given the potential harm in treating a BRAF-WT
patient with RAF/MEK inhibitors. Recent reports indicating the
feasibility of molecular profiling from CSF (24–27) have
mostly focused on primary CNS tumors. With regard to cfDNA
sequencing from CSF for melanoma, previous reports have focused
only on monitoring treatment response in metastatic lesions for
which the genomic status of the primary melanoma lesion was known
(28,29). In contrast, our case displays the
utility of using cfDNA to guide treatment choice in a primary
leptomeningeal melanoma for which no genetic information was
available. This study indicates that rapid targeted sequencing of
cfDNA from the CSF is clinically feasible and should be considered
for guiding treatment in patients in whom a tissue biopsy cannot be
obtained, including those with PLM and leptomeningeal metastatic
disease.
Acknowledgements
We thank Lyle Arnold, PhD and Cecile Rose Vibat, PhD
from Biocept Inc. for providing technical support.
Funding
BI is supported by the National Cancer Institute
(K08CA222663) and the Ludwig Center for Cancer Research at
Harvard.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JCM, KH, TJB, SS, JM and BI took clinical care of
the patient. JT provided pathology slides. JCM, RT and BI wrote the
paper. All authors read, reviewed and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
The family agreed to publication of the case and
material presented here.
Competing interests
VS is an employee of BioCept Inc. The other authors
declare that they have no competing interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hodis E, Watson IR, Kryukov GV, Arold ST,
Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C,
et al: A landscape of driver mutations in melanoma. Cell.
150:251–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Network: Genomic
classification of cutaneous melanoma. Cell. 161:1681–1696. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Flaherty KT, Puzanov I, Kim KB, Ribas A,
McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K and
Chapman PB: Inhibition of mutated, activated BRAF in metastatic
melanoma. N Engl J Med. 363:809–819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Poulikakos PI, Zhang C, Bollag G, Shokat
KM and Rosen N: RAF inhibitors transactivate RAF dimers and ERK
signalling in cells with wild-type BRAF. Nature. 464:427–430. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hodi FS, O'Day SJ, McDermott DF, Weber RW,
Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel
JC, et al: Improved survival with ipilimumab in patients with
metastatic melanoma. N Engl J Med. 363:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Larkin J, Chiarion-Sileni V, Gonzalez R,
Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M,
Rutkowski P, et al: Combined nivolumab and ipilimumab or
monotherapy in untreated melanoma. N Engl J Med. 373:23–34. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Robert C, Schachter J, Long GV, Arance A,
Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al:
Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med.
372:2521–2532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hayward RD: Malignant melanoma and the
central nervous system. A guide for classification based on the
clinical findings. J Neurol Neurosurg Psychiatry. 39:526–530. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosenthal G, Gomori JM, Tobias S, Diment J
and Shoshan Y: Unusual cases involving the CNS and nasal sinuses:
Case 1. Primary leptomeningeal melanoma. J Clin Oncol.
21:3875–3877. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paulus W and Hasselblatt M:
TumorenNeuropathologie. Springer; Berlin-Heidelberg: pp. 481–549.
2012
|
|
12
|
Hsieh YY, Yang ST, Li WH, Hu CJ and Wang
LS: Primary leptomeningeal melanoma mimicking meningitis: A case
report and literature review. J Clin Oncol. 33:e57–e61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van de Nes J, Gessi M, Sucker A, Möller I,
Stiller M, Horn S, Scholz SL, Pischler C, Stadtler N, Schilling B,
et al: Targeted next generation sequencing reveals unique mutation
profile of primary melanocytic tumors of the central nervous
system. J Neurooncol. 127:435–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wick MR, Swanson PE and Rocamora A:
Recognition of malignant melanoma by monoclonal antibody HMB-45. An
immunohistochemical study of 200 paraffin-embedded cutaneous
tumors. J Cutan Pathol. 15:201–207. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tosaka M, Tamura M, Oriuchi N, Horikoshi
M, Joshita T, Sugawara K, Kobayashi S, Kohga H, Yoshida T and
Sasaki T: Cerebrospinal fluid immunocytochemical analysis and
neuroimaging in the diagnosis of primary leptomeningeal melanoma.
Case report. J Neurosurg. 94:528–532. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sagiuchi T, Ishii K, Utsuki S, Asano Y,
Tsukahara S, Kan S, Fujii K and Hayakawa K: Increased uptake of
technetium-99m-hexamethylpropyleneamine oxime related to primary
leptomeningeal melanoma. AJNR Am J Neuroradiol. 23:1404–1406.
2002.PubMed/NCBI
|
|
17
|
Hoffman HJ and Freeman A: Primary
malignant leptomeningeal melanoma in association with giant hairy
nevi. J Neurosurg. 26:62–71. 1967. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salgado CM, Basu D, Nikiforova M, Bauer
BS, Johnson D, Rundell V, Grunwaldt LJ and Reyes-Múgica M: BRAF
mutations are also associated with neurocutaneous melanocytosis and
large/giant congenital melanocytic nevi. Pediatr Dev Pathol.
18:1–9. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Flaherty KT, Robert C, Hersey P, Nathan P,
Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, et
al: Improved survival with MEK inhibition in BRAF-mutated melanoma.
N Engl J Med. 367:107–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Medina TM and Lewis KD: The evolution of
combined molecular targeted therapies to advance the therapeutic
efficacy in melanoma: A highlight of vemurafenib and cobimetinib.
Onco Targets Ther. 9:3739–3752. 2016.PubMed/NCBI
|
|
21
|
Hamid O, Robert C, Daud A, Hodi FS, Hwu
WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, et al:
Safety and tumor responses with lambrolizumab (anti-PD-1) in
melanoma. N Engl J Med. 369:134–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Margolin K, Ernstoff MS, Hamid O, Lawrence
D, McDermott D, Puzanov I, Wolchok JD, Clark JI, Sznol M, Logan TF,
et al: Ipilimumab in patients with melanoma and brain metastases:
An open-label, phase 2 trial. Lancet Oncol. 13:459–465. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wilgenhof S and Neyns B: complete
cytologic remission of V600E BRAF-mutant melanoma-associated
leptomeningeal carcinomatosis upon treatment with dabrafenib. J
Clin Oncol. 33:e109–e111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan W, Gu W, Nagpal S, Gephart MH and
Quake SR: Brain tumor mutations detected in cerebral spinal fluid.
Clin Chem. 61:514–522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Springer S, Zhang M, McMahon KW,
Kinde I, Dobbyn L, Ptak J, Brem H, Chaichana K, Gallia GL, et al:
Detection of tumor-derived DNA in cerebrospinal fluid of patients
with primary tumors of the brain and spinal cord. Proc Natl Acad
Sci USA. 112:9704–9709. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
De Mattos-Arruda L, Mayor R, Ng CK,
Weigelt B, Martínez-Ricarte F, Torrejon D, Oliveira M, Arias A,
Raventos C, Tang J, et al: Cerebrospinal fluid-derived circulating
tumour DNA better represents the genomic alterations of brain
tumours than plasma. Nat Commun. 6:88392015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pentsova EI, Shah RH, Tang J, Boire A, You
D, Briggs S, Omuro A, Lin X, Fleisher M, Grommes C, et al:
Evaluating cancer of the central nervous system through
next-generation sequencing of cerebrospinal fluid. J Clin Oncol.
34:2404–2415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Pan W, Connolly ID, Reddy S, Nagpal
S, Quake S and Gephart MH: Tumor DNA in cerebral spinal fluid
reflects clinical course in a patient with melanoma leptomeningeal
brain metastases. J Neurooncol. 128:93–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Momtaz P, Pentsova E, Abdel-Wahab O,
Diamond E, Hyman D, Merghoub T, You D, Gasmi B, Viale A and Chapman
PB: Quantification of tumor-derived cell free DNA(cfDNA) by digital
PCR (DigPCR) in cerebrospinal fluid of patients with BRAFV600
mutated malignancies. Oncotarget. 7:85430–85436. 2016. View Article : Google Scholar : PubMed/NCBI
|