Introduction
Breast cancer is the most common cancer diagnosed in
women (1). Previous studies
involving gene expression profiling and molecular pathological
analyses have demonstrated that approximately 10–12% of breast
cancer patients are diagnosed with triple-negative breast cancer
(TNBC), and 80% of TNBCs demonstrate the basal-like gene expression
pattern (2). TNBCs are
characterized by the absence of estrogen and progesterone
receptors, as well as human epidermal growth factor receptor-2.
Therefore, endocrine therapy is not recommended as a treatment
option for patients with TNBC, and conventional chemotherapy is
currently the only treatment option. In the clinic, poor prognosis,
high recurrence rates, and drug resistance in patients with TNBC
are considered to be the negative outcomes of conventional
chemotherapy. Therefore, current studies have focused on
identifying novel targets that may be applied for the prediction
and treatment of breast cancer. For instance, luminal-like breast
cancers that present with upregulation of the glucosaminyl
(N-acetyl) transferase-1 gene are more aggressive than basal-like
breast cancers (3). In addition,
O-glycosylated mucin 1 has been used as a biomarker for breast
cancer (4).
Over 50% of all secreted and cellular proteins
undergo glycosylation as a post-translational modification.
Glycans, located on glycoproteins, participate in a number of
biological processes, including immune responses, signal
transduction pathways, invasion, metastasis and drug tolerance
(5,6). Glycan overexpression generally
results in the generation of a dense glycosylation network, which
covers the cell surface and protects the tumor from detection and
drug exposure (7). O- and
N-glycosylation are the two main types of glycosylation in cells
(8). O-glycosylation on the
surface of epithelial cells in various organs, including the colon,
breast and stomach, has a higher molecular weight than
N-glycosylation (9).
O-glycosylation is a complex process that involves several
glycosyltransferase enzymes. Cancer-associated O-glycans have been
attributed, at least in part, to alterations in the expression of
key glycosyltransferases (10).
O-glycans begin with a Tn antigen, which is catalyzed by the
polypeptide N-acetylgalactosaminyltransferases (ppGalNAc-Ts), and
end with sialic acids, fucose and sulfate (11). Cancer-associated O-glycans may be
highly sialylated and less sulfated. Sialylation enzymes are
further classified into four families based on their substrates and
tissue distribution specificities, including sialyltransferase
(ST)-3 Gal (α2,3-ST), ST6Gal (α2,6-ST), ST6GalNAc and ST8Sia
(α2,8-ST) (12,13). In breast cancer, α2,6 sialylation
contributes to cell-to-cell adhesion of MDA-MB-435 cells (14). In addition, overexpression of
α2,3-ST results in the production of core 1-based sialylated
glycans in MDA-MB-231 and T47D cells (15). Furthermore, ST3Gal I may be a tumor
promoter in breast cancer, as indicated by mammary tumor
development in ST3Gal I/PyMT mice (16).
Cell type-specific glycosylation maintains and
stabilizes the conformation of peptides and proteins, and affects
their physical and chemical properties. Matrix metalloproteinase-14
(MMP14) is a collagenolytic enzyme that is involved in various
pathological processes, such as cancer invasion and metastasis
(17). MMP14 is located on the
cell surface and consists of a short transmembrane domain, a
cytoplasmic tail, and four potential O-glycosylation sites at its
hinge region. Previous studies have identified four potential
target sites on MMP14 (Thr291, Thr299, Thr300, and Ser301)
(18). MMP14 is regulated at the
transcriptional and post-transcriptional levels by multiple
coordinated mechanisms. Incomplete glycosylation stimulates
extensive autocatalytic degradation and self-inactivation of MMP14.
In addition, terminal sialylation is an important functional moiety
of the glycoprotein region of MMP14 (19).
Taking into account the results of previous studies,
the present study investigated the role of ppGalNAc-T1 and
ppGalNAc-T2 in the initiation of O-linked glycosylation, and
explored their effects on the terminal α2,3-sialylation of MMP14.
The results of the present study indicated that ppGalNAc-T1 serves
a more important role than ppGalNAc-T2 in mediating the initiation
of GalNAc-O-Ser/Thr (Tn antigen). In addition, abnormal O-linked
glycosylation initiation caused by the inhibition of
glycosyltransferase via RNA interference (RNAi), led to a decrease
in α2,3 terminal sialylation, which was correlated to the
self-proteolysis of MMP14. Furthermore, a low density of cell
surface α2,3-sialic acid contributed to an increase in
intracellular drug uptake, as well as antineoplastic and antitumor
drug effects. These effects resulted in decreased invasion and
metastatic capabilities of MDA-MB-231 cells.
Materials and methods
Cell line
The highly invasive MDA-MB-231 cell line, the poorly
invasive luminal A MCF-7 cell line and the normal-like MCF-12A cell
line, were purchased from the American Type Culture Collection
(Manassas, VA, USA) and were used for the purposes of the present
study. All breast tumor cells were maintained at 80% confluence in
Dulbecco's Modified Eagle's medium (DMEM, Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.), 2 mM
glutamine (Sigma-Aldrich; Merck-Millipore, Darmstadt, Germany), 10
mM HEPES buffer (Sigma-Aldrich; Merck-Millipore), and penicillin
(100 IU/ml)-streptomycin (100 g/ml) (Beyotime Institute of
Biotechnology, Haimen, China) at 37°C in a humidified atmosphere
(5% CO2).
RNAi assay
siRNA-ppGalNAc-T1 and siRNA-ppGalNAc-T2 were
purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China) and
were generated by subcloning multiple ppGalNAc-T1 and ppGalNAc-T2
cDNAs, respectively (Table I).
MDA-MB-231 breast tumor cells were seeded onto a 6-well plate at a
density of 2×105 cells and cultured until they reached
80% confluence for transfection. Cells were transfected with 5 ml
siRNA-ppGalNAc-T1 and/or siRNA-ppGalNAc-T2 vectors, using
Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The 5 µl empty vector,
pEGFP-C1 (GenePharma Co., Ltd. Shanghai, China), was transfected
into the MDA-MB-231 cells and used as a control. Transfection
efficiency was determined by fluorescence microscopy (Zeiss GmbH,
Jena, Germany) according to the fluorescence intensity of the
control group. The optimal transfection system volume was
determined using the GAPDH interference effect with different times
and ratios.
 | Table I.Details of siRNA oligonucleotides
used for the purposes of this study. |
Table I.
Details of siRNA oligonucleotides
used for the purposes of this study.
| siRNA | Base sequence
(5′-3′) |
|---|
|
siRNA-GalNAc-T1-homo-1327 | F:
CCUGGUACCUAGAGAAUAUTT |
|
| R:
AUAUUCUCUAGGUACCAGGTT |
|
siRNA-GalNAc-T1-homo-586 | F:
GGCCUUUAGAGAGUUAUGUTT |
|
| R:
ACAUAACUCUAAAGGCCTT |
|
siRNA-GalNAc-T1-homo-2563 | F:
CCAGUUACAAUGCUCAAAUTT |
|
| R:
AUUUGAGCAUUGUAACUGGTT |
|
siRNA-GalNAc-T2-homo-1065 | F:
GAGGAGAGAACCUAGAGAUTT |
|
| R:
AUCUCUAGGUUCUCUCCUCTT |
|
siRNA-GalNAc-T2-homo-649 | F:
GCCAGUUCUUAGAAAUGAUTT |
|
| R:
AUCAUUUCUAAGAACUCGCTT |
|
siRNA-GalNAc-T2-homo-519 | F:
CACCCAUCAUCGAUGUCAUTT |
|
| R:
AUGACAUCGAUGAUGGGUGTT |
| GAPDH negative
control | F:
GUAUGACAACAGCCUCAAGTT |
|
| R:
CUUGAGGCUGUUGUCAUACTT |
Gene expression analysis by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 1.5×106
MDA-MB-231 cells and 1.5×106 MDA-MB-231 cells that
transfected with siRNA using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and treated with DNase I (Ambion; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA concentration and quality was measured using an ultraviolet
spectrometer at a wavelength ratio of 260/280 nm. cDNA was
synthesized from 1 µg total RNA using an RNA reverse transcription
diagnostic kit (Takara Bio, Inc., Otsu, Japan). PCR reaction
mixtures (30 µl) were amplified with 40 cycles of denaturation at
95°C for 60 sec, annealing at 56°C for 60 sec and extension at 72°C
for 120 sec. In addition, RT-qPCR reaction mixtures (25 µl) were
amplified using 40 cycles of denaturation at 95°C for 60 sec,
annealing at 95°C for 15 sec and extension at 60°C for 60 sec.
RT-qPCR was performed using an ABI PRISM 7500 Sequence Detection
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
SYBR Green Realtime PCR Master Mix (Toyobo Co., Ltd., Osaka,
Japan). Gene expression was quantified using the 2−∆ΔCq
method (20) following
normalization to the expression of GAPDH. The primer
sequences used for RT-qPCR analysis are presented in Table II.
 | Table II.Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table II.
Sequences of primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Human gene | Base sequence
(5′-3′) |
|---|
| GAPDH | F:
TGCTGAGTATGTCGTGGAGT |
|
| R:
AGTTGTCATATTTCTCGTGG |
| MMP2 | F:
AGATACCCTCAAGAAGATGC |
|
| R:
AGCATCATCCACGGTTTCAG |
| MMP9 | F:
AGTCTGGATAAGTTGGGTCT |
|
| R:
AGATGTCGTGTGAGTTCCAG |
| MMP14 | F:
AAACATCAAAGTCTGGGAAGG |
|
| R:
ACTTGGGATACCCTGGCTCT |
| MMP15 | F:
ATCCAGAACTACACTGAGAAG |
|
| R:
TGGAAGCCAGAGGCAAAGAG |
| TGFβ-1 | F:
ACCAAAGACATCTCACACAG |
|
| R:
ACCAAGGTAACGCCAGGAAT |
| VEGF | F:
ATGGATGTCTACCAGCGAAG |
|
| R:
ATGGTGATGTTGCTCTCTGAC |
| ppGalNAc-T1 | F:
GCCAGGATCAAACATGACAG |
|
| R:
GAGCCTGCCATGTACTCAAA |
| ppGalNAc-T2 | F:
GAGAAAGCACAAAGCATGGA |
|
| R:
ATCGTCCCTCCAACATAAGC |
| ppGalNAc-T3 | F:
ATCCAGAGGTGTATGTGCCA |
|
| R:
GGTTTGCCTCCTTGATTGTT |
| ppGalNAc-T4 | F:
CGTCCTCACTTTCCTGGATT |
|
| R:
CTGCTGTTTCATCTCTCCCA |
| ppGalNAc-T7 | F:
TGGGACCAGAATTCAAACAA |
|
| R:
TGCATTCTTCTTGGCGTAAG |
| ppGalNAc-T10 | F:
GAAGTTCTGCTTTGATGCCA |
|
| R:
CACTGACAGGGTGGTACAGG |
| ppGalNAc-T11 | F:
GGCTATACCAGGTGTCGGTT |
|
| R:
AATGCCACTGCTGTGAAGAG |
Western blot analysis
MDA-MB-231 cells (1.5×106) and MDA-MB-231
cells (1.5×106) that transfected with siRNA were
harvested and homogenized with lysis buffer, consisting of 50 mM
Tris-HCl (pH 7.3), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 1%
protease inhibitor cocktail (Beyotime Institute of Biotechnology).
Total protein (15 µg) from each well was boiled for 10 min in 17%
Laemmli buffer (Beyotime Institute of Biotechnology) and separated
via 10% SDS-PAGE, before samples were transferred to a
polyvinylidene difluoride (PVDF) membrane using a semi-dry
technique. The membranes were blocked for ~1.5 h with 5% skim milk
in Tris-buffered saline (TBS), containing 10 mM Tris, 150 mM NaCl
(pH 7.9) and 0.05% Tween-20 (Beyotime Institute of Biotechnology).
The membranes were then incubated with the appropriate primary
antibodies [Anti-MMP14 antibody (cat. no. ab51074) was purchased
from Abcam Co., Ltd., Cambridge, UK. GAPDH antibody (cat. no.
AF0006) was purchased from Beyotime Institute of Biotechnology.
Dilution, 1:1,000] at 4°C overnight. Following washing with TBS
Tween-20 buffer, the cells were incubated with the appropriate
horseradish peroxidase (HRP)-conjugated IgG-AP secondary antibody
(Beyotime Institute of Biotechnology. Dilution, 1:1,000) at room
temperature for 90 min. After washing in TBS Tween-20 buffer,
detection and autoradiography were performed using a
Chemiluminescence Western Blotting kit (Roche Diagnostics GmbH,
Mannheim, Germany), according to the manufacturer's instructions.
Blots were quantified using Quantity One software version 4.62
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Lectin blot analysis
Whole-cell extracts were harvested and homogenized
as aforementioned. Total protein (15 µg) from each well was boiled
for 10 min in Laemmli buffer, separated via 10% SDS-PAGE, and then
transferred onto a PVDF membrane using a semi-dry technique. The
membranes were blocked for ~1.5 h with 3% bovine serum albumin
(BSA) in phosphate-buffered saline (PBS) at room temperature,
before they were incubated with biotinylated Maackia Amurensis
Lectin II (MALII; dilution, 1:500) in PBS with Tween-20 buffer
(PBST) for 1 h at room temperature. After washing in PBST buffer,
the membranes were incubated with HRP-streptavidin (dilution,
1:1,000; Beyotime Institute of Biotechnology) for 1 h at room
temperature and then washed with PBST buffer. Detection and
autoradiography were performed using a Chemiluminescence Western
Blotting kit (Roche Diagnostics GmbH), according to the
manufacturer's protocol.
Analysis of lectin labeling by flow
cytometry
Cells were digested with 0.25% trypsin (Gibco;
Thermo Fisher Scientific, Inc.) and harvested in 1.5 ml Eppendorf
tubes. Cell density was adjusted to 2×106/ml with lectin
buffer (10 mM Hepes, 0.15 M NaCl, 0.08% sodium azide, 0.1 mM
Ca2+ and 0.01 mM Mn2+, pH 7.5), before they
were stained with 10 µg/ml MALII (Sigma-Aldrich; Merck-Millipore)
in PBS (containing 0.5% BSA and 0.05% sodium azide) at 37°C for 120
min. The cells were then washed three times with PBS and incubated
with phycoerythrin-avidin (Sigma-Aldrich; Merck-Millipore) at 37°C
for 60 min. Following incubation, the cells were collected and
fluorescence intensity was measured using a FACScan flow cytometer
(BD Biosciences, Franklin Lakes, NJ, USA) and analyzed with BD
CellQuest software version 7.5.3 (BD Biosciences).
Immunoprecipitation
1.5×106 MDA-MB-231 cells and
1.5×106 MDA-MB-231 cells that transfected with siRNA
Cells were harvested and homogenized with lysis buffer as described
in Western blot analysis. The primary rabbit monoclonal antibody
against MMP14 (cat. no. ab51074, Abcam, Cambridge, UK) was added to
the cell lysate and incubated at 4°C overnight. A total of 30 ml
protein A agarose beads (Beyotime Institute of Biotechnology) was
incubated with the antigen-antibody complex at 4°C for 2–4 h. The
immunoprecipitation products were washed five times with cold PBS.
Finally, the beads were washed with 5 µl PBS, boiled, and gently
centrifuged at 2,000 × g for 2 min at room temperature, and the
recovered samples were run on an 8% SDS-PAGE for lectin blotting
using the aforementioned methods.
In vitro cytotoxicity assay
Drug resistance was measured by using the Cell
Counting Kit-8 (CCK-8; C0038, Beyotime Institute of Biotechnology)
assay. MDA-MB-231 cells and 1.5×106 MDA-MB-231 cells
that transfected siRNA-ppGalNAc-T1 and siRNA-ppGalNAc-T2, were
plated onto 96-well plates (Corning Life Sciences, Tewksbury, MA,
USA) at a density of 2×105 and incubated with 0.1, 1, 10
and 100 ng/µl docetaxel (DOC; Jiangsu Aosaikang Pharmaceutical Co.,
Ltd., Nanjing, China) and 0.5, 2.5, 5 and 10 ng/µl gemcitabine
hydrochloride (GEM; Qilu Pharmaceutical Co., Ltd., Jinan, China)
anticancer drugs for 48 h. Cells were then treated with 100 ml
CCK-8 (Beyotime Institute of Biotechnology) and incubated at 37°C
and 5% CO2 for 4 h. The absorbance value, at a
wavelength of 450 nm, was measured using a microplate reader
(Thermo Fisher Scientific, Inc.,). Each group contained three wells
and was prepared in triplicate. The drug concentrations that
resulted in 50% growth inhibition (the half maximal inhibitory
concentration, IC50 value) were determined.
Transwell invasion assays
In order to evaluate the invasion ability of the
cells transfected with siRNA-ppGalNAc-T1 and siRNA-ppGalNAc-T2,
Transwell plates (Corning Life Sciences) with a filter diameter of
6.5 mm and a pore size of 8.0 µm were employed. According to the
manufacturer's instructions, cells (density, 1×105) were
plated into the upper chambers of 8-µm Transwells in serum-free
medium, and incubated for 24 h to generate serum-starved cells.
Following serum starvation, the culture medium was decanted and 500
µl DMEM was added to the upper chamber, while 500 µl DMEM plus 10%
FBS was introduced to the lower compartment. The cells and Matrigel
(included in the Transwell plates) were incubated at 37°C for 24 h,
before washing twice with pre-heated PBS. The filters were
subsequently fixed with 1 ml 1% paraformaldehyde for 15 min at room
temperature. Following removal of the paraformaldehyde solution
with PBS, the cells and Matrigel were stained with 500 µl eosin
staining solution (Beyotime Institute of Biotechnology) for 20 min
at room temperature, and then rinsed twice with distilled water.
Cell penetration through the membrane was quantified by counting
the number of cells that had diffused through the membrane in 10
fields of view (magnification, ×200) using a light microscope.
Experiments were performed in triplicate.
Statistical analysis
Results are expressed as the mean ± standard
deviation of triplicate experiments. Statistically significant
differences were determined using SPSS version 16.0 software (IBM
SPSS, Armonk, NY, USA). The independent samples t-test was
performed to compare the data collected from independent samples.
P<0.05 was considered to indicate a statistically significant
difference.
Results
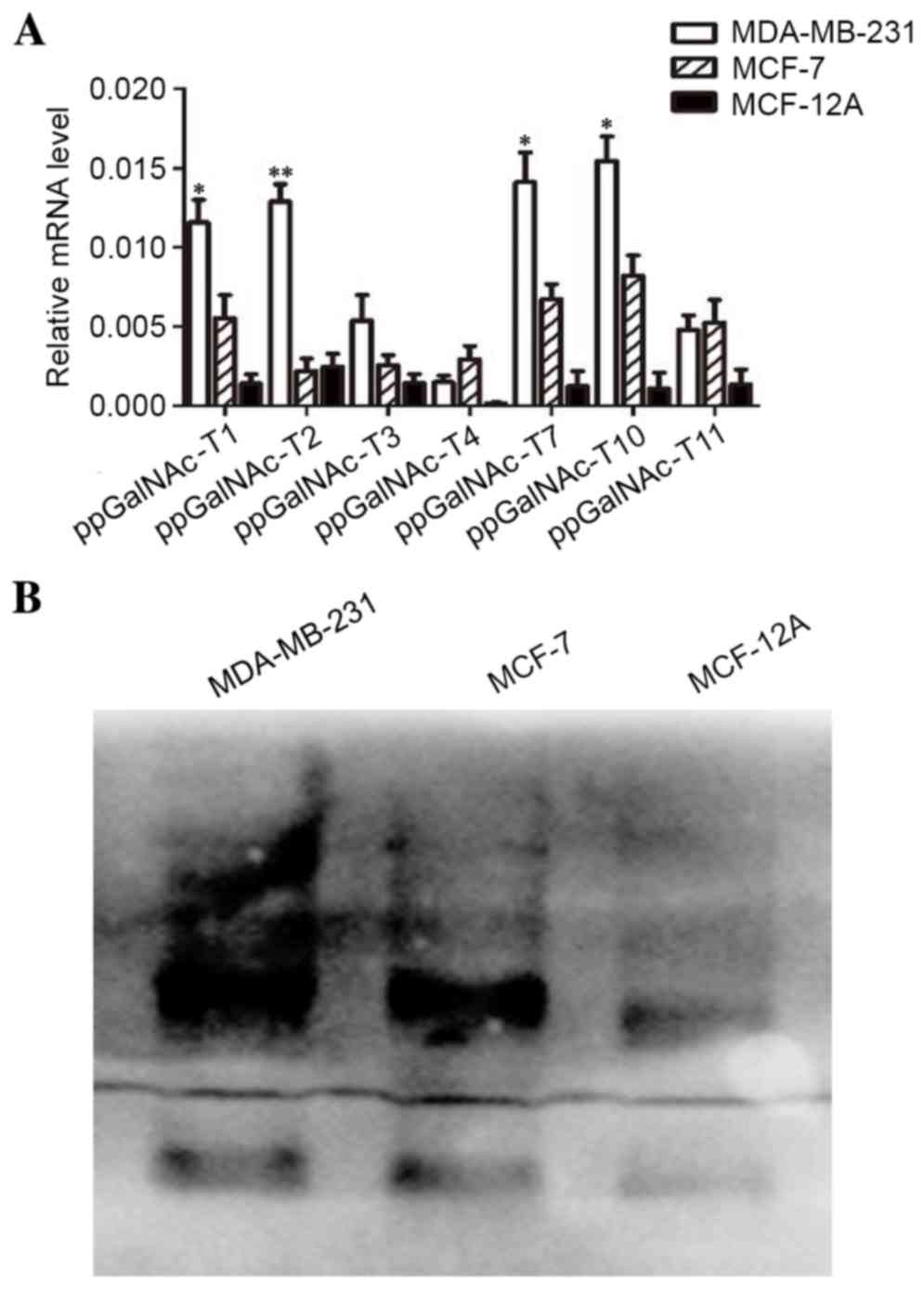
Differential expression of
glycosyltransferases and α2,3-sialic acid in highly and poorly
invasive breast cancer cells
In order to determine the expression level of
glycosyltransferase enzymes in breast cancer cells, the expression
levels of ppGalNAc-T1, T2, T3, T4,
T7, T10 and T11 in highly invasive [estrogen
receptor (ER)-negative] MDA-MB-231 cells and poorly invasive
(ER-positive) MCF-7 cells, as well as in normal-like MCF-12A
mammary cells. The mRNA expression level of these genes was
detected by RT-qPCR analysis. As shown in Fig. 1A, the expression of
ppGalNAc-T1, T2, T7 and T10 in
MDA-MB-231 cells was significantly higher when compared with MCF-7
and MCF-12A cells (T1: P-value, MDA-MB-231 vs. MCF-7=0.035,
MDA-MB-231 vs. MCF-12A=0.0096; T2: MDA-MB-231 vs. MCF-7=0.022,
MDA-MB-231 vs. MCF-12A=0.0082; T7: MDA-MB-231 vs. MCF-7=0.028,
MDA-MB-231 vs. MCF-12A=0.0075; T10: MDA-MB-231 vs. MCF-7=0.031,
MDA-MB-231 vs. MCF-12A=0.0062). In addition, the expression of α2,3
sialic acid was detected by lectin blot analysis. As shown in
Fig. 1B, α2,3-sialic acid was
expressed in breast cancer cells and its expression in MDA-MB-231
cells was markedly higher when compared with MCF-7 and MCF-12A
cells. Upregulation of glycosyltransferase enzymes promotes
O-glycosylation, which may, in turn, lead to the increased
production of terminal α2,3-sialic acids in highly invasive breast
cancer cells (21). Considering
the strong catalytic activity of glycosyltranferase enzymes on
peptide substrates of ppGalNAc-T1 and -T2, the present study
assessed the importance of ppGalNAc-T1 and -T2 in O-linked
glycosylation initiation. In addition, the inhibitory effects of
downstream sugar chain extension on tumor invasion and the
metastatic potential of breast cancer cells was examined.
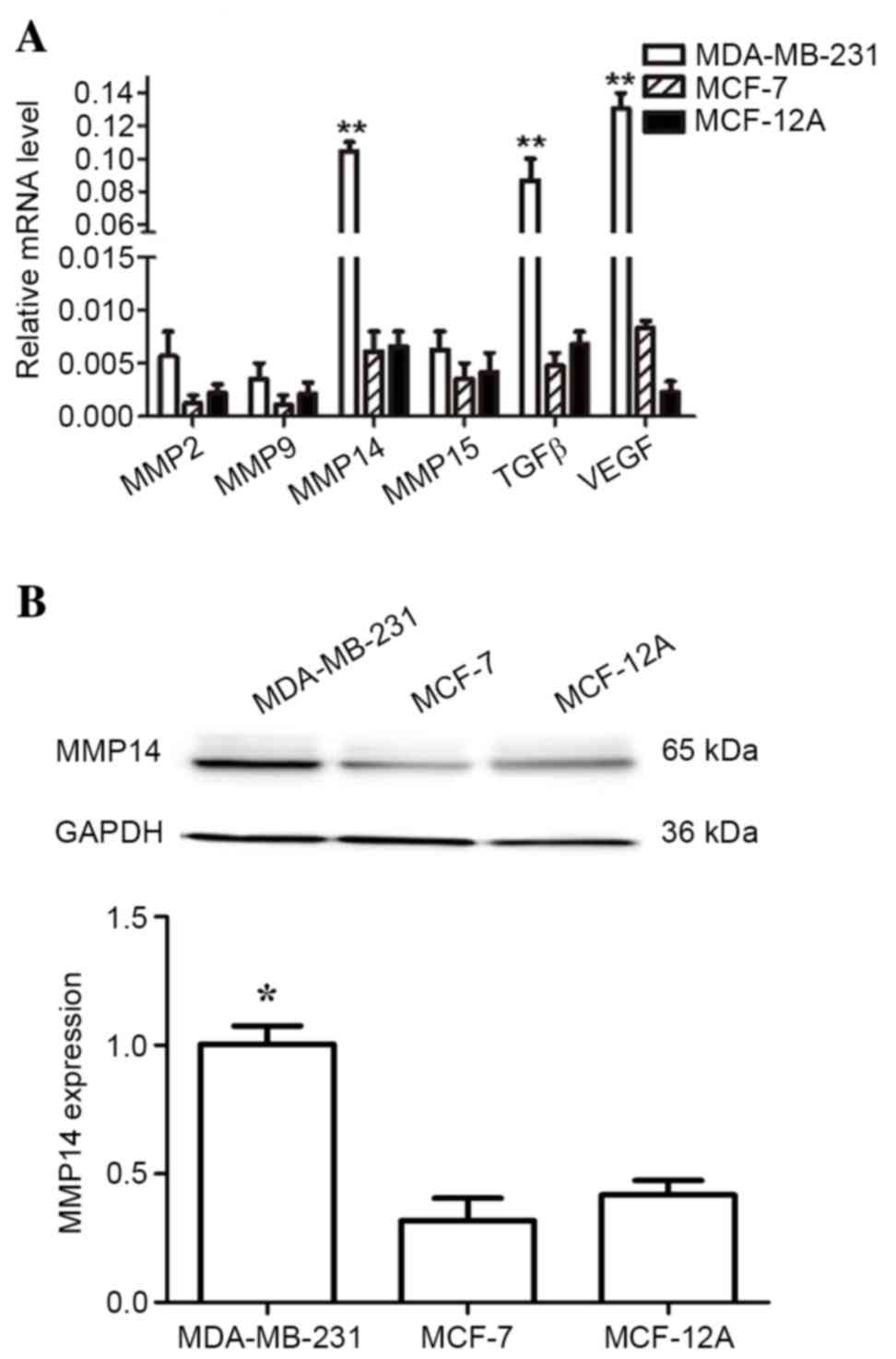
Differential expression of MMPs,
vascular endothelial growth factor (VEGF) and transforming growth
factor β (TGFβ) in breast cancer cells at various degrees of
malignancy
Of the identified members of the MMP family, MMP2 is
an enzyme that has been reported to directly participate in the
degradation of the extracellular matrix (ECM). By contrast, MMP14
is involved in tumor invasion and metastasis through activation of
pro-MMP2 (22). In order to
investigate the expression of metastasis-associated genes in breast
cancer cells, the present study measured the expression of the
MMP2, MMP9, MMP14, MMP15, TGFβ
and VEGF genes in MDA-MB-231, MCF-7, and MCF-12A cell lines
by RT-qPCR analysis. Fig. 2A shows
that MMP14, TGFβ, and VEGF expression levels
were significantly increased in highly invasive MDA-MB-231 cells,
when compared with MCF-7 and MCF-12A cells. Upregulation of
MMP14, TGFβ, and VEGF is associated with tumor
invasion, metastasis and angiogenesis (23). No alterations in the expression
levels of MMP2 among these cell lines were observed. Out of
all MMP proteins examined, only MMP14 was significantly upregulated
in MDA-MB-231 breast cancer cells at the mRNA and protein levels
(Fig. 2A and B). These results
suggest that MMP14 may serve an important role in the invasion and
metastasis of breast cancer cells.
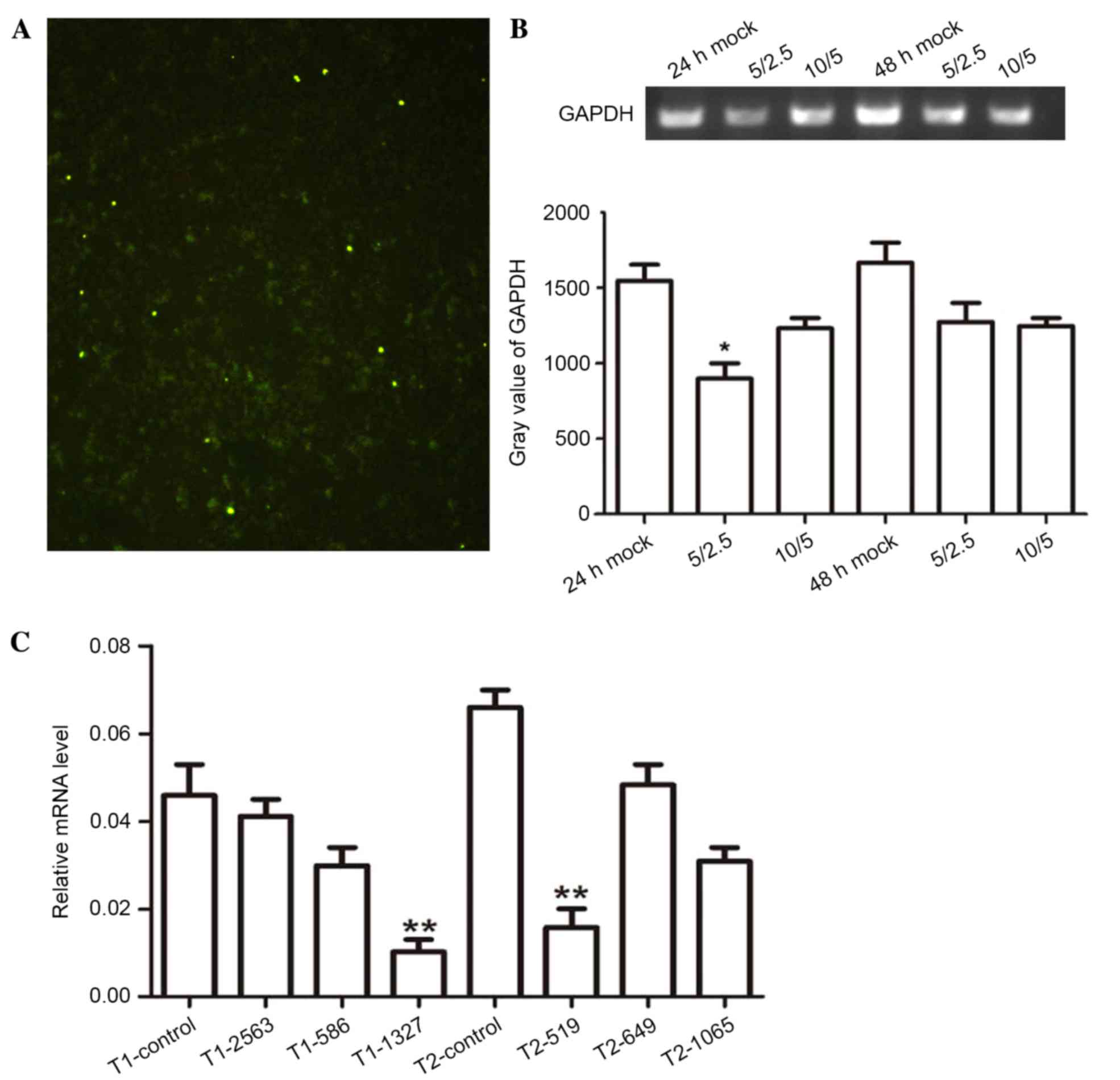
Silencing of ppGalNAc-T1 and
ppGalNAc-T2 in MDA-MB-231 cells by RNAi
To further explore the role of ppGalNAc-T1 and
ppGalNAc-T2 in breast tumors, highly invasive MDA-MB-231 cells were
transiently transfected with siRNA-ppGalNAc-T1 and for
siRNA-ppGalNAc-T2. The transfection efficiency was 72% and was
determined by fluorescence microscopy (Fig. 3A). The optimal transfection system
volume was determined using the GAPDH interference effect (Fig. 3B). ppGalNAc-T1 and
ppGalNAc-T2 mRNA levels in MDA-MB-231 cells were then
measured using RT-qPCR analysis. The expression levels of
ppGalNAc-T1 and ppGalNAc-T2 were significantly
downregulated following transfection with siRNA-ppGalNAc-T1-1327
and siRNA-ppGalNAc-T2-519 vectors, respectively, when compared with
control vectors (Fig. 3C,
P=0.0071, P=0.0093). These results demonstrated the optimal
concentration and interference fragments required to silence the
expression of ppGalNAc-T1 and ppGalNAc-T2 in
MDA-MB-231 cells. The results presented were used to further
examine the role of ppGalNAc-T1 and ppGalNAc-T2 in breast cancer
cells.
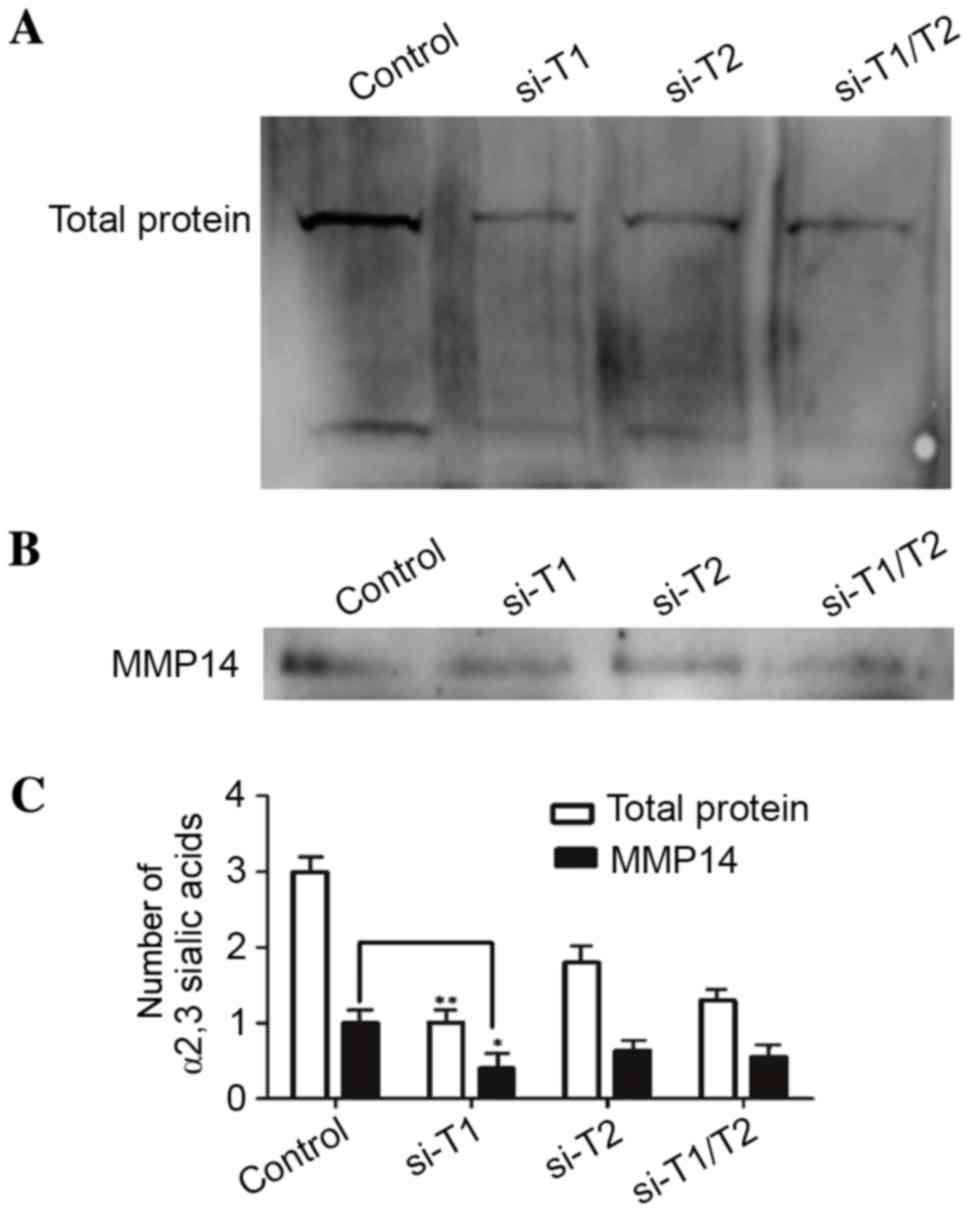
Effect of silencing ppGalNAc-T1 and
ppGalNAc-T2 on the expression of α2,3-sialic acid in MDA-MB-231
cells
ppGalNAc-Ts catalyze the attachment of the first
GalNAc monosaccharide to the polypeptide during the initiation of
O-linked glycosylation. The final step of O-linked glycosylation is
terminal sialic acid acidification, and the metabolic flux through
the sialic acid synthesis pathway is associated with substrate
availability (11). Thus, the
downregulated expression of ppGalNAc-T1 and
ppGalNAc-2 by RNAi may reduce terminal sialic acidification.
Fig. 4A demonstrates that the
terminal sialic acid in the RNAi groups was downregulated when
compared with the control group. In addition, downregulation of
ppGalNAc-T1 in MDA-MB-231 cells resulted in a lower density
of sialylated proteins when compared to that of the
siRNA-ppGalNAc-T2 groups and the co-interference groups. The
findings presented in the current study indicated that the sialic
acid structure of O-glycans in glycoproteins generally decreased
due to the reduction in substrate following the downregulation of
ppGalNAc-T1. ppGalNAc-T1 serves an important role in
O-glycosylation of MMP14 due to the presence of potential
O-glycosylation sites at the hinge region of MMP14, which was
demonstrated by the results of the lectin blot analysis of α2,3
sialic acids, including the 65 kDa protein of MMP14 proteins
(Figs. 1B and 2B). Based on these results, the authors
hypothesized that an α2,3 sialylation event had occurred at the
hinge region of MMP14. Fig. 4B
demonstrated the detection of α2,3 sialic acids on MMP14 using an
immunoprecipitation assay, and the expression of α2,3 sialic acids
in the RNAi groups was downregulated. The siRNA-ppGalNAc-T1 group
exhibited a lower density of α2,3 sialic acids when compared with
the siRNA-ppGalNAc-T2 and co-transfection groups. These results
suggest that α2,3 sialylation occurs in the hinge region of MMP14,
and terminal sialylation is regulated by the initiation of
O-glycosylation.
Effect of silencing ppGalNAc-T1 and
ppGalNAc-T2 on the expression of MMPs, VEGF and TGFβ
Fig. 2 demonstrated
that MMP14, VEGF, and TGFβ were upregulated in MDA-MB-231 cells
when compared with MCF-7 and MCF-12A cells. MMP14 is a key enzyme
involved in tumor invasion and angiogenesis through activation of
pro-MMP2 and regulating VEGF (24). Incomplete glycosylation of this
glycoprotein stimulates extensive autocatalytic degradation and
self-inactivation of MMP14 (19).
Due to the decrease in terminal α2,3 sialic acids on MMP14 by
transient transfection of siRNA constructs against ppGalNAc-T1 and
ppGalNAc-T2, the expression of MMP14, MMP2 and
VEGF were measured by RT-qPCR and western blot analyses.
Fig. 5A shows that MMP9,
VEGF, and TGFβ mRNA expression was downregulated.
MMP14 may activate MMP2 at post-transcription level; therefore, the
MMP2 mRNA expression remained unchanged. Fig. 5B shows that MMP14 protein
expression was downregulated in the RNAi groups, which was
consistent with the immunoprecipitation results presented in
Fig. 4B. In addition, alterations
in the protein expression patterns of MMP2 and VEGF were similar to
that of MMP14 in all experimental groups. Thus, decreasing α2,3
sialylation by silencing ppGalNAc-T1 may lead to extensive
autocatalytic degradation and self-inactivation of MMP14.
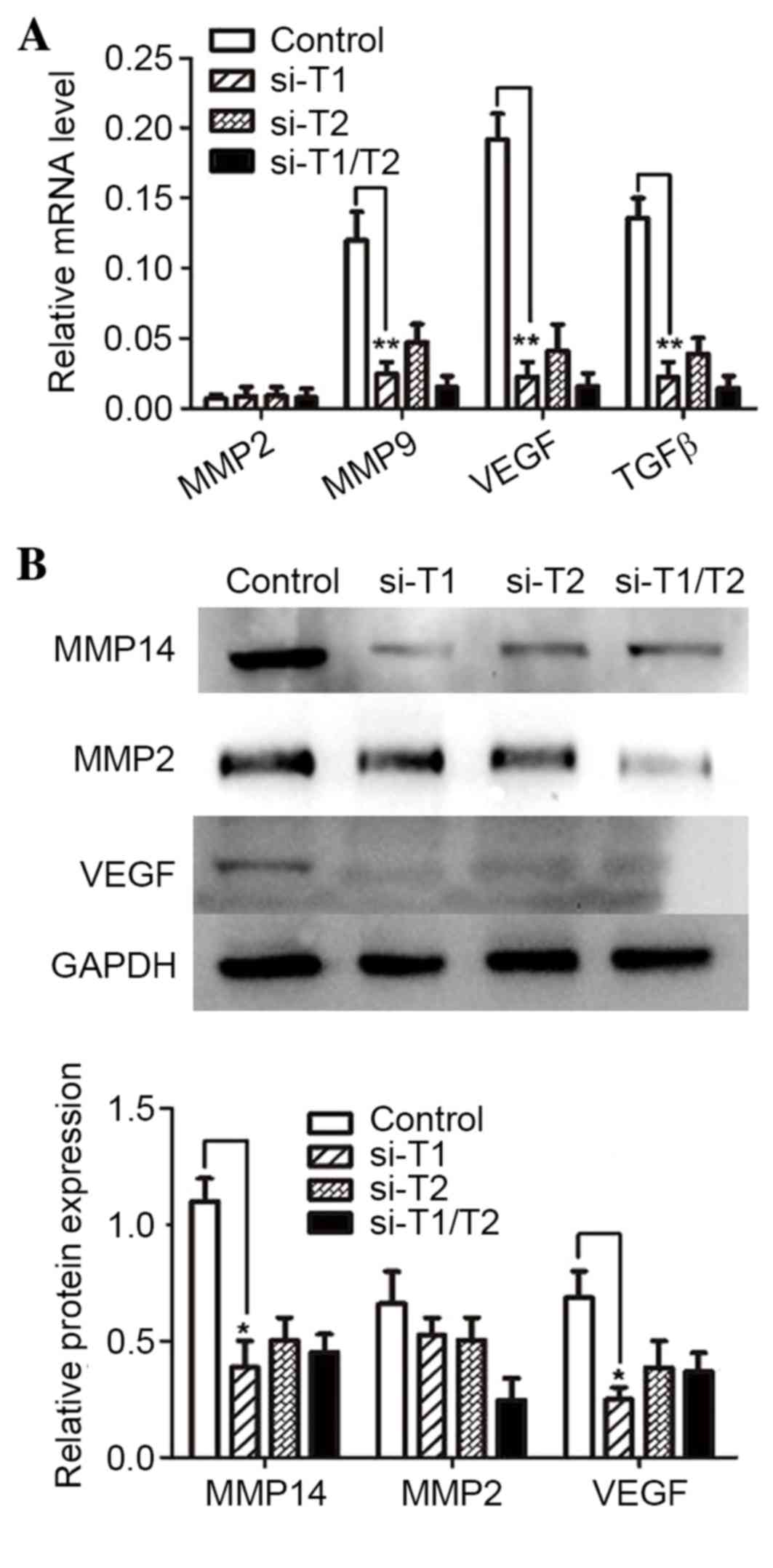 | Figure 5.Effect of silencing
ppGalNAc-T1 and ppGalNAc-T2 on the expression of
MMP2, MMP14, TGFβ and VEGF. (A) The expression levels of
MMP2, MMP14, TGFβ and VEGF mRNA in
control, siRNA-ppGalNAc-T1 (si-T1), siRNA-ppGalNAc-T2 (si-T2) and
co-transfection (si-T1/T2) groups as determined by reverse
transcription-quantitative polymerase chain reaction analysis. (B)
MMP2, MMP14 and VEGF protein expression was measured by western
blot analysis. Results are presented as the mean ± standard
deviation of three independent experiments, each measured in
triplicate. *P<0.05 and **P<0.01 vs. control. ppGalNAc-T,
polypeptide N-acetylgalactosaminyltransferase; MMP, matrix
metalloproteinase; VEGF, vascular endothelial growth factor. |
Knockdown of ppGalNAc-T1 and
ppGalNAc-T2 increases chemosensitivity to DOC and GEM
Chemotherapy resistance is a major obstacle for the
successful treatment of TNBC with chemotherapeutic agents. The
effect of ppGalNAc-T expression on the sensitivity of TNBC cells to
anticancer drugs remains elusive. In order to examine its effect in
more detail in the present study, the expression of ppGalNAc-T1 and
ppGalNAc-T2 was inhibited by RNAi in MDA-MB-231 cells prior to
treatment with DOC (0.1–100 µg/ml) or GEM (0.5–10 µg/ml) for 48 h.
As shown in Fig. 6A,
downregulation of ppGalNAc-T1 or ppGalNAc-T2 resulted in a greater
reduction in the proliferation of MDA-MB-231 cells treated with DOC
and GEM, when compared cells treated with DOC and GEM alone
(P-values, 1 µg/ml DOC, MDA-MB-231-si-T2: MDA-MB-231=0.003,
MDA-MB-231-si-T1: MDA-MB-231=0.042; 10 µg/ml DOC, MDA-MB-231-si-T2:
MDA-MB-231=0.0026, MDA-MB-231-si-T1: MDA-MB-231=0.038; 100 µg/ml
DOC, MDA-MB-231-si-T2: MDA-MB-231=0.041, MDA-MB-231-si-T1:
MDA-MB-231=0.040; P-values, 0.5 µg/ml GEM, MDA-MB-231-si-T2:
MDA-MB-231=0.031; 2.5 µg/ml GEM, MDA-MB-231-si-T1:
MDA-MB-231=0.002; 5 µg/ml GEM, MDA-MB-231-si-T1: MDA-MB-231=0.034,
MDA-MB-231-si-T2: MDA-MB-231=0.001; 100 µg/ml GEM,
MDA-MB-231-si-T2: MDA-MB-231=0.046, MDA-MB-231-si-T1:
MDA-MB-231=0.044). The IC50 values of the drugs were
significantly lower in the ppGalNAc-T1 siRNA
(ICDOC50=7.3, P=0.0005 vs. untreated MDA-MB-231 group;
ICGEM50=6.2, P=0.021 vs. untreated MDA-MB-231 group) or
ppGalNAc-T2 siRNA (ICDOC50=3.8, P=0.0002 vs. untreated
MDA-MB-231 group; ICGEM50=5.7, P=0.014 vs. untreated
MDA-MB-231 group) group compared with those in the untreated
MDA-MB-231 group (ICDOC50=19.2; ICGEM50=8.1).
Therefore, downregulation of ppGalNAc-T1 or ppGalNAc-T2 in
MDA-MB-231 cells resulted in increased chemosensitivity to
anti-tumor drugs.
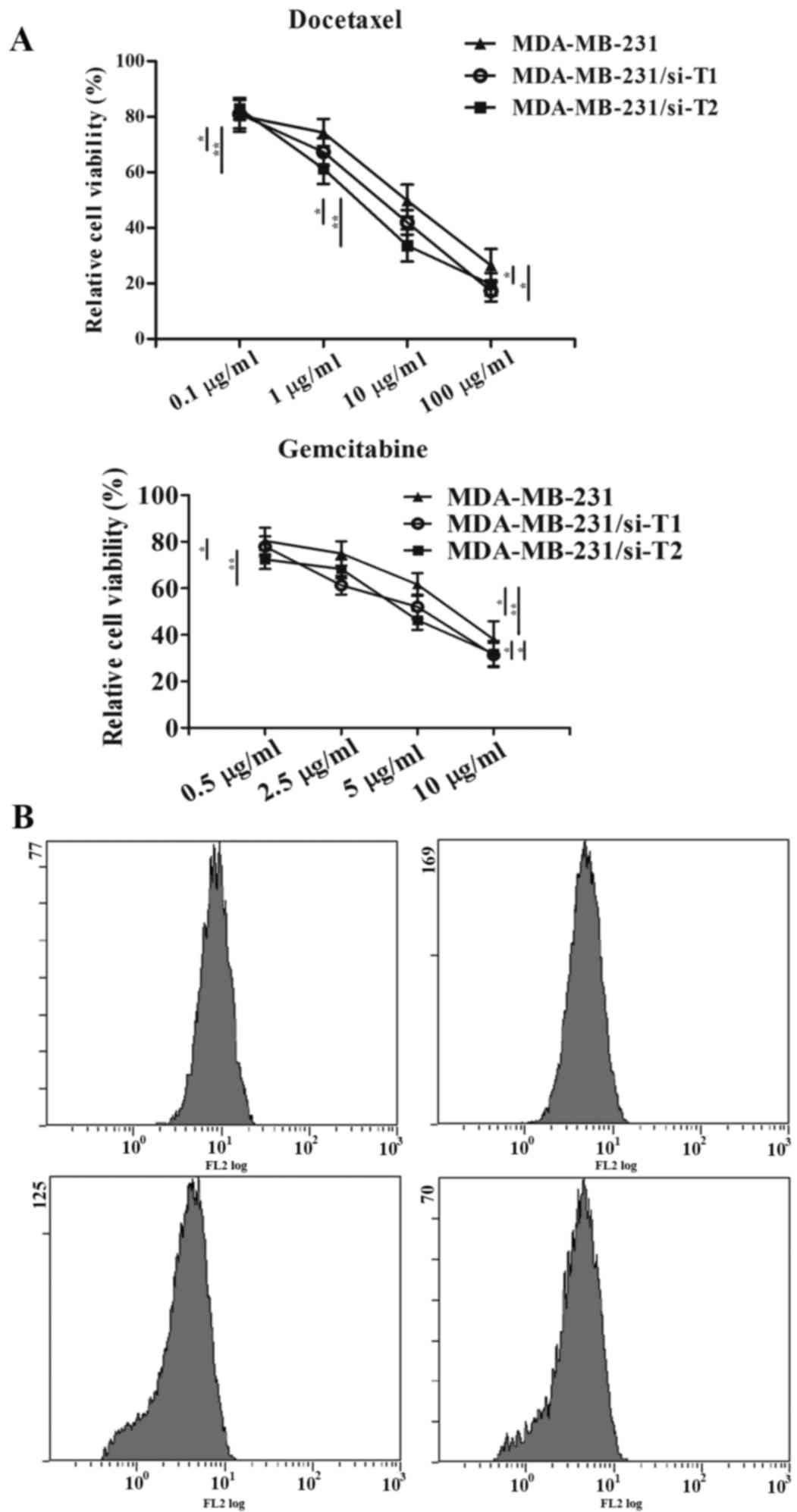 | Figure 6.Effect of ppGalNAc-T1 and ppGalNAc-T2
knockdown on the chemosensitivity and terminal α2,3 sialic acid
levels in MDA-MB-231 cells. (A) Cell viability was detected by
conducting an CCK-8 assay. Untreated controls, siRNA-ppGalNAc-T1
(si-T1) or siRNA-ppGalNAc-T2 (si-T2) groups were treated with 0.1,
1, 10 and 100 µg/ml DOC or 0.5, 2.5, 5 and 10 µg/ml gemcitabine.
*P<0.05, **P<0.01. (B) Terminal α2,3 sialic acid levels were
measured by flow cytometry. Untreated controls, si-T1 and si-T2
cells were treated with 10 µg/ml DOC for 24 h. Data are expressed
as the mean ± standard deviation of three independent experiments.
ppGalNAc-T, polypeptide N-acetylgalactosaminyltransferase; DOC,
docetaxel; CCK-8, cell counting kit-8. |
Flow cytometry assays were performed to evaluate the
inhibitory effect of ppGalNAc-T1 and ppGalNAc-T2 on terminal α2,3
sialylation. Cells were treated with 10 µg/ml DOC for 48 h.
Fig. 6B demonstrated a marked
reduction in cell surface α2,3 sialic acids between the control
group and the DOC treatment group. In addition, knockdown of
ppGalNAc-T1 and ppGalNAc-T2 inhibited sialylation further following
DOC treatment. These results indicate that ppGalNAc-T1 and
ppGalNAc-T2 may be involved in mediating drug resistance in TNBC
cells by regulating the generation of terminal α2,3 sialic acids by
O-glycosylation.
Effect of silenced ppGalNAc-T1 and
ppGalNAc-T2 expression on the invasion of breast cancer cells
The results presented so far, demonstrated that
MMP14 expression and associated α2,3 sialylation were affected by
the downregulation of ppGalNAc-T1 and ppGalNAc-T2. MMP14 is a
crucial membrane enzyme that participates in tumor invasion and
migration (24). Therefore, the
effect of ppGalNAc-T1 and ppGalNAc-T2 expression on the capacity of
breast carcinoma cells to metastasize was investigated in the
present study. Transwell invasion chambers coated with Matrigel
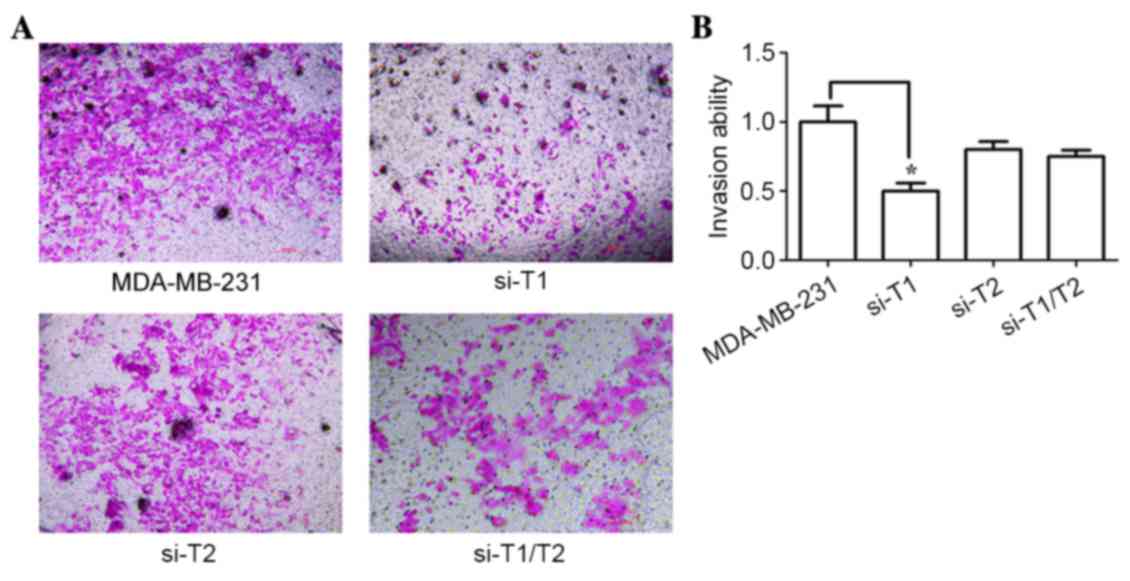
were used. Fig. 7 shows that the
RNAi groups exhibited a significant reduction in invasion
capabilities when compared with that of the control groups (P=0.013
<0.05). In addition, the inhibitory effect of the
siRNA-ppGalNAc-T1 group was significantly higher when compared with
that of the siRNA-ppGalNAc-T2 and co-interference groups (P=0.022;
P=0.031). These results suggest that ppGalNAc-T1 may regulate the
metastatic behavior of breast cancer cells by regulating the
initiation of O-glycosylation. In addition, decreasing α2,3
sialylation through silencing of ppGalNAc-T1 may lead to extensive
autocatalytic degradation and self-inactivation of MMP14.
Furthermore, the glycosyltransferase ppGalNAc-T1 may serve a more
important role in the glycosylation-initiation process than
ppGalNAc-T2.
Discussion
Information regarding the pathogenesis of TNBC
remains scarce, and requires further study. Previous clinical data
has demonstrated that high histological grade, advanced disease,
high recurrence rate and drug resistance influence the prognosis of
TNBC patients (25–27). As there are currently no standard
treatments for TNBC, researchers are conducting investigations to
address this issue using a number of different methods, including
the analysis of gene and receptor expression levels,
immunomodulatory activities and signaling pathways (28,29).
Glycomics is an emerging field of TNBC research that is currently
receiving extensive attention (30).
Aberrant glycoconjugates on the cell surface are a
common feature of tumor cells (31). O-linked alterations have been
detected by high performance liquid chromatography and
immunolocalization in various types of cancer cell lines, including
MCF-7, MDA-MB-231 and T47D (15,16).
O-glycan biosynthesis and its associated regulatory mechanisms are
relatively complex and poorly understood.
O-linked glycosylation is initiated by ppGalNAc-T by
catalyzing the addition of a single GalNAc monosaccharide to a
serine or threonine residue on the polypeptide. Based on in
silico analysis, a total of 24 unique human ppGalNAc-Ts genes
have been identified thus far, and ppGalNAc-T isoforms display
tissue-specific expression patterns in adult mammals, as well as
unique spatial and temporal patterns of expression during murine
development (32–34). In the present study, MDA-MB-231
cells were observed to express high levels of ppGalNAc-T1,
T2, T3, T7, T10 when compared to that
in MCF-7 and MCF-12A cells. ppGalNAc-T1 and ppGalNAc-T2 have been
previously demonstrated to exhibit strong catalytic activity on
bare peptide substrates (35).
ppGalNAc-T7 (36) and ppGalNAc-T10
(37) require prior glycosylation
by GalNAc for optimal activity. In addition, ppGalNAc-T1 and
ppGalNAc-T2 are expressed in a wide range of normal human tissues,
and at low levels in breast cancer (38). These enzymes possess housekeeping
functions and are ubiquitously expressed at a low level. Silencing
the expression of ppGalNAc-T1 inhibited the proliferation and tumor
growth of bladder cancer (39). A
previous study indicated that ppGalNAc-T2 regulates cellular
metastatic behavior in gastric cancer (40). Therefore, investigation of the role
of ppGalNAc-T1 and ppGalNAc-T2 on O-linked glycosylation
initiation, and its inhibitory effects on downstream sugar chain
extension, may facilitate an improved understanding of tumor
invasion and the metastatic potential of breast cancer.
Extension and termination reactions in
O-glycosylation are influenced by upstream enzymes and substrates.
For instance, core 1 is converted to the branched core 2 structure
by C2GnT (41), or to the
sialylated core 1 structure by α3-ST (42). The present study indicated that
α2,3 sialic acid is downregulated by exogenous ppGalNAc-T1 and
ppGalNAc-T2 siRNA. Downregulation of terminal sialylation was
associated with the inhibition of GalNAc-O-Ser/Thr (Tn antigen) by
interference. An increased level of sialic acids has been detected
in various cancer cells and were demonstrated to participate in
tumor-associated truncation of O-glycans (43). Increased sialic acid levels may
increase the viscosity of mucin, cover the glycoside antigen site
or inhibit drug exposure. The results of the present study
demonstrated that MDA-MB-231 cells transfected with
siRNA-ppGalNAc-T1 and siRNA-ppGalNAc-T2 exhibited low levels of
α2,3 sialic acids, and demonstrated higher levels of
chemosensitivity when compared with untreated MDA-MB-231 cells.
This suggested that a low level of α2,3 sialic acid may contribute
to an improvement of the efficacy of chemotherapy by enhancing drug
sensitivity. It has been previously reported that incomplete
glycosylation stimulates extensive autocatalytic degradation and
self-inactivation of MMP14 (19).
In addition, the sialylated terminal is the most important
functional moiety of the glycoprotein region of MMP14 (19). In the present study, the α2,3
sialic acids on MMP14 were identified by immunoprecipitation. This
clearly indicated that α2,3 sialic acid is involved in the
O-glycosylation of MMP14, and a lower density of α2,3 sialic acids
on MMP14 was detected in the siRNA-ppGalNAc-T1-treated groups.
MMP14 is an important membrane-type MMP involved in the breakdown
of the ECM in normal and disease processes, as well as tumor
invasion, metastasis and angiogenesis (44). In addition, MMP14 was observed to
be upregulated in highly invasive MDA-MB-231 cells. Due to its
potential O-glycosylation sites at the hinge region, together with
the important role of MMP14 in tumor invasion and migration in
breast cancer (45), the mRNA and
protein levels of MMP14, as well as MMP2 and VEGF, were
investigated in the present study. It has been widely accepted that
MMP2 is activated by MMP14 in vitro and in vivo
(46), and VEGF is upregulated by
MMP14 in breast carcinoma (47).
In the present study, downregulation of VEGF was comparable to the
observed downregulation of MMP14 in the RNAi groups, which was more
notable in the siRNA-ppGalNAc-T1 group when compared with that of
other groups. However, downregulation of MMP2 was not particularly
evident in the siRNA-ppGalNAc-T1 and siRNA-ppGalNAc-T2 groups,
whereas a notable reduction was observed in the co-transfected
groups. Considering the association between MMP2 and MMP14
(22), this result requires
further investigation in future studies. The invasion capabilities
of MDA-MB-231 cells in RNAi groups was reduced, and the inhibitory
effect in the siRNA-ppGalNAc-T1 group was significantly higher when
compared with that of the siRNA-ppGalNAc-T2 and co-transfection
groups.
In conclusion, the downregulation of ppGalNAc-T1 and
ppGalNAc-T2 in MDA-MB-231 cells decreased the terminal α2,3 sialic
acid levels on MMP14, which may have been responsible for the
observed decrease in invasion capabilities and increased
chemosensitivity in vitro. In addition, a significant
downregulation in VEGF expression, was observed when compared with
the RNAi group, which was more prominent in the siRNA-ppGalNAc-T1
group when compared with that of the other groups. Therefore, the
results of the present study suggest that ppGalNAc-T1 may serve a
more important role than ppGalNAc-T2 in mediating the initiation of
GalNAc-O-Ser/Thr (Tn antigen). In addition, abnormal O-linked
glycosylation initiation caused by the inhibition of
glycosyltransferase via RNAi, led to a decrease in α2,3 terminal
sialylation, which may have been associated with the
self-proteolysis of MMP14 in MDA-MB-231 cells. Furthermore, the
downregulation of MMP14 may have led to the observed decrease in
VEGF expression.
Acknowledgements
The current study was supported by the National
Natural Science Foundation of China (grant no. 31170772) and the
Suzhou Municipal Natural Science Foundation (grant nos. SYS201572
and zxy2013036).
References
|
1
|
Davies EL: Breast cancer. Medicine.
44:42–46. 2016. View Article : Google Scholar
|
|
2
|
Lin NU, Claus E, Sohl J, Razzak AR,
Arnaout A and Winer EP: Sites of distant recurrence and clinical
outcomes in patients with metastatic triple-negative breast cancer:
High incidence of central nervous system metastases. Cancer.
113:2638–2645. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim J, Villadsen R, Sørlie T, Fogh L,
Grønlund SZ, Fridriksdottir AJ, Kuhn I, Rank F, Wielenga VT,
Solvang H, et al: Tumor initiating but differentiated luminal-like
breast cancer cells are highly invasive in the absence of
basal-like activity. In: Proc Natl Acad Sci USA. 109. pp.
6124–6129. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haddon L and Hugh J: MUC1-mediated
motility in breast cancer: A review highlighting the role of the
MUC1/ICAM-1/Src signaling triad. Clin Exp Metastasis. 32:393–403.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saitoh O, Wang WC, Lotan R and Fukuda M:
Differential glycosylation and cell surface expression of lysosomal
membrane glycoproteins in sublines of a human colon cancer
exhibiting distinct metastatic potentials. J Biol Chem.
267:5700–5711. 1992.PubMed/NCBI
|
|
6
|
Devi KS, Behera B, Mishra D and Maiti TK:
Immune augmentation and Dalton's Lymphoma tumor inhibition by
glucans/glycans isolated from the mycelia and fruit body of
Pleurotus ostreatus. Int Immunopharmacol. 25:207–217. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kalra AV and Campbell RB: Mucin
overexpression limits the effectiveness of 5-FU by reducing
intracellular drug uptake and antineoplastic drug effects in
pancreatic tumours. Eur J Cancer. 45:164–173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ecker M, Mrsa V, Hagen I, Deutzmann R,
Strahl S and Tanner W: O-mannosylation precedes and potentially
controls the N-glycosylation of a yeast cell wall glycoprotein.
EMBO Rep. 4:628–632. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brockhausen I: Mucin-type O-glycans in
human colon and breast cancer: Glycodynamics and functions. EMBO
Rep. 7:599–604. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Varki A, Kannagi R and Toole BP:
Glycosylation Changes in CancerEssentials of Glycobiology. 2nd.
Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR,
Hart GW and Etzler ME: Cold Spring Harbor Laboratory Press; Cold
Spring Harbor, NY: 2009, View Article : Google Scholar
|
|
11
|
Nakada H: Map 2: Biosynthetic Pathways of
O-Glycans. handbook of glycosyltransferases and related genes.
1667–1671. 2014. View Article : Google Scholar
|
|
12
|
Büll C, Stoel MA, den Brok MH and Adema
GJ: Sialic acids sweeten a tumor's life. Cancer Res. 74:3199–3204.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schultz MJ, Swindall AF and Bellis SL:
Regulation of the metastatic cell phenotype by sialylated glycans.
Cancer Metastasis Rev. 31:501–518. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin S, Kemmner W, Grigull S and Schlag PM:
Cell surface alpha 2,6 sialylation affects adhesion of breast
carcinoma cells. Exp Cell Res. 276:101–110. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sproviero D, Julien S, Burford B,
Taylor-Papadimitriou J and Burchell JM: Cyclooxygenase-2 enzyme
induces the expression of the α-2,3-sialyltransferase-3 (ST3Gal-I)
in breast cancer. J Biol Chem. 287:44490–44497. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Picco G, Julien S, Brockhausen I, Beatson
R, Antonopoulos A, Haslam S, Mandel U, Dell A, Pinder S,
Taylor-Papadimitriou J and Burchell J: Over-expression of ST3Gal-I
promotes mammary tumorigenesis. Glycobiology. 20:1241–1250. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tagashira M and Toma K: Effect of Peptide
Glycosylation on the Conformation of the Peptide Backbone.
Cheminform. 37:362006. View Article : Google Scholar
|
|
18
|
Wu YI, Munshi HG, Sen R, Snipas SJ,
Salvesen GS, Fridman R and Stack MS: Glycosylation broadens the
substrate profile of membrane type 1 matrix metalloproteinase. J
Biol Chem. 279:8278–8289. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Remacle AG, Chekanov AV, Golubkov VS,
Savinov AY, Rozanov DV and Strongin AY: O-glycosylation regulates
autolysis of cellular membrane type-1 matrix metalloproteinase
(MT1-MMP). J Biol Chem. 281:16897–16905. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gómez H, Rojas R, Patel D, Tabak LA, Lluch
JM and Masgrau L: A computational and experimental study of
O-glycosylation. Catalysis by human UDP-GalNAc polypeptide: GalNAc
transferase-T2. Org Biomol Chem. 12:2645–2655. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shaverdashvili K, Wong P, Ma J, Zhang K,
Osman I and Bedogni B: MT1-MMP modulates melanoma cell
dissemination and metastasis through activation of MMP2 and RAC1.
Pigment Cell Melanoma Res. 27:287–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Loomans HA and Andl CD: Intertwining of
Activin A and TGFβ Signaling: Dual roles in cancer progression and
cancer cell invasion. Cancers (Basel). 7:70–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hui P, Xu X, Xu L, Hui G, Wu S and Lan Q:
Expression of MMP14 in invasive pituitary adenomas: Relationship to
invasion and angiogenesis. Int J Clin Exp Pathol. 8:3556–3567.
2015.PubMed/NCBI
|
|
25
|
Nielsen TO, Hsu FD, Jensen K, Cheang M,
Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler
L, et al: Immunohistochemical and clinical characterization of the
basal-like subtype of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guiu S, Michiels S, André F, Cortes J,
Denkert C, Di Leo A, Hennessy BT, Sorlie T, Sotiriou C, Turner N,
et al: Molecular subclasses of breast cancer: How do we define
them? The IMPAKT 2012 Working Group Statement. Ann Oncol.
23:2997–3006. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triplenegative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bilir B, Kucuk O and Moreno CS: Wnt
signaling blockage inhibits cell proliferation and migration, and
induces apoptosis in triple-negative breast cancer cells. J Transl
Med. 11:2802013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brooks M: Breast cancer screening and
biomarkers. Methods Mol Biol. 472:307–321. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paszek MJ, DuFort CC, Rossier O, Bainer R,
Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L,
et al: The cancer glycocalyx mechanically primes integrin-mediated
growth and survival. Nature. 511:319–325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang L, Tian T, Kelly G and Hagen T:
UDP-N-Acetyl-Alpha-D Galactosamine: Polypeptide
N-Acetylgalactosaminyltransferases (ppGalNAc-Ts). Handbook of
Glycosyltransferases and Related Genes. 495–505. 2014. View Article : Google Scholar
|
|
33
|
Ten Hagen KG, Fritz TA and Tabak LA: All
in the family: The UDP-GalNAc: Polypeptide
N-acetylgalactosaminyltransferases. Glycobiology. 13:1R–16R. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tian E and Ten Hagen KG: Expression of the
UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase family is
spatially and temporally regulated during Drosophila development.
Glycobiology. 16:83–95. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Perrine CL, Ganguli A, Wu P, Bertozzi CR,
Fritz TA, Raman J, Tabak LA and Gerken TA: Glycopeptide-preferring
polypeptide GalNAc transferase 10 (ppGalNAc T10), involved in
mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding
site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol
Chem. 284:20387–20397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bennett EP, Hassan H, Hollingsworth MA and
Clausen H: A novel human UDP-N-acetyl-D-galactosamine: Polypeptide
N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for
partial GalNAc-glycosylated acceptor substrates. FEBS Lett.
460:226–230. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng L, Tachibana K, Zhang Y, Guo JM,
Kahori Tachibana K, Kameyama A, Wang H, Hiruma T, Iwasaki H,
Togayachi A, et al: Characterization of a novel human UDP-GalNAc
transferase, pp-GalNAc-T10. FEBS Lett. 531:115–121. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Brooks SA, Carter TM, Bennett EP, Clausen
H and Mandel U: Immunolocalisation of members of the polypeptide
N-acetylgalactosaminyl transferase (ppGalNAc-T) family is
consistent with biologically relevant altered cell surface
glycosylation in breast cancer. Acta Histochem. 109:273–284. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ding MX, Wang HF, Wang JS, Zhan H, Zuo YG,
Yang DL, Liu JY, Wang W, Ke CX and Yan RP: ppGalNAc T1 as a
potential novel marker for human bladder cancer. Asian Pac J Cancer
Prev. 13:5653–5657. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hua D, Shen L, Xu L, Jiang Z, Zhou Y, Yue
A, Zou S, Cheng Z and Wu S: Polypeptide
N-acetylgalactosaminyltransferase 2 regulates cellular
metastasis-associated behavior in gastric cancer. Int J Mol Med.
30:1267–1274. 2012.PubMed/NCBI
|
|
41
|
Fukuda M:
Beta-1,3-Galactosyl-O-Glycosyl-Glycoprotein
Beta-1,6-N-Acetylglucosaminyltransferase 1 (GCNT1) (C2GnT-L) and
Beta-1,3-Galactosyl-O-Glycosyl-Glycoprotein
Beta-1,6-N-Acetylglucosaminyltransferase 3 (GCNT4) (C2GnT-T).
Handbook Of Glycosyltransferases and Related genes. 355–366.
2014.
|
|
42
|
Skrincosky D, Kain R, El-Battari A, Exner
M, Kerjaschki D and Fukuda M: Altered Golgi localization of core 2
beta-1,6-N-acetylglucosaminyltransferase leads to decreased
synthesis of branched O-glycans. J Biol Chem. 272:22695–22702.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pearce OM and Läubli H: Sialic acids in
cancer biology and immunity. Glycobiology. 26:111–128. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Woskowicz AM, Weaver SA, Shitomi Y, Ito N
and Itoh Y: MT-LOOP-dependent localization of membrane type I
matrix metalloproteinase (MT1-MMP) to the cell adhesion complexes
promotes cancer cell invasion. J Biol Chem. 288:35126–35137. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Têtu B, Brisson J, Wang CS, Lapointe H,
Beaudry G, Blanchette C and Trudel D: The influence of MMP-14,
TIMP-2 and MMP-2 expression on breast cancer prognosis. Breast
Cancer Res. 8:R282006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sato H, Takino T, Okada Y, Cao J,
Shinagawa A, Yamamoto E and Seiki M: A matrix metalloproteinase
expressed on the surface of invasive tumour cells. Nature.
370:61–65. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Deng YP, Li W and Li YL, Xu H, Liang SS,
Zhang LH and Li YL: MT1-MMP up-regulates VEGF expression in human
breast carcinoma MCF-7 cells and induces tumor angiogenesis.
Zhonghua Zhong Liu Za Zhi. 31:727–731. 2009.(In Chinese).
PubMed/NCBI
|