Introduction
In the last decade, research in the field of
cytotoxic chemotherapy for non-small-cell lung cancer (NSCLC) has
considerably slowed down, mainly due to the clinical development of
targeted therapies for use in patients whose disease harbors driver
genetic alterations, as well as immunotherapy for patients without
actionable mutations (1).
Nevertheless, the majority of newly diagnosed advanced NSCLCs do
not harbor actionable genetic mutations, and only 15–20% derive
clinical benefit from immunotherapy, which still makes conventional
chemotherapy a fundamental treatment option for a number of
patients. In this context, the identification of patients who will
benefit from cytotoxic treatment would help physicians to deliver
effective treatment to sensitive patients, while preventing others
from suffering the side effects of inactive drugs. In recent years,
several predictive markers of sensitivity to chemotherapy have been
investigated with this purpose, including excision repair
cross-complementation 1 (ERCC1), thymidylate synthase
(TS), ribonucleotide reductase M1 (RRM1), and breast
cancer susceptibility 1 (BRCA1), which are DNA synthesis and
repair genes that can potentially predict sensitivity to platinum
agents, pemetrexed, gemcitabine, and taxanes, respectively
(2,3).
Kirsten rat sarcoma viral oncogene homolog
(KRAS) mutation represents the most common genetic
alteration in NSCLC, being found in approximately 20–30% of
patients (4). Although KRAS
acts as a driver mutation in NSCLC, it is not yet an actionable
target, since clinical trials with targeted therapies aimed at
blocking the RAS pathway have invariably led to disappointing
results. On the other hand, recent data have unveiled a negative
predictive role for KRAS mutation with regard to cytotoxic
treatment, particularly platinum-based chemotherapy, which still
represents the standard of care for a few KRAS-mutant
advanced NSCLCs (5,6).
Against this background, we investigated the mRNA
expression levels of ERCC1, TS, RRM1, and BRCA1 in
KRAS-mutant advanced NSCLC patients in order to provide a
plausible explanation to the clinical observation that has linked
KRAS mutation to poor sensitivity to cytotoxic
chemotherapy.
Materials and methods
Study design and patients
Patients diagnosed with epidermal growth factor
receptor (EGFR) wild type (WT) advanced NSCLC at the Medical
Oncology of Perugia Hospital from January 2006 and November 2016
were eligible for this study. EGFR and KRAS mutation
tests were performed on tumor tissue (either primary or metastic,
if both tissues were available metastatic cancer specimen was
preferred) following physician's request in patients who were
eligible to receive cytotoxic treatment for advanced disease. The
mRNA expression levels of ERCC1, TS, RRM1, and BRCA1 were assessed
through reverse transcription-polymerase chain reaction (RT-PCR) in
patients with available tumor tissue. If there was not enough
tissue for the evaluation of all markers, the analysis was
sequentially conducted according to the following order: ERCC1, TS,
RRM1, and BRCA1. In case further tissue was available, anaplastic
lymphoma kinase (ALK) gene status was performed.
This retrospective study was approved by local
Ethics Committee (Comitato Etico Aziende Sanitarie Umbria),
waiving patient consent.
Assessment of EGFR and KRAS mutation
status
Formalin-fixed paraffin-embedded (FFPE) tumor blocks
were reviewed for quality and tumor content. Tumor cells (≥70%)
were macrodissected, and genomic DNA was isolated using QIAmp DNA
extraction kit and automatically purified by BioRobot EZ1
instrument (Qiagen S.p.A., Milan, Italy) according to the
manufacturer's instructions. Nested polymerase chain reactions
(PCRs) were carried out using primers to amplify exons 18 to 21 of
EGFR and exons 2 to 3 of KRAS. To facilitate
sequencing, internal primers incorporated an M13Tag. PCR products
were purified with Exonuclease 1 and Shrimp Alkaline Phosphatase
(ExoSAP-IT) a 37°C for 15 min followed by heating at 80°C for 15
min to stop the enzymatic reaction. After purification, the PCR
products were sequenced with forward and reverse M13 primers and
Big Dye Terminator v1.1 Cycle Sequencing Kit. Sequencing fragments
were detected by capillary electrophoresis using 3500 Genetic
Analyzer (Applied Biosystems, Foster City, CA, USA).
Electropherograms were analyzed for the presence of mutations using
SeqScape v2.7 Software. In all cases, samples harboring mutations
were reamplified and resequenced using the same experimental
conditions.
Assessment of the mRNA expression
levels of ERCC1, TS, RRM1 and BRCA1
RNA was extracted and purified from five consecutive
8-µM slides of microdissected FFPE tumor tissue, using RNeasy FFPE
Kit on QIAcube instrument (Qiagen, Milan, Italy). Expression levels
of ERCC1, TS, RRM1, and BRCA1 were evaluated on 100 ng RNA of each
tumor sample and compared to synthetic healthy lung RNA, using
QuantiFast Probe Duplex Assays and QuantiFast Probe RT-PCR Plus Kit
(Qiagen, Milan, Italy). This kit offers an integrated genomic DNA
removal step to avoid false-positive signals. Reverse transcription
PCR and RT-PCR took place in the same tube (One Step RT-PCR).
QuantiFast Probe Assays were predesigned to enable amplification
and detection of specific gene targets (ERCC1 cat. no. QF00270641;
TS cat. no. QF00102375; RRM1 cat. no. QF00452382; BRCA1 cat. no.
QF0043126 by Qiagen). These assays were based on dual-labeled
hydrolysis probe detection using two different dyes, FAM binds the
specific gene of interest and MAX binds selected reference gene
(duplex PCR). The reaction probe mix was aliquoted into specific
PCR tubes for Rotor-Gene Q Instrument and the cDNA samples were
then added. Cycling conditions for one-step RT-PCR included:
denaturation at 95°C for 20 min and the PCR conditions included an
initial denaturation at 95°C for 5 min followed by 45 cycles of
denaturation at 95°C for 15 sec and annealing at 60°C for 30
sec.
All runs included a calibrator sample and a one
no-template control, and all samples were measured in triplicate.
Comparative Cq method was used for gene expression quantification
using β-actin as internal reference gene and commercial RNA control
(mRNA from lung, Stratagene, La Jolla, CA, USA) as calibrator.
Final expression values were determined as follows: 2−(ΔCt
sample-ΔCt calibrator), where ΔCt values of the sample and
the calibrator are estimated by subtracting the Cq value of the
target gene from the median of the reference genes values as
reported by Livak and Schmittgen (7).
Assessment of ALK gene
rearrangement
ALK immunohistochemistry based on a 4-tiered
score (clone D5-F3, cat. no. 3633, Cell Signaling Technology,
dilution 1:250, incubation time: 30 min/room temperature) according
to a laboratory developed test was used as screening method for the
assessment of ALK status. For each stain positive controls,
represented by inflammatory myofibroblastic tumour, were used.
Based on the mutual exclusiveness of driver mutations, only
patients who were KRAS WT were tested for ALK. In
case of any ALK IHC positivity, specimens underwent
confirmatory FISH (Break Apart Vysis Probe kit). ALK
FISH-positive patients were excluded from the present analysis.
Statistical analysis
Statistical analysis was conducted using the SPSS
statistical software package (version 24; SPSS, Inc., Chicago, IL,
USA). Chi-square tests or Fisher exact tests were used to analyze
correlations between EGFR mutation or ALK
rearrangement status and clinico-pathologic variables. Data related
to mRNA expression levels of ERCC1, TS, RRM1 and BRCA1 were
presented as mean and standard deviation. Student's t test was used
for comparison of two groups. All statistical tests were conducted
at a 2-sided level of significance of P<0.05.
Results
Patients characteristics
From January 2006 and November 2016 184 EGFR
WT advanced NSCLC patients were evaluable for the analysis of at
least one DNA repair gene, of which 92 were KRAS-mutants.
Table I lists patients
characteristics. Overall, the median age was 62 years, 60.3% of
patients were male, and 89.7% of patients were current/former
smokers. Virtually all patients had adenocarcinoma histology. Among
KRAS-mutants, the majority had a KRAS codon 12
mutation (88%), the most common being G12C (44.4% of cases). When
KRAS-mutant patients were compared with those who were
KRAS WT, significantly more individuals in the
KRAS-mutant group were current/former smokers (P=0.003) and
males (P=0.023) (Table I). No other
statistically significant differences were observed.
 | Table I.Patients characteristics. |
Table I.
Patients characteristics.
| Variable | All patients |
KRAS-mutant | KRAS wild
type | P-value |
|---|
| Number of patients,
n | 184 | 92 | 92 | – |
| Median age, year
(range) | 62 (23–85) | 63 (42–82) | 62 (23–85) | 0.589 |
| Sex, n (%) |
|
Male | 111 (60.3) | 63 (68.5) | 48 (52.2) | 0.023 |
|
Female | 73 (39.7) | 29 (31.5) | 44 (47.8) |
|
| Performace status,
n (%) |
|
0–1 | 174 (94.5) | 88 (95.7) | 86 (93.5) | 0.474 |
| ≥2 | 10 (5.5) | 4 (4.3) | 6 (6.5) |
|
| Stage IV |
| De
novo | 129 (70.1) | 65 (70.7) | 64 (69.6) | 0.871 |
|
Recurrent | 55 (29.9) | 27(29.3) | 28 (30.4) |
|
| Smoking history, n
(%) |
|
Nevera | 19 (10.3) | 4 (4.3) | 15 (16.3) | 0.003 |
|
Current/former | 165 (89.7) | 88 (95.7) | 77 (83.7) |
|
| Histology, n
(%) |
|
Adenocarcinoma | 178 (96.7) | 91 (98.9) | 87 (94.6) | 0.090 |
|
Squamous cell carcinoma | 6 (3.3) | 1 (1.1) | 5 (5.4) |
|
| ALK gene
status |
|
Negative | 57 (31.0) | – | 57 (62.0) | – |
| Not
assessed | 127 (69.0) | 92
(100.0)b | 35
(38.0)c |
|
| KRAS
mutations, n (%) |
| Codon
12 |
| 81
(88.0) |
|
|
|
G12C |
| 36 (44.5) |
|
|
|
G12V |
| 18 (22.2) |
|
|
|
G12D |
| 10 (12.3) |
|
|
|
G12A |
| 8 (9.9) |
|
|
|
G12F | – | 6 (7.4) | – | – |
|
G12R |
| 3 (3.7) |
|
|
| Codon
13 |
| 6 (6.6) |
|
|
|
G13C |
| 3 (50.0) |
|
|
|
G13D |
| 3 (50.0) |
|
|
| Codon
59 |
| 1 (1.1) |
|
|
| Codon
61 |
| 4 (4.3) |
|
|
ERCC1, TS, RRM1 and BRCA1. Overall, TS and RRM1
expression levels were significantly lower in patients with
non-squamous NSCLC as compared with squamous patients (P=0.032 and
P=0.017, respectively) (Table II).
Similarly, never smokers had significantly lower levels of TS
expression vs. smokers (P=0.021). No significant differences were
noted for any characteristics according to the type of KRAS
mutation (data not shown). Likewise, no significant differences
were noted with regard to the distribution of ERCC1, TS, RRM1, and
BRCA1 in relation to the type of KRAS mutation (G12C vs.
other) (Table III).
 | Table II.Patients characteristics and
expression levels of ERCC1, TS, RRM1, and BRCA1. |
Table II.
Patients characteristics and
expression levels of ERCC1, TS, RRM1, and BRCA1.
| Variable | ERCC1 (mean) | TS (mean) | RRM1 (mean) | BRCA1 (mean) |
|---|
| Sex, n (%) |
|
Male | 2.41 | 5.42 | 12.15 | 11.07 |
|
Female | 2.73 | 4.9 | 13.82 | 11.17 |
|
P-value | NS | NS | NS | NS |
| Performace status,
n (%) |
|
0–1 | 2.59 | 5.31 | 12.72 | 11.07 |
| ≥2 | 1.16 | 2.6 | 13.73 | 10.3 |
|
P-value | NS | NS | NS | NS |
| Stage IV |
| De
novo | 2.12 | 4.88 | 11.85 | 11.16 |
|
Recurrent | 3.37 | 5.87 | 14.78 | 10.99 |
|
P-value | NS | NS | NS | NS |
| Smoking history, n
(%) |
|
Never | 2.66 | 5.4 | 13.02 | 11.55 |
|
Current/former | 1.36 | 2.88 | 10.75 | 7.14 |
|
P-value | NS | P=0.021 | NS | NS |
| Histology, n
(%) |
|
Adenocarcinoma | 2.53 | 5.07 | 12.39 | 10.87 |
|
Squamous cell carcinoma | 2.68 | 9.28 | 24.98 | 17.65 |
|
P-value | NS |
P=0.032a |
P=0.017a | NS |
 | Table III.Median and mean expression levels of
TS, ERCC1, RRM1, and BRCA1 according to KRAS codon 12
mutations. |
Table III.
Median and mean expression levels of
TS, ERCC1, RRM1, and BRCA1 according to KRAS codon 12
mutations.
| Variable | All patients | G12C | Other codon 12
mutation | P-value |
|---|
| ERCC |
|
|
|
|
| N | 81 | 36 | 45 | – |
| Mean ±
SD | 3.37±6.93 | 4.98±10 | 2.08±2.03 | 0.062 |
| Median
(range) | 1.69
(0.43–55.02) | 1.91
(0.43–11.4) | 1.38
(0.53–55.02) | 0.074 |
| TS |
|
|
|
|
| N | 79 | 35 | 44 | – |
| Mean ±
SD | 4.25±3.38 | 3.92±3.05 | 4.52±3.45 | 0.423 |
| Median
(range) | 1.73
(0.04–14.7) | 3.03
(0.04–11.8) | 3.48
(0.25–14.7) | 0.415 |
| RRM1 |
|
|
|
|
| N | 77 | 34 | 43 | – |
| Mean ±
SD | 12.87±14.83 | 14.64±19.2 | 11.47±8.99 | 0.343 |
| Median
(range) | 8.84
(1.04–93.98) | 8.89
(1.04–93.98) | 8.02
(1.35–38.29) | 0.516 |
| BRCA1 |
|
|
|
|
| N | 72 | 33 | 39 | – |
| Mean ±
SD | 9.54±8.45 | 9.74±8.87 | 9.9±8.27 | 0.863 |
| Median
(range) | 6.66
(0.01–35.21) | 5.73
(0.01–32.18) | 7.05
(0.45–35.21) | 0.921 |
Table IV lists the
mRNA expression levels of ERCC1, TS, RRM1, and BRCA1 in all
patients and according to KRAS mutation status. ERCC1 levels
ranged from 0.07 to 55.02, with a mean of 2.54 and median of 1.55;
as for TS, the levels ranged from 0.04 to 35.0, with a mean of 5.21
and median of 3.78. RRM1 levels ranged from 0.001 to 93.9, with a
mean of 12.82 and a median of 6.86; BRCA1 levels ranged from 0.01
to 64.3, with a mean of 11.11 and median of 7.62. When analyzed
according to KRAS mutation status, KRAS-mutant
patients had significantly higher mean levels of ERCC1 as compared
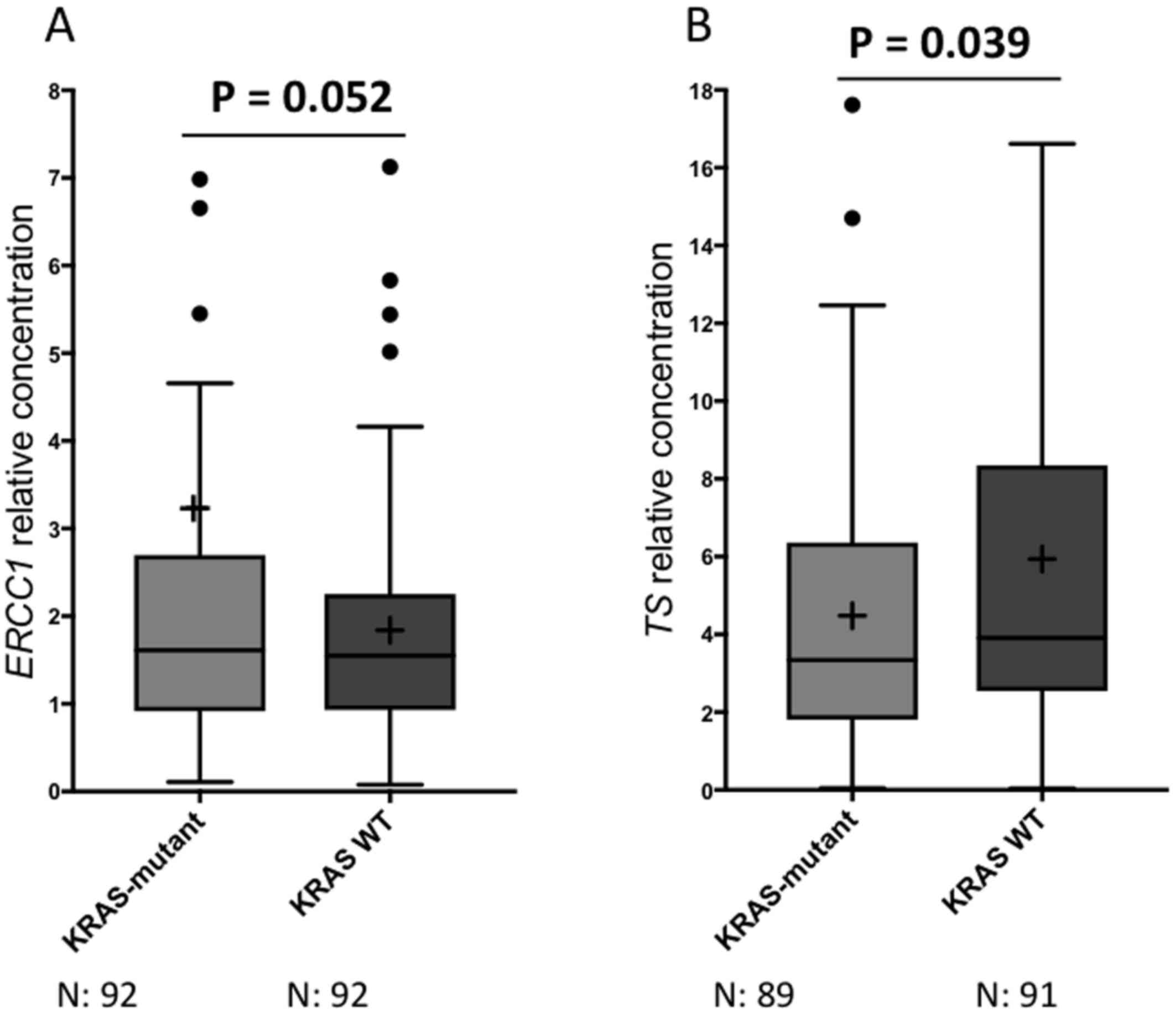
with KRAS WT patients (3.23±6.63 vs. 1.84±1.24; P=0.052,
Table IV, Fig. 1A). On the other hand, mean expression
levels of TS were significantly lower in patients who were
KRAS-mutant as compared with KRAS WT patients
(4.48±3.75 vs. 5.94±6.4; P=0.039, Table
IV, Fig. 1B). No statistically
significant differences were noted for the median expression levels
of ERCC1 and TS according to KRAS mutation status (Table IV). Similarly, no significant
differences were observed neither for mean nor for median levels of
RRM1 and BRCA1 according to KRAS mutation status.
 | Table IV.Median and mean expression levels of
TS, ERCC1, RRM1, and BRCA1. |
Table IV.
Median and mean expression levels of
TS, ERCC1, RRM1, and BRCA1.
| Variable | All patients |
KRAS-mutant | KRAS WT | P-value |
|---|
| ERCC1 |
|
|
|
|
| N | 184 | 92 | 92 | – |
| Mean ±
SD | 2.54±4.82 | 3.23±6.63 | 1.84±1.24 | 0.052 |
| 95%
CI | 1.83–3.24 | 1.86–4.6 | 1.58–2.1 |
|
| Median
(range) | 1.55
(0.07–55.02) | 1.61
(0.1–55.02) | 1.54
(0.07–7.12) | 0.623 |
| TS |
|
|
|
|
| N | 180 | 89 | 91 | – |
| Mean ±
SD | 5.21±4.75 | 4.48±3.75 | 5.94±6.4 | 0.039 |
| 95%
CI | 4.51–5.91 | 3.79–5.35 | 4.87–7.51 |
|
| Median
(range) | 3.78
(0.04–35.0) | 3.34
(0.05–17.6) | 3.91
(0.04–35.3) | 0.133 |
| RRM1 |
|
|
|
|
| N | 176 | 87 | 89 | – |
| Mean ±
SD | 12.82±12.05 | 12.82±14.02 | 12.82±9.82 | 0.991 |
| 95%
CI | 11.03–14.61 | 9.83–15.81 | 10.75–14.89 |
|
| Median
(range) | 6.86
(0.001–93.9) | 8.84
(1.04–93.9) | 9.1
(0.001–56.8) | 0.332 |
| BRCA1 |
|
|
|
|
| N | 171 | 82 | 89 | – |
| Mean ±
SD | 11.11±10.03 | 10.16±8.98 | 11.99±10.88 | 0.234 |
| 95%
CI | 9.6–12.63 | 8.18–12.13 | 9.7–14.29 |
|
| Median
(range) | 7.62
(0.01–64.3) | 7.58
(0.01–44.5) | 7.99
(0.1–64.3) | 0.223 |
Discussion
The aim of this study was to associate the mRNA
expression levels of ERCC1, TS, RRM1 and BRCA1 with KRAS
mutation status in patients with advanced NSCLC. In light of the
mutual exclusiveness existing between EGFR mutation and
KRAS mutation, and the evidence that EGFR-mutant
patients belong to a biologically distinct subset of patients, we
considered it was important to exclude from the analysis
EGFR-mutant patients, thus focusing only on individuals
whose tumor had a documented EGFR WT status.
Interestingly, we showed for both ERCC1 and TS a
similar range of expression levels and median values as compared
with a previous report from Maus et al, which analyzed the
distribution of ERCC1, TS, and RRM1 in >2,000 NSCLC specimens
(8). Likewise, similarly to the Maus
et al study, we found that these markers were expressed at a
lower level in adenocarcinoma histology as compared with squamous
cell carcinoma, which was statistically significant only for TS and
RRM1 in our study (Table II).
However, it should be noted that the present study was not powered
for addressing differences according to histology owing to the
small number of squamous cell carcinomas that were included.
Therefore, these results should be interpreted very cautiously.
Importantly, we observed that KRAS-mutant
patients had significantly higher mean ERCC1 expression levels as
compared with KRAS WT patients (P=0.052, Table IV, Fig.
1A). Accordingly, our group and others have previously reported
that KRAS-mutant advanced NSCLCs perform poorly on
platinum-based chemotherapy (9,10). This
finding has been further corroborated by two metanalyses, in which
KRAS-mutant advanced NSCLCs treated with platinum-based
chemotherapy appeared to experience significantly lower response
rates and progression-free survival as compared with the
KRAS WT counterpart (5,6).
Therefore, the higher mean levels of ERCC1 that have been found in
KRAS-mutant patients provide a molecularly plausible
explanation for the poor sensitivity to platinum-based chemotherapy
observed in patients whose tumor harbors a KRAS
mutation.
On the other hand, we reported significantly lower
mean levels of TS expression in KRAS-mutant patients as
compared with KRAS WT patients (P=0.039, Table IV, Fig.
1B). This finding might imply an increased sensitivity to
pemetrexed-based chemotherapy in KRAS-mutant advanced
NSCLCs, which would be of clinical relevance owing to the fact that
KRAS mutation are mainly found in adenocarcinoma patients,
and pemetrexed is approved for clinical use in this histological
subset only (4,11). Previously, Moran and colleagues have
already reported that KRAS-mutant cell line depend on
enhanced folate metabolism in functional experiments (12). Accordingly, a small retrospective
analysis suggested that KRAS mutation might predict
sensitivity to pemetrexed (13).
Moreover, several clinical studies have already reported that TS
levels may represent a predictive biomarker for antifolate agents
in NSCLC (2). Against this
well-established background, we found that patients with
KRAS-mutant NSCLC had lower levels of TS, which might
explain the enhanced sensitivity to pemetrexed reported in
literature. Although preliminary, these findings suggest that the
KRAS-mutant subset of patients, for whom no targeted
therapies are available, may exhibit a particular sensitivity to
pemetrexed, which could represent the basis for the design of
future clinical studies aimed to further address this issue.
Importantly, KRAS-mutant NSCLC is a
heterogeneous disease, as many type of different type amino-acid
substitutions result into several types of KRAS gene
mutations. On this basis, in order to exclude an imbalance in terms
of expression levels of either ERCC1 and TS, we performed an
analysis in KRAS-mutant codon 12 patients according to the
type of KRAS mutation (other KRAS codon 12 mutations
excluded due to the low number). However, we were not able to
identify any differential mRNA expression levels for any markers
(Table III). Therefore, we
conclude that ERCC1 and TS expression levels were homogeneously
distributed in KRAS-mutant codon 12 patients, regardless of
the KRAS mutation variant.
Of note, our findings further enlarge the evidence
indicating that each molecularly defined subgroup of NSCLC is
associated with a different of expression of DNA synthesis and
repair genes. In fact, some authors have previously reported that
EGFR-mutant NSCLCs express lower ERCC1 expression levels as
compared with EGFR WT patients, which might account for the
increased sensitivity to platinum-based chemotherapy in
EGFR-mutant NSCLCs (14–16).
Likewise, lower TS expression levels were observed in
ALK-positive NSCLCs, which results into greater benefit from
pemetrexed-based chemotherapy in ALK-positive patients
(17–19). Therefore, we provide evidence that,
despite being a heterogeneous disease, also KRAS-mutant
NSCLC is associated with a peculiar pattern of expression of DNA
synthesis and repair genes, mainly ERCC1 and TS, which, in turn,
could account for a different sensitivity to platinum agents and
pemetrexed, respectively.
Certainly, our study is affected by some
limitations, including the retrospective design, the relatively
small sample size of patients, the lack of clinical outcome
information, and the absence of confirmation by additional
functional experiments. In addition, ALK status was assessed
only in 62% of KRAS WT patients. However, as ALK
rearrangements have been associated with low levels of TS, it is
very unlikely that this could have affected the results of this
study in terms of TS levels (17).
On the other hand, it could be argued that immunohistochemistry
(IHC) rather than RT-PCR would provide a direct measure of the
acting element, namely the protein. However, there are several
arguments against using an IHC-based technique, including the need
for highly specific antibodies, the lack of standardized tissue
fixation and staining protocols, difficulty in finding a consensual
standard for microscopic evaluation, and universal cutoff value
(20). For the same reason, some
researchers have previously attempted to develop a more reliable
and reproducible IHC method based on a fully automated and
quantitative immunofluorescence technique (AQUA) (21).
In conclusion, to the best of our knowledge, this is
the first study that has evaluated the expression of DNA synthesis
and repair genes according to KRAS mutation status in
advanced NSCLC patients, which suggests significantly higher mRNA
expression levels for ERCC1 and lower for TS in KRAS-mutant
patients, but no difference for either RRM1 and BRCA1. As for
ERCC1, these results provide a rationale behind the poor
sensitivity to platinum-based chemotherapy of KRAS-mutant
advanced NSCLCs. On the other hand, whether lower TS expression
levels translate into enhanced clinical efficacy of
pemetrexed-based chemotherapy in KRAS-mutant patients
remains to be determined in prospective clinical trials.
Acknowledgements
Not applicable.
Funding
The study was supported by Associazione Italiana per
la Ricerca contro il Cancro (AIRC) (grant no. 15713 AIRC 2014).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
VL, LC and GM designed the research. FRT, CM, ASid,
ASig, MSR, RC, SB and GB performed the experiments. BR, DG and GM
analyzed the data. BR and GM wrote the paper. VL and GM critically
revised the manuscript for important intellectual content.
Ethics approval and consent to
participate
Comitato Etico Aziende Sanitarie (CEAS) Umbria
approved the study (approval no. 4796/15/AV).
Patient consent for publication
CEAS Umbria approval waived patient consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ,
Dobelbower M, et al: Non-small cell lung cancer, version 5.2017,
NCCN clinical practice guidelines in oncology. J Natl Compr Canc
Netw. 15:504–535. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Olaussen KA and Postel-Vinay S: Predictors
of chemotherapy efficacy in non-small-cell lung cancer: A
challenging landscape. Ann Oncol. 27:2004–2016. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reguart N, Cardona AF, Carrasco E, Gomez
P, Taron M and Rosell R: BRCA1: A new genomic marker for
non-small-cell lung cancer. Clin Lung Cancer. 9:331–339. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ricciuti B, Leonardi GC, Metro G, Grignani
F, Paglialunga L, Bellezza G, Baglivo S, Mencaroni C, Baldi A,
Zicari D and Crinò L: Targeting the KRAS variant for treatment of
non-small cell lung cancer: Potential therapeutic applications.
Expert Rev Respir Med. 10:53–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Fang W, Yan Y, Wang M, Kang S,
Sheng J, Zhan J, Chen N, Hong S, Yang Y, et al: The efficacy of
first-line chemotherapy is associated with KRAS mutation status in
patients with advanced non-small cell lung cancer: A meta-analysis.
Med Oncol. 32:612015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pan W, Yang Y, Zhu H, Zhang Y, Zhou R and
Sun X: KRAS mutation is a weak, but valid predictor for poor
prognosis and treatment outcomes in NSCLC: A meta-analysis of 41
studies. Oncotarget. 7:8373–8388. 2016.PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maus MK, Mack PC, Astrow SH, Stephens CL,
Zeger GD, Grimminger PP, Hsiang JH, Huang E, Li T, Lara PN, et al:
Histology-related associations of ERCC1, RRM1 and TS biomarkers in
patients with non-small-cell lung cancer: Implications for therapy.
J Thorac Oncol. 8:582–586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Metro G, Chiari R, Bennati C, Cenci M,
Ricciuti B, Puma F, Flacco A, Rebonato A, Giannarelli D, Ludovini
V, et al: Clinical outcome with platinum-based chemotherapy in
patients with advanced nonsquamous EGFR wild-type non-small-cell
lung cancer segregated according to KRAS mutation status. Clin Lung
Cancer. 15:86–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marabese M, Ganzinelli M, Garassino MC,
Shepherd FA, Piva S, Caiola E, Macerelli M, Bettini A, Lauricella
C, Floriani I, et al: KRAS mutations affect prognosis of
non-small-cell lung cancer patients treated with first-line
platinum containing chemotherapy. Oncotarget. 6:34014–34022. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomasini P, Barlesi F, Mascaux C and
Greillier L: Pemetrexed for advanced stage nonsquamous non-small
cell lung cancer: Latest evidence about its extended use and
outcomes. Ther Adv Med Oncol. 8:198–208. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moran DM, Trusk PB, Pry K, Paz K,
Sidransky D and Bacus SS: KRAS mutation status is associated with
enhanced dependency on folate metabolism pathways in non-small cell
lung cancer cells. Mol Cancer Ther. 13:1611–1624. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Levy B, Drilon A, Chachoua A, Seetharamu
N, Richardson S, Lucido D, Legasto A, Grossbard M and Becker D:
KRAS mutations predict sensitivity to pemetrexed-based
chemotherapy. Lung Cancer Manag. 2:275–280. 2013. View Article : Google Scholar
|
|
14
|
Gandara DR, Grimminger P, Mack PC, Lara PN
Jr, Li T, Danenberg PV and Danenberg KD: Association of epidermal
growth factor receptor activating mutations with low ERCC1 gene
expression in non-small cell lung cancer. J Thorac Oncol.
5:1933–1938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalikaki A, Koutsopoulos A, Hatzidaki D,
Trypaki M, Kontopodis E, Stathopoulos E, Mavroudis D, Georgoulias V
and Voutsina A: Clinical outcome of patients with non-small cell
lung cancer receiving front-line chemotherapy according to EGFR and
K-RAS mutation status. Lung Cancer. 69:110–115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang S, Wang Z, Guo J, Liu J, Li C, Liu L,
Shi H, Liu L, Li H, Xie C, et al: Correlation between EGFR mutation
status and response to first-line platinum-based chemotherapy in
patients with advanced non-small cell lung cancer. Onco Targets
Ther. 7:1185–1193. 2014.PubMed/NCBI
|
|
17
|
Ren S, Chen X, Kuang P, Zheng L, Su C, Li
J, Li B, Wang Y, Liu L, Hu Q, et al: Association of EGFR mutation
or ALK rearrangement with expression of DNA repair and synthesis
genes in never-smoker women with pulmonary adenocarcinoma. Cancer.
118:5588–5594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu CW, Wang G, Wang WL, Gao WB, Han CJ,
Gao JS, Zhang LY, Li Y, Wang L, Zhang YP, et al: Association
between EML4-ALK fusion gene and thymidylate synthase mRNA
expression in non-small cell lung cancer tissues. Exp Ther Med.
9:2151–2154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee JO, Kim TM, Lee SH, Kim DW, Kim S,
Jeon YK, Chung DH, Kim WH, Kim YT, Yang SC, et al: Anaplastic
lymphoma kinase translocation: A predictive biomarker of pemetrexed
in patients with non-small cell lung cancer. J Thorac Oncol.
6:1474–1480. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Besse B, Olaussen KA and Soria JC: ERCC1
and RRM1: Ready for prime time? J Clin Oncol. 31:1050–1060. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Metro G, Zheng Z, Fabi A, Schell M,
Antoniani B, Mottolese M, Monteiro AN, Vici P, Rivera Lara S,
Boulware D, et al: In situ protein expression of RRM1, ERCC1, and
BRCA1 in metastatic breast cancer patients treated with
gemcitabine-based chemotherapy. Cancer Invest. 28:172–180. 2010.
View Article : Google Scholar : PubMed/NCBI
|