Introduction
Apoptosis is a spontaneous cell death subroutine,
that is triggered by various physiological and pathological stimuli
and that has distinct morphological characteristics and
energy-dependent mechanisms. Apoptosis has a crucial role in a
variety of processes, including normal cell population maintenance,
the appropriate development of the immune system, embryonic
development and chemical-induced cell death. Inappropriate
apoptosis (either too little or too much) may lead to
neurodegenerative diseases, autoimmune disorders, ischemic damage
and various types of cancer (1).
Previous studies have made substantial progress in the biochemical
and genetic exploration of apoptosis, resulting in the
identification of many key apoptotic genes, signaling pathways and
specific biochemical features (2),
the molecular mechanisms that underlie apoptosis remain to be fully
elucidated.
Apoptosis is believed to be strictly regulated by
various key apoptotic genes, including those in the caspase family,
the B cell leukemia/lymphoma 2 (Bcl-2) family, the poly(ADP-ribose)
polymerase (PARP) family, oncogenes and tumor suppressor genes.
Poly ADP-ribosylation (PARylation), a type of reversible protein
modification, is performed by the enzymes in the PARP family and
poly(ADP-ribose) (PAR)-degrading enzymes, such as PAR
glycohydrolase (3), ADP-ribosyl
hydrolase 3 (4) and members of the
nucleoside diphosphate linked moiety X family of proteins (5). It has been previously estimated that
~90% of PAR polymers are formed via the catalysis of PARP-1, which
was positively correlated with PARP activity. PARP-1 is activated
primarily by DNA breaks; however, it may also be activated by
alternative mechanisms, such as phosphorylation. Activated PARP-1
and PARylation regulate various cellular machineries, including
those involved in cell death (6).
A previous study suggested that PARylation and PARP-1 mediate
several cell death subroutines, including necrosis (necroptosis),
parthanatos, autophagy and apoptosis (7). Previous studies have demonstrated
that inhibiting PARP activity or knocking out/silencing PARP-1 may
sensitize cells to the cytotoxic effects of ionizing radiation or
DNA-damaging agents, such as DNA alkylates, cisplatin and
topoisomerase poisons (8,9). However, whether PARylation by PARP-1
affects the apoptotic process remains to be determined. In response
to mild DNA damage, PARylation facilitates DNA repair and therefore
survival, whereas more severe genotoxic stimuli may activate the
apoptotic pathway. Severe DNA damage may lead to in apoptosis or
necrosis, which are attributed to the depletion of donor
nicotinamide adenine dinucleotide (NAD+)/ATP by the
excessive activation of PARP-1 (10).
Hydroquinone (HQ) is a major active metabolite of
benzene and is commonly used as a substitute for benzene in in
vitro experiments. Benzene is a confirmed human carcinogen that
was proposed to be associated with myelodysplastic syndromes, acute
myeloid leukemia, non-Hodgkin lymphoma and childhood leukemia
(11,12). However, previous in vivo and
in vitro studies have determined that the pathways through
which HQ contributes to benzene-induced leukemia may also include
oxidative stress, DNA damage, cell cycle regulation and apoptosis
(13–15), the underlying molecular mechanisms
involved in HQ toxicity remain unclear. Our previous study revealed
that HQ leads to abnormal cell cycle progression that is
accompanied with the upregulation of PARP-1 in TK6 cells (16). A previous study, based on the
specificity of the synthesized substrate NAD+ used
affinity purification and tandem mass spectrometry and revealed
that zona occludens 2 (ZO-2) is a target of PARP-1 in HEK 293T
nuclear lysates (17). ZO-2, a
160-kDa protein (18), belongs to
the family of membrane-associated guanylate kinase (MAGUK)
homologs, which localize on the cytoplasmic surface of
intercellular contacts in epithelial and endothelial cells and have
also been observed to be distributed in the nucleus. ZO-2 affects
various cellular processes, including cell proliferation,
apoptosis, stress tolerance and barrier integrity (19,20).
It is of note that the importance of ZO-2 in lymphoblastoid cells
remains to be determined. The present study used TK6 lymphoblastoid
cells and PARP-1-silenced TK6 cells to investigate cell death and
cell fate specification following prolonged exposure to HQ. The
present study also aimed to determine the role of pleiotropic
PARP-1 in these processes and to identify the association between
PARP-1 and ZO-2.
Materials and methods
Chemicals and reagents
HQ, DAPI and mouse anti-human PAR (cat. no. MAB3192)
primary antibody were purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany). Rabbit anti-human Bcl-2 (cat. no. ab32124),
caspase-3 (cat. no. ab408) primary antibodies and a
2′,7′-dichlorofluorescin diacetate (DCFDA)-Cellular Reactive Oxygen
Species Detection assay kit were purchased from Abcam (Cambridge,
MA, USA). Rabbit anti-human PARP-1 antibody (cat. no. CST 9542),
ZO-2 (cat. no. CST 2847), Bax (cat. no. CST 5023P), GAPDH (cat. no.
CST 2118), mouse anti-human α-tubulin (cat. no. CST 3873) primary
antibodies, and Alexa Fluor 555-conjugated (CST 4409s) and
488-conjugated (cat. no. CST 4412s) secondary antibodies were
acquired from Cell Signaling Technology (Danvers, MA, USA). Mouse
anti-human PARP-1 antibodies (cat. no. sc8007) and horseradish
peroxidase-conjugated goat anti-rabbit (cat. no. sc2004) or goat
anti-mouse IgG (cat. no. sc2005) antibodies were obtained from
Santa Cruz Biotechnology (Dallas, TX, USA). Primers, TRIzol,
ECL-PLUS chemiluminescence assay and RevertAid First Strand cDNA
Synthesis kits were obtained from Thermo Fisher Scientific, Inc.
(Waltham, MA, USA). CellTiter-Glo Luminescent Cell Viability and
Caspase-Glo 3/7 assay kits were obtained from Promega Corporation
(Madison, WI, USA). FastStart Universal SYBR Green Master mix was
obtained from Roche Diagnostics (Mannheim, Germany). Annexin V-FITC
Apoptosis Detection kit was obtained from Keygen Biotech, Co., Ltd.
(Nanjing, China).
Cell cultures and chemical
treatments
The TK6 lymphoblastoid cell line was provided by
Professor Lishi Zhang (Sichuan University, Chengdu, China). The
cell properties, culture conditions and chemical treatment methods
used in the present study have been described in our previous study
(21). The PARP-1-silenced TK6
cells, which stably express PARP-1-short hairpin (sh)RNA have been
termed as shPARP-1, were provided by Professor Tang (Guangdong
Medical University, Dongguan, China) (22). Empty TK6 cells were used as control
cells. The experimental cells were treated with 10 µM HQ for 72 h,
and the control cells were treated with PBS.
Western blot analysis
Total proteins were extracted from prepared cells
using cell lysis buffer (Cell Signaling Technology, Inc.), and
their concentrations were determined using bicinchoninic acid (BCA)
assays (Beijing ComWin Biotech Co., Ltd., Beijing, China). Equal
quantities (25 µg) of protein were separated using SDS-PAGE on a
10% gel and then transferred to PVDF membranes. The membranes were
blocked with 5% fat free milk for 50 min at room temperature. The
target proteins and the loading control proteins were quantified on
the same membrane by dividing it into pieces according to the known
molecular weights of pre-stained protein standards. The membranes
were incubated at 4°C overnight with primary antibodies against
ZO-2 (1:2,000), PARP-1 (1:5,000), Bcl-2 (1:2,000), caspase-3
(1:2,000) or Bax (1:3,000). They were then incubated at room
temperature for 1 h with the corresponding species-specific
secondary antibodies. GAPDH (1:5,000) or α-tubulin (1:10,000) were
used as internal control. The immunoreactivity of the bands on the
membranes was visualized using ECL-PLUS chemiluminescence
reactions. Three repeats of the densitometric analysis were
performed using Quantity One software, version 4.6.2 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated, analyzed and transcribed
into cDNA as previously described (16). PCR reactions were performed using
FastStart Universal SYBR Green Master on a PikoReal 96 Real-Time
PCR system (Thermo Fisher Scientific, Inc.). The following forward
(F) and reverse (R) primers were used: ZO-2-F,
5′-CCGTTGCTGGTAATGAAACTCCT-3′ and ZO-2-R,
5′-TTATCTTGCTCCTCACTGCTCTCTC-3′. The qPCR conditions were as
follows: Initial cycle was 95°C for 10 min, followed by 40 cycles
at 95°C for 15 sec and 58°C for 40 sec. Dissociation curves were
used to confirm specificity. Expression levels were calculated
using the 2−∆∆Cq method (23) with GAPDH as selected internal
control.
Flow cytometry
The collected cells were suspended in binding buffer
and then stained with Annexin V-FITC Apoptosis Detection kit and
propidium iodide according to the manufacturer's protocol. The
cells were incubated at room temperature for 15 min in the dark and
then immediately analyzed using a BD FACSCanto II flow cytometer
(excitation=488 nm; emission=530 nm) (BD Biosciences, San Jose, CA,
USA). A minimum of 10,000 cells per sample were acquired and
analyzed using FlowJo software, version 7.6.1 (Tree Star, Inc.,
Ashland, OR, USA).
Quantification of caspase-3/7 activity
and ATP production
Cell numbers were counted using Countstar IC1000
(Ruiyu Biotech Co., Ltd, Shanghai, China). Caspase-3/7 activity and
ATP levels were quantified using Caspase-Glo 3/7 assay kits and
CellTiter-Glo Luminescent Cell Viability assays according to the
manufacturer's protocol in each kit. For the Caspase-3/7 assays,
100 µl of suspended cells were plated in 96-well plates and 100 µl
Caspase-Glo 3/7 reagent was added. The mixture was agitated for 2
min and then incubated for 10 min at room temperature. For the ATP
assays, 100 µl suspended cells were plated in 96-well plates and
100 µl CellTiter-Glo reagent was then added. The mixtures were
agitated for 2 min and then incubated for 5 min at room
temperature. The generated luminescent signal was read using the
luminescent detection method on a BioTek Synergy2 microplate reader
(BioTek Instruments, Inc., Winooski, VT, USA). The luminescent
signal readout was normalized according to the respective cell
number and was recorded as the caspase-3/7 or ATP luminescence
value.
Indirect immunofluorescence
staining
A previously described protocol used to prepare
cells for staining (16). Cells
were sequentially incubated with either anti-PARP-1 and ZO-2
primary antibodies or anti-PAR and ZO-2 primary antibodies at a
dilution of 1:500 at room temperature for 1 h and then Alexa Fluor
555-conjugated and Alexa Fluor 488-conjugated secondary antibodies
at a dilution of 1:1,000 at room temperature for 30 min. Cell
nuclei were counterstained using DAPI. The cells were smeared on
coverslips and mounted in 90% glycerol in PBS. They were
subsequently imaged using an Olympus FLUOVIEW FV1000 confocal
laser-scanning microscope (Olympus Corporation, Tokyo, Japan).
Quantification of ROS production
Reactive oxygen species (ROS) production was
determined using a DCFDA-Cellular Reactive Oxygen Species Detection
assay kit according to the manufacturer's protocol. Cells
(5×105) were treated with 10 µM HQ for 72 h. The
collected cells were then washed in PBS and stained with 20 µM
DCFDA in 1X buffer for 30 min at 37°C in the dark. The stained
cells were then washed, resuspended in 400 µl 1X buffer, and
equally seeded in four wells of an opaque 96-well microplate.
Staining intensities were immediately determined using a BioTek
Synergy2 microplate reader set for an excitation wavelength of 485
nm and an emission wavelength of 535 nm.
Statistical analysis
Data are presented as the mean ± standard deviation
of at least three independent experiments. One-way analysis of
variance was used for comparisons of means among multiple groups,
followed by the Student-Newman-Keuls post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
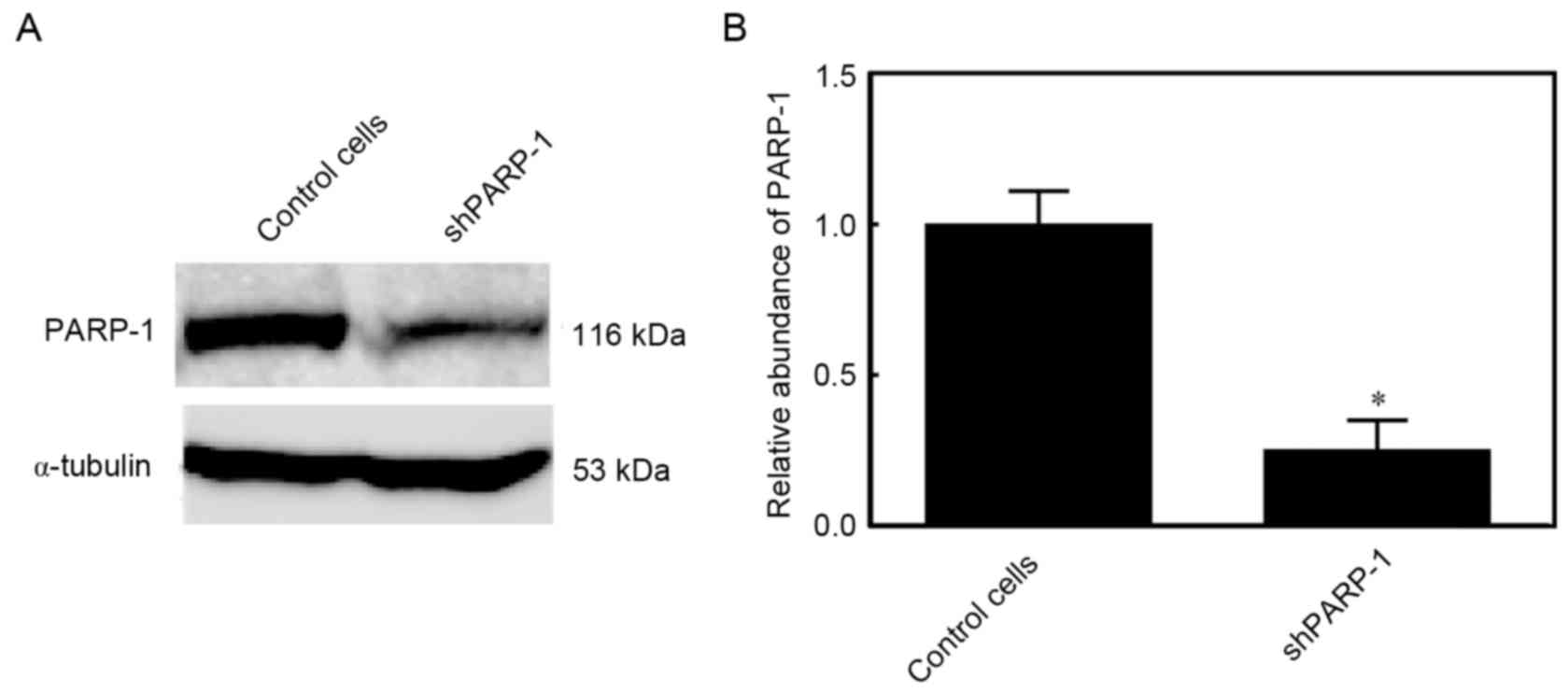
Expression of PARP-1 in
PARP-1-silencing TK6 cells
PARP-1 is a pleiotropic molecule that has an
important role in various cell death processes. PARP-1-silenced TK6
cells were generated to elucidate the underlying mechanisms
involving PARP-1. In these cells, the PARP-1 protein expression was
reduced by 75%, indicating that PARP-1 was efficiently silenced
(P<0.05; Fig. 1).
Inhibiting PARP-1 attenuates
HQ-induced apoptosis
In order to determine the level of apoptosis induced
by HQ treatment and the role of PARP-1 in this process, control
cells and shPARP-1 cells were treated with PBS or 10 µM HQ,
respectively, for 72 h and then subjected to flow cytometry
analysis. A higher number of apoptotic cells was observed in the
HQ-treated control cells than in the PBS group. The number of
apoptotic cells was lower in the PBS-treated shPARP-1 cells
compared with the PBS-treated control cells. Additionally, the
HQ-treated shPARP-1 cells also had a reduced number of apoptotic
cells compared with the HQ-treated control cells, indicating that
prolonged exposure to low levels of HQ induced apoptosis and that
this effect was attenuated by PARP-1 knockdown (Fig. 2A and B). Changes in the apoptotic
rate may lead to altered cell numbers; therefore, cell counting was
performed. It was determined that the number of cells in the
HQ-treated control cell group was lower when compared with the
number of cells in the control + PBS group. The number of cells in
the shPARP-1 group was higher compared with the control cells + HQ
group (Fig. 2C). These findings
suggested that HQ mediated growth inhibition by inducing apoptosis
in TK6 cells.
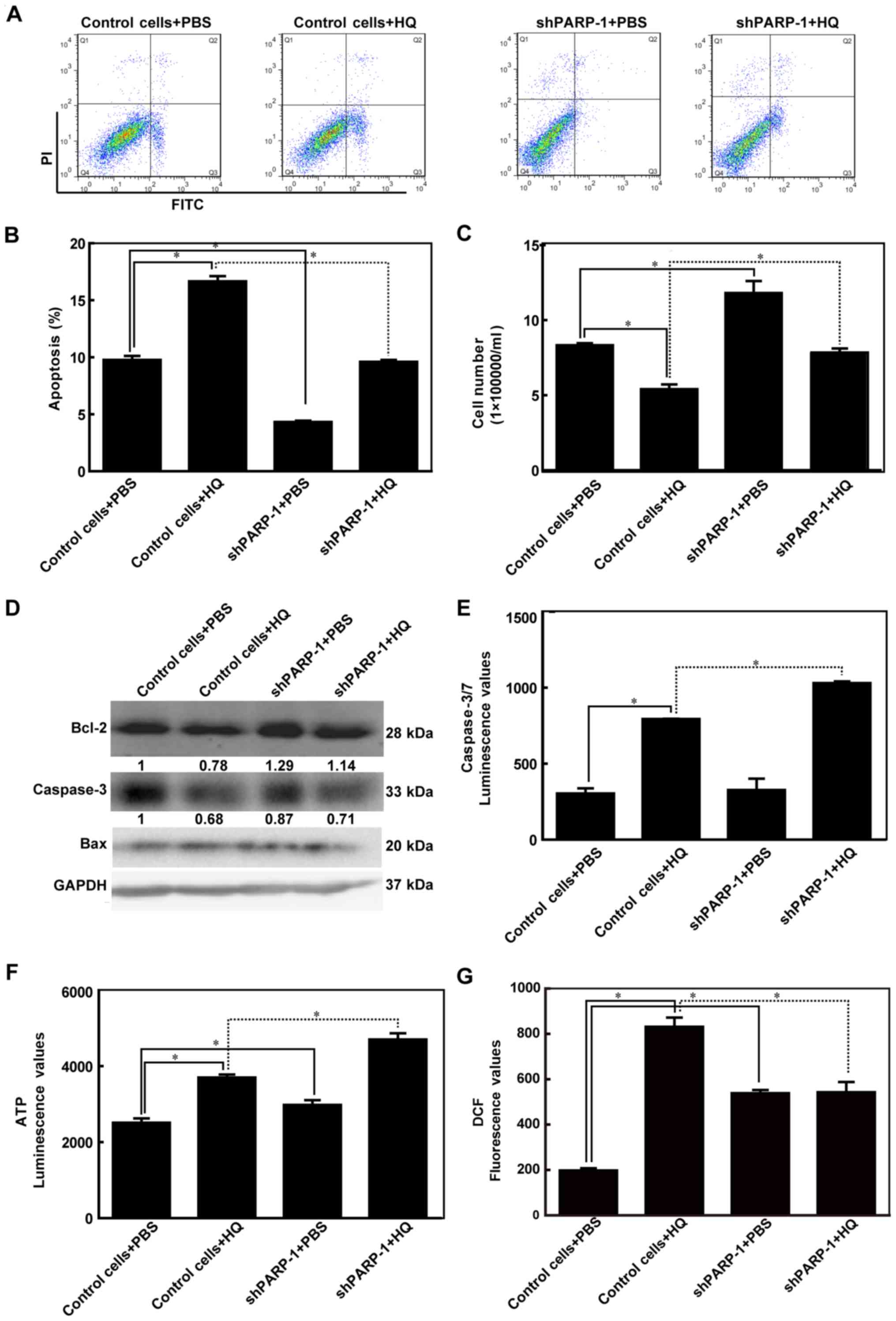 | Figure 2.PARP-1 attenuated HQ-induced
apoptosis. Empty TK6 cells were used as control cells. Cells were
treated with PBS or 10 µM HQ for 72 h. (A) Representative images of
flow cytometry using Annexin V-FITC and PI double-stained cells.
(B) Cell apoptosis rates are presented as the mean ± standard
deviation for at least three independent experiments. (C) Number of
cells was counted using Countstar (IC1000). (D) Protein expression
levels of the Bcl-2, caspase-3 and Bax proteins were detected using
western blot analysis. (E) Caspase-3/7 luminescence values, which
indicate caspase-3/7 activity, were detected using Caspase-Glo-3/7
assay kits. (F) ATP luminescence values, which indicate ATP level,
were measured using CellTiter-Glo Luminescent Cell Viability assay
kits. (G) Reactive oxygen species production was measured using
DCFDA. *P<0.05. HQ, hydroquinone; PARP-1, poly (ADP-ribose)
polymerase-1; shPARP-1, short hairpin PARP-1; Bcl2, B cell
leukemia/lymphoma 2; Bax, BCL2 associated X; DCFDA,
2′,7′-dichlorofluorescin diacetate; FITC, fluorescein
isothiocyanate; PI, propidium iodide. |
To investigate the underlying mechanism of
HQ-induced apoptosis, Bcl-2, caspase-3 and Bax were detected using
western blot analysis. Bcl-2 was present at lower levels in
HQ-treated control cells compared with the PBS-treated cells and at
higher levels in the HQ-treated shPARP-1 cells compared with the
HQ-treated control cells. Caspase-3 was present at lower levels in
the HQ-treated control cells. However, no significant difference in
terms of caspase-3 expression was observed between the HQ-treated
shPARP-1 cells and the HQ-treated control cells. Bax expression
remained constant in both HQ treatment and PARP-1 knockdown
(Fig. 2D). These findings
suggested that HQ induced the downregulation of Bcl-2 and caspase-3
in TK6 cells. PARP-1 inhibition antagonized the downregulation of
Bcl-2 that was induced by HQ treatment.
Caspase-3/7 are key regulators of apoptosis and the
activity of these two enzymes indirectly reflects the downstream
cascade reaction of cell apoptosis. Consistent with the increase
that was observed in the proportion of apoptotic cells in the
control cells treated with HQ, an increase in caspase-3/7 activity
was also observed in these cells. However, higher caspase-3/7
activity was also observed in shPARP-1 + HQ cells compared with the
control cells + HQ and this was inconsistent with the results
observed in flow cytometry (Fig.
2E). These findings indicated that inhibition of PARP-1
attenuated HQ-induced apoptosis, this effect may not be achieved
via the caspase signaling pathway.
ATP levels were also quantified in order to
determine the mechanisms involved in HQ-induced apoptosis as PARP-1
also participates in energy metabolism. More ATP was produced in
the control cells treated with HQ; however, the level of ATP was
higher in the HQ-treated shPARP-1 cells compared with the
HQ-treated control cells (Fig.
2F). These data indicated that the energy-saving effect that
was induced by silencing PARP-1 may be involved in protecting
against HQ-induced apoptosis. These data also support a previous
study that determined that PARP-1 affects ATP depletion (10). Additionally, ROS production was
significantly increased in the HQ-treated control cells compared
with the PBS only group (P<0.05; Fig. 2G). A considerably reduced level of
ROS production was observed in the HQ-treated shPARP-1 cells
compared with the HQ-treated control cells (Fig. 2G), indicating that inhibition of
PARP-1 expression may attenuate HQ-induced apoptosis by reducing
ROS production.
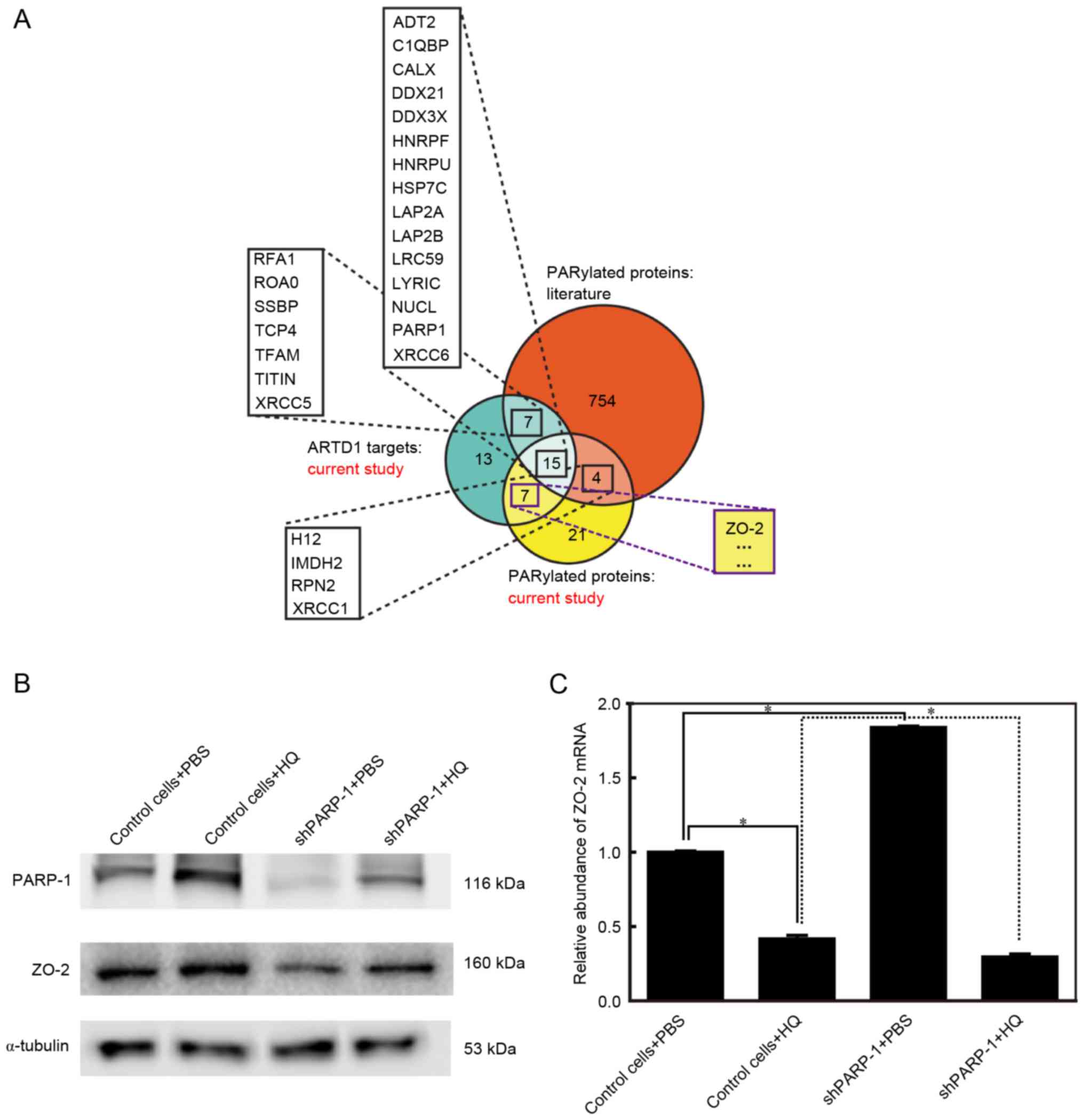
PARP-1 protein is required for the
abundance of the ZO-2 protein
A previous study described a rational design that
includes orthogonal NAD+ analogue-engineered PARP pairs
that may be used to identify the direct protein targets of
individual PARPs (17). In this
design, ZO-2 was found to be PARylated by PARP-1 (Fig. 3A) (17). Additionally, ZO-2 was reported to
be involved in apoptosis (19).
The present study aimed to further investigate the association
between PARP-1, ZO-2 and apoptosis and detected the ZO-2 and PARP-1
protein expression levels using western blot analysis. PARP-1 was
upregulated in the control cells treated with 10 µM HQ for 72 h,
consistent with the findings of our previous study (21). PARP-1 was effectively inhibited in
shPARP-1 cells. PARP-1 levels were lower in shPARP-1 cells treated
with HQ compared with HQ-treated control cells; however, shPARP-1
cells treated with HQ expressed higher levels of PARP-1 than
shPARP-1 cells treated with PBS (Fig.
3B). The expression levels of ZO-2 changed in parallel to the
changes observed in PARP-1 (Fig.
3B). ZO-2 protein expression was upregulated in control cells
treated with HQ, whereas in the shPARP-1 cells, where PARP-1
expression was reduced, ZO-2 levels were lower than the levels in
the control cells in the group treated with PBS and HQ (Fig. 3B). These findings indicated that
PARP-1 and ZO-2 are involved in HQ-induced apoptosis and that the
expression of ZO-2 is dependent on PARP-1. To reveal the mechanism
by which ZO-2 is dependent on PARP-1, the mRNA expression of ZO-2
was detected using RT-qPCR. It is of note that mRNA levels of ZO-2
were the reverse to those observed for the ZO-2 protein (Fig. 3C), indicating that ZO-2 protein
levels were post-translationally regulated by PARP-1.
ZO-2 is modified by PARylation
PARylation, a post-translational modification, is
primarily catalyzed by PARP-1. The present study determined whether
ZO-2 is modified by PARylation during HQ-induced apoptosis by
analyzing the co-localization of ZO-2 and PAR (or PARP-1) using
immunofluorescence confocal microscopy. It was revealed that ZO-2
was clearly integrating with PAR (Fig.
4, upper panel) and with PARP-1 respectively (Fig. 4, lower panel), indicating that ZO-2
was PARylated via an interaction with PARP-1. The abundance of ZO-2
in the nucleus was dependent on the abundance of PARP-1 (Fig. 4). These data are consistent with
the findings from the western blot analysis.
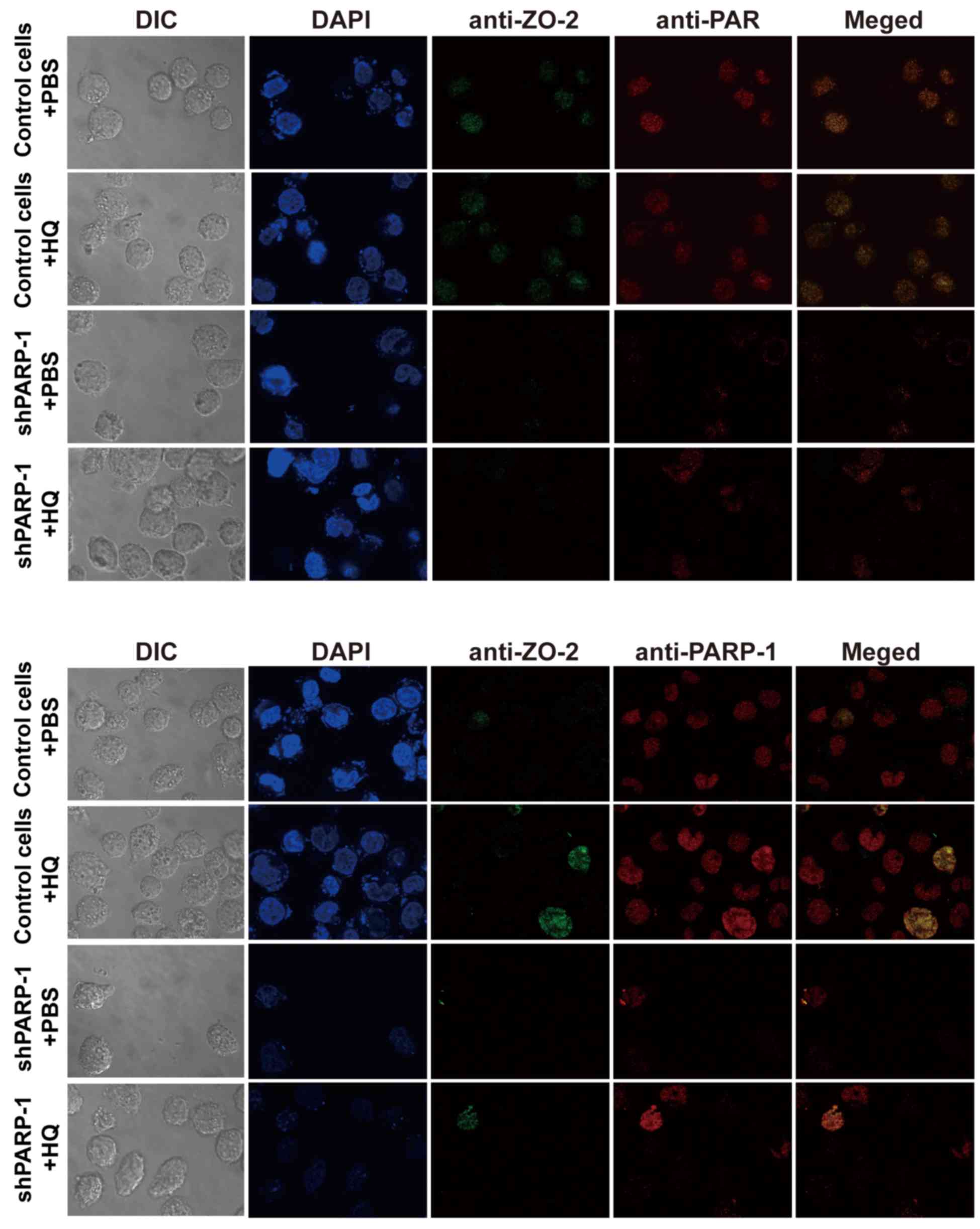 | Figure 4.ZO-2 may be modified by PARylation.
Cells were treated with 10 µM HQ for 72 h and then stained using
indirect immunofluorescence for ZO-2 (green), PAR (red, upper
panel), PARP-1 (red, lower panel) and DAPI (blue) and the ZO-2
protein was modified by PARylation (upper panel) via an interaction
with PARP-1 (lower panel). Magnification, ×1,000. ZO-2, zona
occludens 2; DIC, differential interference contrast; HQ,
hydroquinone; PARP-1, poly (ADP-ribose) polymerase-1; shPARP-1,
short hairpin PARP-1; PAR, poly (ADP-ribose). |
Discussion
Previous studies have revealed that different doses
of HQ may induce apoptosis (24,25).
Treatment with a low dose of HQ for 24 h induced HQ-mediated
apoptosis in murine fetal livers and bone marrow hematopoietic
cells (24). Treatment with a high
dose of HQ for 24 h promoted apoptosis in bone marrow-derived
mesenchymal stem cells (25).
Treatment with 50 or 100 µM HQ for 14 h promoted apoptosis in W7.2
rfau cells (26). In the present
study, treating cells with 10 µM HQ for 72 h promoted apoptosis.
The balance between the pro-apoptotic (Bcl2 associated agonist of
cell death and Bax) and anti-apoptotic (Bcl-2 and Bcl-extra large)
members of the Bcl-2 family is critical for controlling
mitochondria-induced apoptosis (27). Activated caspases may also act as
biochemical markers of apoptotic cascade reactions. The present
study, consistent with the findings of the flow cytometry
experiments, determined that caspase-3/7 activity was activated,
caspase-3 protein levels were upregulated and ATP production was
upregulated in TK6 cells treated with HQ, indicating that
HQ-induced apoptosis involved caspase-dependent intrinsic
apoptosis.
The present study demonstrated that in TK6 cells,
PARP-1 knockdown via RNA interference (RNAi) partially blocked
apoptosis in cells exposed to a low dose of HQ for a prolonged time
period. A recent study has also revealed that knockdown of PAPR-1
prevents apoptosis induced by HQ (28). Additionally, the present study
determined that the mechanisms underlying this protection may
involve the activation of Bcl-2 and the production of ATP and ROS.
HQ-induced apoptosis was reduced from 16.66±0.47 to 9.61±0.17% when
PARP-1 was knocked down, indicating that PARP-1 regulates
HQ-induced apoptosis. PARP-1 knockdown cells that were treated with
HQ, Bcl-2 was upregulated and caspase-3 levels were not changed. It
is of note, HQ-induced caspase-3/7 activity and ATP production were
enhanced by PARP-1 knockdown. These findings suggest that prolonged
exposure to a low dose of HQ did not block the production of energy
and that inhibition of PARP-1 in TK6 cells maintained ATP levels.
Inhibition of PARP-1 prevented the depletion of NAD+ and
ATP and partially restored the ability of cells to perform DNA,
RNA, and protein synthesis. Therefore, in TK6 cells, PARP-1
knockdown via RNAi partially blocked apoptosis in cells treated a
low dose of HQ for a prolonged period of time and that the
mechanisms underlying this protection may involve the activation of
Bcl-2 and the production of ATP. Furthermore, HQ treatment
increased the production of ROS, which was consistent with the
findings previously reported in ARPE-19 human retinal pigment
epithelial cells, R-28 rat retinal neurosensory cells, human
microvascular endothelial cells and MIO-M1 human retinal Müller
cells (29–31). A previous study has revealed that
may HQ induce apoptosis through a ROS pathway (29). In the present study, inhibition of
PARP-1 attenuated the production of ROS that was induced by HQ
treatment, indicating that inhibiting PARP-1 may attenuate
HQ-induced apoptosis partly via the ROS pathway.
PARylation facilitates DNA repair and therefore cell
survival in response to mild DNA damage, whereas more severe
genotoxic stimuli activate the apoptotic pathway, and the most
severe DNA damage may lead to the excessive activation of PARP and
the depletion of the cellular NAD+ and ATP stores.
Depleting NAD+ and ATP blocks apoptosis and leads to
necrosis (10,32). The present study determined that
apoptosis was induced by prolonged exposure to a low dose of HQ and
that this effect may be partially reversed by inhibition of PARP-1;
however, it is difficult identify PARP-1 as a survival or a
cytotoxic factor that contributes to cell survival, death or
deterioration following exposure to different doses of HQ for
different periods of time in specific cell types. Therefore, more
experiments are required to determine the precise mechanisms by
which HQ induces toxicity and to identify in which of these
mechanisms PARP-1 is involved.
Activated PARP-1 and PARylation regulate various
cellular processes, such as replication, transcription, DNA repair
and ATP metabolism, and they mediate various cellular phenomena,
such as proliferation, differentiation, senescence and cell death
(33,34). The presence of ZO-2 in the nucleus
was determined more than a decade ago; however, its role in
lymphoblastoid cells that lack tight junctions remains to be fully
elucidated. In the nucleus, ZO-2 reportedly interacts with several
proteins, including Jun, Fos, c-Myc and PKA (35–37).
Co-localization between ZO-2 and PAR (or PARP-1 protein) was
demonstrated by the present study using immunofluorescence confocal
microscopy, indicating that ZO-2 is PARylated via an interaction
with PARP-1. These findings were consistent with its protein
expression detected using western blot analysis and the
co-localization revealed that treatment with HQ led to the
upregulation of ZO-2 in control cells and its downregulation in
shPARP-1 cells. To the best of our knowledge, the present study is
the first to determine that PARylation may modify the ZO-2 protein
and to reveal that ZO-2 is not distributed in the nucleus in
lymphoblastoid cells that lack tight junctions.
ZO-2 induces the expression of various
apoptosis-associated genes, including Bcl-2, interleukin-6, REL
proto-oncogene, NF-κB subunit and translocator protein (38). It interacts with the
transcriptional activator Yes kinase-associated protein 2 (YAP2)
and enhances the nuclear localization and pro-apoptotic function of
YAP (39). When PARP-1 expression
was inhibited in the present study, ZO-2 was downregulated in
parallel with changes observed in apoptosis in HQ-treated cells.
Therefore, ZO-2 acted as a promoter of HQ-induced apoptosis and
these data confirmed that ZO-2 may act as a tumor-suppressing
protein.
In summary, the findings of the present study
indicated that prolonged exposure to a low dose of HQ induced TK6
cells to undergo apoptosis and that inhibition of PARP-1 attenuated
cellular apoptosis by activation of Bcl-2, inducing energy-saving
mechanisms and reducing ROS. During this HQ-induced process, ZO-2
localized into the nucleus in lymphoblastoid cells and ZO-2 was
modified by PARP-1 via PARylation. Therefore, when investigating
the molecular mechanisms of HQ-induced toxicity and the
contribution of HQ to benzene-induced leukemia, it is important to
consider pleiotropic PAPR-1 and its inhibitors during clinical
treatment.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81202231 to
Dr Linhua Liu; grant no. 81273116 to Professor Huanwen Tang; a key
program 81430079 to Professor Wen Chen, and grant no. 81372962 to
Professor Yongmei Xiao), the Science and Technology Program of the
Guangdong Bureau of Science and Technology, China (grant no.
2013B021800069 to Dr Linhua Liu), the Guangdong Provincial Natural
Science Foundation, China (grant no. S2013010015153 to Professor
Huanwen Tang, grant no. 2014KQNCX102 to Dr Xiaoxuang Ling), the Key
Project of Science and Technology Program of Dongguan Bureau of
Science and Technology, China (grant no. 2012108101011 to Professor
Huanwen Tang), the Science and Technology Program of Zhanjiang
Bureau of Science and Technology, China (grant no. 2013B01082 to
Professor Huanwen Tang), and the Science Foundation of Guangdong
Medical University, China (grant no. M2013004 to Professor Huanwen
Tang).
Glossary
Abbreviations
Abbreviations:
|
HQ
|
hydroquinone
|
|
PARP-1
|
poly(ADP-ribose) polymerase-1
|
|
PARylation
|
poly ADP-ribosylation
|
|
PAR
|
poly (ADP-ribose)
|
|
NAD+
|
donor nicotinamide adenine
dinucleotide
|
|
ZO-2
|
zona occludens 2
|
|
ROS
|
reactive oxygen species
|
References
|
1
|
Elmore S: Apoptosis: A review of
programmed cell death. Toxicol Pathol. 35:495–516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fulda S: Cross talk between cell death
regulation and metabolism. Methods Enzymol. 542:81–90. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oka S, Kato J and Moss J: Identification
and characterization of a mammalian 39-kDa poly(ADP-ribose)
glycohydrolase. J Biol Chem. 281:705–713. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Davidovic L, Vodenicharov M, Affar EB and
Poirier GG: Importance of poly(ADP-ribose) glycohydrolase in the
control of poly(ADP-ribose) metabolism. Exp Cell Res. 268:7–13.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gibson BA and Kraus WL: New insights into
the molecular and cellular functions of poly(ADP-ribose) and PARPs.
Nat Rev Mol Cell Biol. 13:411–424. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Virag L, Robaszkiewicz A, Rodriguez-Vargas
JM and Oliver FJ: Poly(ADP-ribose) signaling in cell death. Mol
Aspects Med. 34:1153–1167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Galluzzi L, Vitale I, Abrams JM, Alnemri
ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry
WS, Fulda S, et al: Molecular definitions of cell death
subroutines: Recommendations of the nomenclature committee on cell
death 2012. Cell Death Differ. 19:107–120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
De Vos M, Schreiber V and Dantzer F: The
diverse roles and clinical relevance of PARPs in DNA damage repair:
Current state of the art. Biochem Pharmacol. 84:137–146. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Curtin NJ and Szabo C: Therapeutic
applications of PARP inhibitors: Anticancer therapy and beyond. Mol
Aspects Med. 34:1217–1256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Virág L and Szabó C: The therapeutic
potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev.
54:375–429. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McHale CM, Zhang L and Smith MT: Current
understanding of the mechanism of benzene-induced leukemia in
humans: Implications for risk assessment. Carcinogenesis.
33:240–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slater ME, Linabery AM, Spector LG,
Johnson KJ, Hilden JM, Heerema NA, Robison LL and Ross JA: Maternal
exposure to household chemicals and risk of infant leukemia: A
report from the children's oncology group. Cancer Causes Control.
22:1197–1204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
North M, Tandon VJ, Thomas R, Loguinov A,
Gerlovina I, Hubbard AE, Zhang L, Smith MT and Vulpe CD:
Genome-wide functional profiling reveals genes required for
tolerance to benzene metabolites in yeast. PLoS One. 6:e242052011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gaskell M, McLuckie KI and Farmer PB:
Genotoxicity of the benzene metabolites para-benzoquinone and
hydroquinone. Chem Biol Interact 153–154. 1–270. 2005.
|
|
15
|
Smith MT: The mechanism of benzene-induced
leukemia: A hypothesis and speculations on the causes of leukemia.
Environ Health Perspect. 104 Suppl 6:S1219–S1225. 1996. View Article : Google Scholar
|
|
16
|
Liu L, Ling X, Tang H, Chen J, Wen Q and
Zou F: Poly(ADP-ribosyl)ation enhances H-RAS protein stability and
causes abnormal cell cycle progression in human TK6 lymphoblastoid
cells treated with hydroquinone. Chem Biol Interact. 238:1–8. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carter-O'Connell I, Jin H, Morgan RK,
David LL and Cohen MS: Engineering the substrate specificity of
ADP-ribosyltransferases for identifying direct protein targets. J
Am Chem Soc. 136:5201–5204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gumbiner B, Lowenkopf T and Apatira D:
Identification of a 160-kDa polypeptide that binds to the tight
junction protein ZO-1. Proc Natl Acad Sci USA. 88:pp. 3460–3464.
1991; View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Traweger A, Toepfer S, Wagner RN,
Zweimueller-Mayer J, Gehwolf R, Lehner C, Tempfer H, Krizbai I,
Wilhelm I, Bauer HC and Bauer H: Beyond cell-cell adhesion:
Emerging roles of the tight junction scaffold ZO-2. Tissue
Barriers. 1:e250392013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Walsh T, Pierce SB, Lenz DR, Brownstein Z,
Dagan-Rosenfeld O, Shahin H, Roeb W, McCarthy S, Nord AS, Gordon
CR, et al: Genomic duplication and overexpression of TJP2/ZO-2
leads to altered expression of apoptosis genes in progressive
nonsyndromic hearing loss DFNA51. Am J Hum Genet. 87:101–109. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu L, Ling X, Liang H, Gao Y, Yang H,
Shao J and Tang H: Hypomethylation mediated by decreased DNMTs
involves in the activation of proto-oncogene MPL in TK6 cells
treated with hydroquinone. Toxicol Lett. 209:239–245. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang H, Liang H, Chen J and Xu Y:
Construction of PARP-1 gene silencing cell lines by
lentiviral-mediated RNA interference technology. J Environ Health.
31:288–291, 377. 2014.
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Z, Wang C, Zhu J, Bai Y, Wang W, Zhou
Y, Zhang S, Liu X, Zhou S, Huang W, et al: The possible role of
liver kinase B1 in hydroquinone-induced toxicity of murine fetal
liver and bone marrow hematopoietic stem cells. Environ Toxicol.
31:830–841. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang J, Zhao M, Li X, Ma L, Zhang J, Shi
J, Li B, Fan W and Zhou Y: The cytotoxic effect of the benzene
metabolite hydroquinone is mediated by the modulation of MDR1
expression via the NF-κB signaling pathway. Cell Physiol Biochem.
37:592–602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Siew EL, Chan KM, Williams GT, Ross D and
Inayat-Hussain SH: Protection of hydroquinone-induced apoptosis by
downregulation of Fau is mediated by NQO1. Free Radic Biol Med.
53:1616–1624. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cosentino K and Garcia-Sáez AJ:
Mitochondrial alterations in apoptosis. Chem Phys Lipids.
181:62–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sha Y, Zhou W, Yang Z, Zhu X, Yang X, Li
TD and Zhu D: Down-regulation of poly (ADP-ribose) polymerase 1
leads to change of hydroquinone cytotoxicity in TK6 cells. Toxicol
Mech Methods. 25:467–477. 2015.PubMed/NCBI
|
|
29
|
Ramirez C, Pham K, Franco MF, Chwa M, Limb
A, Kuppermann BD and Kenney MC: Hydroquinone induces oxidative and
mitochondrial damage to human retinal Müller cells (MIO-M1).
Neurotoxicology. 39:102–108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sharma A, Neekhra A, Gramajo AL, Patil J,
Chwa M, Kuppermann BD and Kenney MC: Effects of Benzo(e)Pyrene, a
toxic component of cigarette smoke, on human retinal pigment
epithelial cells in vitro. Invest Ophthalmol Vis Sci. 49:5111–5117.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Patil AJ, Gramajo AL, Sharma A, Chwa M,
Seigel GM, Kuppermann BD and Kenney MC: Effects of benzo(e)pyrene
on the retinal neurosensory cells and human microvascular
endothelial cells in vitro. Curr Eye Res. 34:672–682. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kraus WL: PARPs and ADP-ribosylation: 50
years … and counting. Mol Cell. 58:902–910. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ryu KW, Kim DS and Kraus WL: New facets in
the regulation of gene expression by ADP-ribosylation and
poly(ADP-ribose) polymerases. Chem Rev. 115:2453–2481. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bürkle A and Virág L: Poly(ADP-ribose):
PARadigms and PARadoxes. Mol Aspects Med. 34:1046–1065. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hernandez S, Munguia B Chavez and
Gonzalez-Mariscal L: ZO-2 silencing in epithelial cells perturbs
the gate and fence function of tight junctions and leads to an
atypical monolayer architecture. Exp Cell Res. 313:1533–1547. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Betanzos A, Huerta M, Lopez-Bayghen E,
Azuara E, Amerena J and González-Mariscal L: The tight junction
protein ZO-2 associates with Jun, Fos and C/EBP transcription
factors in epithelial cells. Exp Cell Res. 292:51–66. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Avila-Flores A, Rendon-Huerta E, Moreno J,
Islas S, Betanzos A, Robles-Flores M and González-Mariscal L:
Tight-junction protein zonula occludens 2 is a target of
phosphorylation by protein kinase C. Biochem J. 360:295–304. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gonzalez-Mariscal L, Bautista P, Lechuga S
and Quiros M: ZO-2, a tight junction scaffold protein involved in
the regulation of cell proliferation and apoptosis. Ann N Y Acad
Sci. 1257:133–141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Oka T, Remue E, Meerschaert K, Vanloo B,
Boucherie C, Gfeller D, Bader GD, Sidhu SS, Vandekerckhove J,
Gettemans J and Sudol M: Functional complexes between YAP2 and ZO-2
are PDZ domain-dependent, and regulate YAP2 nuclear localization
and signalling. Biochem J. 432:461–472. 2010. View Article : Google Scholar : PubMed/NCBI
|