Introduction
Juvenile idiopathic arthritis (JIA) is a complex,
autoimmune disease, characterized by persistent chronic arthritis.
At present, the role of genetic and environmental factors in the
development of the disease has been confirmed (1). This disorder encompasses all forms of
chronic arthritis and onset appears before 16 years of age, and
usually persists for more than 6 weeks (1). JIA is the most common rheumatic
disease in children, with different disease manifestations in
populations of different ethnicities or race. Currently, seven
different subtypes of the disease have been distinguished according
to the International League of Associations for Rheumatology (ILAR)
classification (2). However, the
underlying mechanisms leading to a variety of different JIA
phenotypes have not been delineated yet. Nowadays, numerous
suggestions have been proposed to revise this classification
(3).
There is a strong genomic component to disease and a
number of established susceptibility loci have been reported thus
far (4–8). These data have been accumulated by
using a variety of approaches, including candidate gene approaches
and genome-wide association studies (GWAS). However, the
pathophysiology and genetic etiology of the disease remains
elusive. Most of the genetic predisposition to JIA was determined
by the human leukocyte antigen (HLA) (8–10)
and especially the HLA-DRB1 locus (10), with its contribution to be
estimated at a rate of approximately 20% (11). Moreover, several non-HLA genes have
been associated with JIA, including ATXN2, PTPN2, c12orf30, c3orf1,
PTPN22, STAT4, TRAF1-C5, AFF3, CD247, CD226, MBL2, PSTPIP1, RANTES
(CCL5), CTLA4, PRKCQ, PTPRC, TYK2, PRR9_LOR, ILDR1_CD86, WDFY4,
PTH1R, RNF215, AHI1_LINC00271, JAK1, LINC00951, IL2RA, CCR5, COG6,
ANGPT1, HBP1, 6q23/TNFAIP3, CXCR4 (5–7,12–19).
Of these loci, TRAF1, STAT4, TNFAIP3 and PRKCQ are good candidates
considering the known pathogenesis of JIA as they are located
within regions known to be involved in T-cell receptor signaling or
activation pathways (12).
Furthermore, the role of family history regarding a higher
prevalence of other autoimmune diseases among relatives of JIA
patients has been confirmed (11).
No genetic risk factors common for sJIA and other JIA subtypes have
been reported, thus suggesting that sJIA is a unique disease
process (20). A plethora of
biomarkers has been suggested aiming to define the different JIA
subtypes or predict either the course of the disease or treatment
response (in specific therapies) (21).
Not all independent replications in various cohorts
have been successful, as shown by using the ImmunoChip single
nucleotide polymorphism (SNP) array (7) or by investigating an Australian
cohort (15). Different genetic
variations have been reported as risk factors for JIA, but the
difficulty to confirm previously collected results by analyzing
populations of different ethnicities indicates the existence of a
heterogeneity regarding the genetic factors that are involved in
JIA.
Although numerous SNPs have been associated with
JIA, only a few of these studies were replicated and validated. In
the present study, we aimed to validate three SNPs, namely PTPRC
(rs10919563), TYK2 (rs34536443) and PRKCQ (rs4750316), previously
found to be associated with JIA (7,12,22)
and to investigate whether the 27-bp VNTR polymorphism on intron 4
of endothelial nitric oxide synthase (eNOS), which is associated
with various autoimmune diseases, is associated with risk for JIA
in Greece.
Patients and methods
Patient population and study
design
The sample set consisted of 125 JIA patients
followed in two departments (the Rheumatology Unit of the 4th
Medical Department of Internal Medicine and the Rheumatology Unit
of the 1st Medical Department of Pediatrics) of the Aristotelian
University of Thessaloniki Hospital (Thessaloniki, Greece). In this
study, only patients who fulfilled the 2001 revised ILAR criteria
for JIA were enrolled. Written informed consent was obtained from
parents for those patients who were <18 years of age. A total of
221 unrelated healthy individuals, sex- and ethnic-matched selected
from the Transfusion Medicine Department of Hippokration Hospital
of Thessaloniki, served as controls for the genotyping (14). The study was approved by the local
Ethics Committee for medical research and was carried out in
compliance with the Declaration of Helsinki.
Genotyping
A panel of 3 SNPs was genotyped in a Greek cohort of
JIA patients and healthy controls, including PTPRC
rs10919563, PRKCQ rs4750316 and TYK2 rs34536443.
Moreover, the 27-bp VNTR polymorphism on intron-4 of eNOS
was analyzed, considering its previous association with multiple
complex diseases (23). Genomic
DNA was isolated from peripheral blood leukocytes by using the
commercial kit Puregene (Gentra Systems, Minneapolis, MN, USA)
according to the manufacturer's instructions.
Allelic discrimination of PTPRC rs10919563, PRKCQ
rs4750316 and TYK2 rs34536443 SNPs was carried out using premade
TaqMan SNP Genotyping assays from Applied Biosystems (Foster City,
CA, USA), according to the Applied Biosystems protocol (cat. nos.
C_31565763_10, C_32036787_10 and C__60866522_10 for rs10919563,
rs4750316 and rs34536443, respectively). A 96-well plate was
prepared with a mixture on an Applied Biosystems ViiA™ 7 Real-Time
PCR system of 1X TaqMan SNP Genotyping assay, 1X TaqMan Universal
Master mix (both from Applied Biosystems) and 20 ng DNA per well.
After PCR, the plates were read, and the data analyzed using Quant
Studio™ Real-Time PCR software (Applied Biosystems). Each assay was
run with negative controls. For quality control, a random 10% of
the samples were amplified twice to ensure accuracy of the results,
with a reproducibility rate of 100%. Genotyping of eNOS gene intron
4 a/b VNTR polymorphism was performed as described in detail
elsewhere (23). Upon PCR
amplification, the 420-bp band indicated five repeats of the 27-bp
sequence and the 393-bp band represented four repeats,
corresponding to b and a alleles, respectively (24).
Construction of a three-dimensional
(3D) model
A bioinformatics analysis was performed using BlastP
(for sequence analysis) on the Uniprot sequence database. PyMOL
(DeLano Scientific, San Carlos, CA, USA) was used for the direction
of the development of a 3D model. Maestro and Desmond (Schrödinger,
Inc., New York, NY, USA) were used aiming to analyze mutations and
stability of the protein model. The initial 3D model used was based
on the crystal structure of Homo sapiens TYK2.
Statistical analysis
The GraphPad Prism statistical program (GraphPad
Software, San Diego, CA, USA) was used in the framework of the
analyses conducted. A two-tailed P<0.05 was defined as
statistically significant. Odds ratios (OR) and 95% confidence
intervals (CI) were calculated. The possible deviation from
Hardy-Weinberg equilibrium (HWE) was performed by using the program
‘Calculate’ (Copyright TRG, SR, INMD, 2008).
Results
Developing a 3D model of TYK2
protein
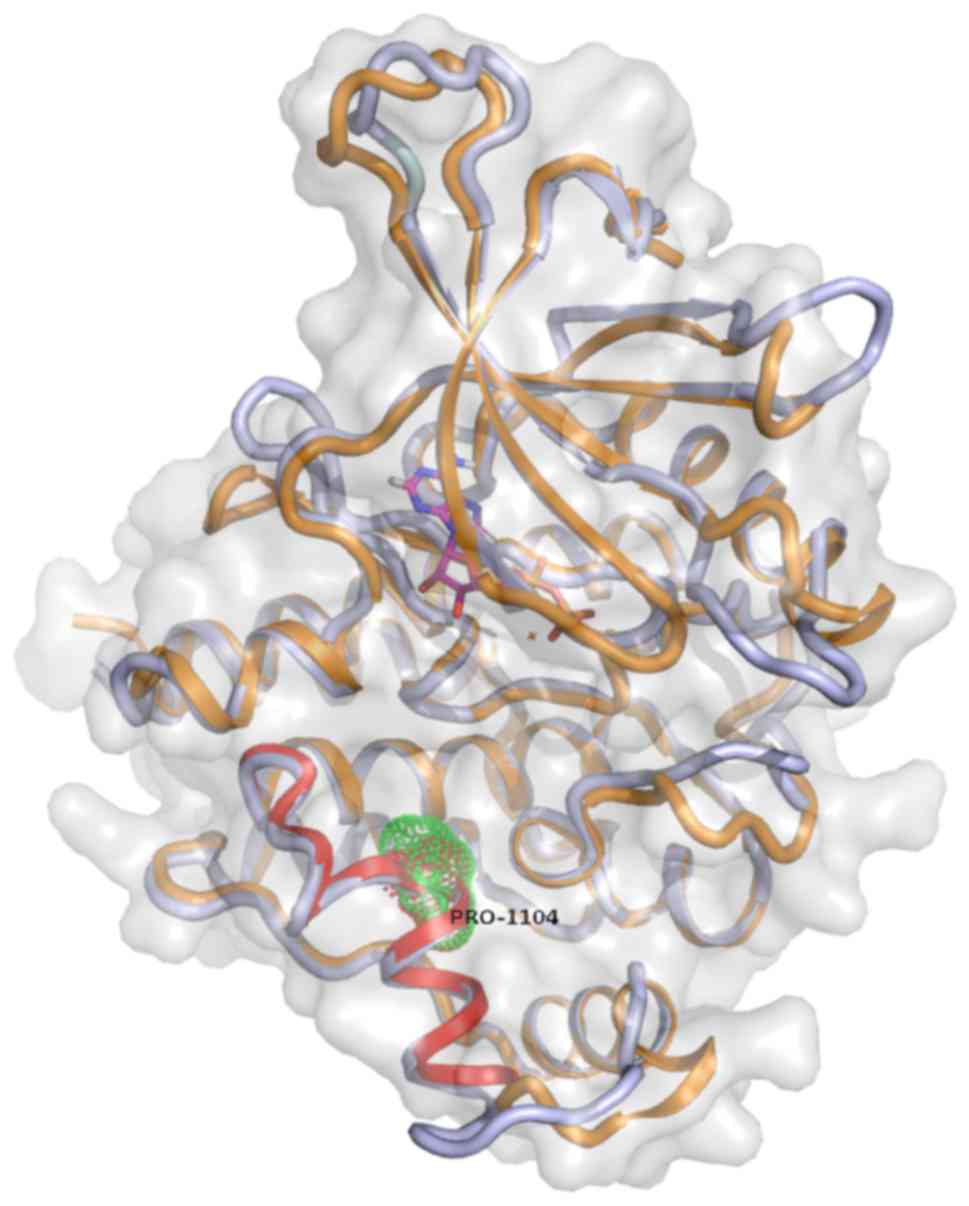
Aiming to gain insight to the potential
structural/functional consequences of rs34536443 SNP, which has
been associated with various rheumatic and other complex human
diseases, we further analyzed the respective region of TYK2 protein
by developing a new 3D model, improving the pre-existing one
developed by Myrthianou et al (26). The rs34536443 SNP leads to the
substitution of a G to a C nucleotide, thus resulting in a
Pro1104Ala mutation in the TYK2 protein. Accordingly, replacement
of the Pro1104 amino acid residue of the protein TYK2 may introduce
a conformational change that may extend the α-helical segment by
five more residues (Fig. 1). Thus,
it is possible that the local 3D structure may be altered and,
consequently, the folding of TYK2 protein may be affected. These
alterations are supposed to affect the protein from the
functionality viewpoint. Subsequent analyses of molecular dynamics
of the mutant TYK2 demonstrated further folding changes at the
level of the structure of the protein (Fig. 1).
Investigating a potential genetic
association of TYK2, PTPRC and eNOS SNPs with JIA
In the context of this case-control study, three
successful markers were analyzed. The allelic and genotype
frequencies of the eNOS VNTR of patients and controls are
shown in Table I. The homozygous
genotype b/b appeared the higher frequency in controls as well as
JIA patients (73.30 vs. 48%, respectively). Importantly, a/a
genotypes had a frequency that was associated with disease
occurrence (p<0.0001, OR=0.15, 95% CI: 0.065–0.37). Moreover,
the frequency of the eNOS intron-4 ‘a’ allele of JIA
patients was associated with the disease, being higher in
comparison to that of the controls (33.2 vs. 14.71%; p<0.0001,
OR=0.34, 95% CI: 0.23–0.49). These data indicated the 27-bp
eNOS VNTR with JIA in Greece.
 | Table I.Genotype and allele frequencies of
the 27-bp eNOS VNTR analyzed in 125 JIA patients and 221 healthy
controls. |
Table I.
Genotype and allele frequencies of
the 27-bp eNOS VNTR analyzed in 125 JIA patients and 221 healthy
controls.
| Variables | JIA (N=125) | Controls
(N=221) | P-value | OR (95% CI) |
|---|
| Genotypes |
|
|
|
|
|
b/b | 60 (48%) | 162 (73.30%) |
|
|
|
b/a | 47 (37.6%) | 49 (22.17%) | 0.05 | 0.4
(0.16–1.02) |
|
a/a | 19 (15.2%) | 8 (3.63%) |
<0.0001 | 0.15
(0.065–0.37) |
| Alleles |
|
|
|
|
| B | 167 (66.8%) | 373 (84.39%) |
|
|
| A | 85 (33.2%) | 65 (14.71%) |
<0.0001 | 0.34
(0.23–0.49) |
Notably, the remaining two SNPs PTPRC (rs10919563)
and TYK2 (rs34536443) were found to be associated with JIA apart
from previous findings that suggested a genetic association between
these SNPs and JIA in different ethnic populations (6,22).
Thus, patients with JIA did not present higher frequencies either
for the GG genotype or G allele of the PTPRC rs10919563 SNP
(p=0.41, OR=0.3, 95% CI: 0.03–2.6; and p=0.49, OR=0.82, 95% CI:
0.52–1.31, respectively) (data not shown). Moreover, concerning the
TYK2 rs34536443 SNP, no statistical significant differences were
observed between patients and controls either at genotype or
allelic level (p=0.34, OR=2.50, 95% CI: 1.53–11.75; and p=0.34,
OR=2.45, 95% CI: 0.53–11.55 for G/C genotype and minor allele C,
respectively) (data not shown). All the gene polymorphisms examined
were found to be under HWE (P>0.01).
In our attempt to analyze a possible association of
the polyarticular and oligoarticular course of JIA with the two
SNPs and the 27-bp VNTR under study, a further genetic analysis was
conducted but no association was observed. Considering the moderate
size of our sample, which precluded any detailed subtype-specific
association analyses, it is understood that such a subtype analysis
must be conducted in a larger cohort, consisted of adequately sized
samples, in order to lead to a high statistical power.
Search for a sex-specific genetic association of
PRKCQ rs4750316 SNP with JIA. A genetic association study was
performed involving the PRKCQ rs4750316 SNP (Table II). However, no significant
association of the minor allele C of rs4750316 with JIA was
observed (p=0.62, OR=1.12, 95% CI: 0.75–1.68). Nevertheless,
preliminary unpublished data collected from different ethnic
populations have previously shown PRKCQ rs4750316 SNP to be
associated with JIA specifically to males. Therefore, a
sex-stratified analysis was conducted in the Greek samples but no
sex-based difference in the frequency of this allele was observed
between patients (p=0.14, OR=1.80, 95% CI: 0.88–3.64 vs. p=0.16,
OR=0.61, 95% CI: 0.32–1.17, respectively). Given that the sex bias
differs in the JIA subtype, these associations should be further
investigated in individual subtypes. However, the small sample size
did not allow for further analyses, depending on JIA subtype at
level of a statistical certainty, to be performed.
 | Table II.Frequencies of the minor allele ‘C’
of PRKCQ rs4750316 SNP in a sex-stratified genetic analysis. |
Table II.
Frequencies of the minor allele ‘C’
of PRKCQ rs4750316 SNP in a sex-stratified genetic analysis.
| Gene | SNP | Samples | N cases/N
controls | OR (95% CI) | P-value |
|---|
| PRKCQ | rs4750316 | All | 144/185 | 1.12
(0.75–1.68) | 0.62 |
|
|
| Males | 36/129 | 1.80
(0.88–3.64) | 0.14 |
|
|
| Females | 108/56 | 0.61
(0.32–1.17) | 0.16 |
Discussion
Accumulated evidence has indicated the existence of
a heterogeneity, depending on the population's ethnicity, of
genetic factors involved in the development of autoimmune diseases.
Deepening of our knowledge regarding the homogeneity or
heterogeneity at the level of alleles that is associated with these
diseases may confer to a more reasonable understanding of plausible
common gene function and pathways. To the best of our knowledge,
the present analysis revealed for the first time that eNOS VNTR
polymorphism is associated with susceptibility to JIA and,
therefore, suggests a role for the risk allele ‘a’ in the
development of different autoimmune diseases. In a previous study,
referring to a Greek population, the implication of eNOS gene 27-bp
VNTR in susceptibility to rheumatoid arthritis (RA) was identified
(23). Moreover, in that study,
the implication of this VNTR to systemic lupus erythematosus (SLE)
was shown (23). In the present
study, it was found that the presence of a/b genotype or ‘a’ allele
may be a disease-susceptibility factor for RA or lupus
glomerulonephritis.
The functional role of nitric oxide (NO) regarding
the cell pathophysiology and function has been previously analyzed.
Thus, it has been suggested that it is a potent mediator in
different biological reactions, which is involved in various
responses based either on inflammation or autoimmunity processes.
NO was suggested to be an inhibiting factor for proinflammatory
gene expression in endothelial cells or an activator of resident
leukocytes (27). Obviously, gene
polymorphisms or other factors that influence the expression of the
enzyme eNOS that catalyzes the synthesis of NO can be considered as
key genes for the control of NO levels activity. For the
maintenance of physiological endothelial function, low levels of NO
are needed (28). Although the
functional significance of eNOS gene VNTR on protein activity and
expression is not well documented yet, some preliminary experiments
have managed to clarify this issue. The a allele has been related
to low NO metabolite levels, and a/a homozygotes were found to
produce lower NO metabolite levels compared to b/b individuals
(29). In addition, it has been
shown that the 27-bp VNTR of eNOS is able to bind nuclear proteins
acting either as an enhancer or repressor, thus promoting or
suppressing transcription efficiency (30). Therefore, apart from the suggested
crucial role of the eNOS VNTR under discussion dealing with the
decreased gene expression of eNOS in allele ‘a’ carriers, the
possibility of its proximity and, therefore, of a linkage
disequilibrium with other variants cannot be underestimated.
An association of eNOS gene with various
multifactorial diseases, including as primary biliary cirrhosis
(31), RA (23), type-1 and −2 diabetes (32) and SLE (23,33)
has been shown thus far. However, a dependence of JIA and eNOS
polymorphisms according to race or ethnicity has been suggested.
Indeed, an association of the eNOS VNTR polymorphism with SLE was
shown in the past in Turkey (33)
but not in Greece (23) or with RA
in Greece (23) but not in Serbia
(34).
Notably, although it was previously shown that PTPRC
rs10919563 (6), PRKCQ rs4750316
(12) and TYK2 rs34536443
(22) SNPs play a role in the
phenotypical manifestations accompanying JIA, the present study did
not confirm association of the SNPs examined with the disease in
Greece.
It is well known that the high female:male sex-ratio
is a characteristic observed in almost all autoimmune diseases,
including JIA. Recently, a sex-specific pattern of association with
JIA has been reported. Particularly, a female-association of PTPN22
rs2476601 was detected with JIA in different ethnic populations,
including the Greek one (35). In
this framework, the rs4750316 SNP of PRKCQ was analyzed as well,
taking into account that it was demonstrated originally to exhibit
an association with JIA in a first analysis conducted in 2010
(12), thus appearing as a
promising JIA candidate gene in the UK cohort. However, this
finding was not confirmed in the same population when an analysis
by ImmunoChip array was performed (7). Notably, an association of this SNP
with JIA was found recently in samples from different ethnicities,
stratified by sex (unpublished data). While in the Greek population
an analysis for rs4750316 SNP did not reach statistical
significance, more solid data are to be obtained by increasing the
sample size and further investigating any new data.
It is worth mentioning that a study performed by
using the Immunochip and next generation sequencing approaches has
shown that allele ‘C’ of rs34536443 of TYK2 was protective for
inflammatory bowel disease, RA and SLE (36) but conferred a risk for psoriasis
(37) and JIA (6). In the framework of an analysis
conducted in a UK cohort by ImmunoChip array (7), TYK2 rs34536443 was not found to be
associated with JIA, a result that coincides with the present data
from the Greek cohort. This finding was noteworthy considering that
TYK2 rs34536443 SNP was found recently, in a study performed in the
Greek population (26), to exhibit
a genetic overlap with PsA and RA that is well known to possess or
share several similar clinical manifestations with JIA. Moreover,
according to the constructed 3D model of TYK2 protein and previous
observations of our group (26),
it is assumed that the Pro1104Gly mutation may affect the protein's
functionality by affecting elements contributing to the structural
conformation of the molecule.
According to the data of the present study, minor
allele frequencies (MAFs) of PRKCQ (rs4750316), TYK2 (rs34536443)
SNPs and eNOS VNTR were lower in healthy individuals from Greece in
comparison to samples analyzed by other investigators working with
different ethnic/racial populations (6,14,22,37,38).
This finding suggests that existing population-specific differences
influence the frequency of these alleles. Thus, there is still the
possibility that an association of TYK2 rs34536443 with JIA may be
detected if the cohort size is increased. However, given the MAF of
the minor allele C of this SNP in the controls from Greece (0.22),
it can be assumed that a high number of patients and controls was
required to be collected (probably higher than 5,000 for each
category) to obtain an 80% power of the study (at p=0.05) (26).
Findings of this study highlight the importance of
conducting comparative studies in different populations,
considering that replication of previously identified markers is
paramount to determine which SNPs are true risk loci, thus
suggesting key disease pathways which require further study.
Additionally, the ultimate task upon a proper functional validation
of these SNPs is the translation of any findings into a better
management of specific groups of patients with JIA.
Acknowledgements
We would like to thank Dr J. Ellis and Dr R.C.
Chiaroni-Clarke (Victoria, Australia) for the constructive
discussions concerning the role of PRKCQ gene polymorphism in
JIA.
References
|
1
|
Ravelli A and Martini A: Juvenile
idiopathic arthritis. Lancet. 369:767–778. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Petty RE, Southwood TR, Manners P, Baum J,
Glass DN, Goldenberg J, He X, Maldonado-Cocco J, Orozco-Alcala J,
Prieur AM, et al International League of Associations for
Rheumatology, : International League of Associations for
Rheumatology classification of juvenile idiopathic arthritis:
Second revision, Edmonton, 2001. J Rheumatol. 31:390–392.
2004.PubMed/NCBI
|
|
3
|
Martini A: It is time to rethink juvenile
idiopathic arthritis classification and nomenclature. Ann Rheum
Dis. 71:1437–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thompson SD, Sudman M, Ramos PS, Marion
MC, Ryan M, Tsoras M, Weiler T, Wagner M, Keddache M, Haas JP, et
al: The susceptibility loci juvenile idiopathic arthritis shares
with other autoimmune diseases extend to PTPN2, COG6, and ANGPT1.
Arthritis Rheum. 62:3265–3276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thompson SD, Marion MC, Sudman M, Ryan M,
Tsoras M, Howard TD, Barnes MG, Ramos PS, Thomson W, Hinks A, et
al: Genome-wide association analysis of juvenile idiopathic
arthritis identifies a new susceptibility locus at chromosomal
region 3q13. Arthritis Rheum. 64:2781–2791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hinks A, Cobb J, Sudman M, Eyre S, Martin
P, Flynn E, Packham J, Barton A, Worthington J, Langefeld CD, et al
Childhood Arthritis Prospective Study (CAPS), ; UK RA Genetics
(UKRAG) Consortium, ; British Society of Paediatric and Adolescent
Rheumatology (BSPAR) Study Group, : Investigation of rheumatoid
arthritis susceptibility loci in juvenile idiopathic arthritis
confirms high degree of overlap. Ann Rheum Dis. 71:1117–1121. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hinks A, Cobb J, Marion MC, Prahalad S,
Sudman M, Bowes J, Martin P, Comeau ME, Sajuthi S, Andrews R, et al
Boston Children's JIA Registry, ; British Society of Paediatric and
Adolescent Rheumatology (BSPAR) Study Group, ; Childhood Arthritis
Prospective Study (CAPS), ; Childhood Arthritis Response to
Medication Study (CHARMS), ; German Society for Pediatric
Rheumatology (GKJR), ; JIA Gene Expression Study, ; NIAMS JIA
Genetic Registry, ; TREAT Study, ; United Kingdom Juvenile
Idiopathic Arthritis Genetics Consortium (UKJIAGC), : Dense
genotyping of immune-related disease regions identifies 14 new
susceptibility loci for juvenile idiopathic arthritis. Nat Genet.
45:664–669. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hinks A, Bowes J, Cobb J, Ainsworth HC,
Marion MC, Comeau ME, Sudman M, Han B, Becker ML, Bohnsack JF, et
al Juvenile Arthritis Consortium for Immunochip, : Fine-mapping the
MHC locus in juvenile idiopathic arthritis (JIA) reveals genetic
heterogeneity corresponding to distinct adult inflammatory
arthritic diseases. Ann Rheum Dis. 76:765–772. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thomson W, Barrett JH, Donn R, Pepper L,
Kennedy LJ, Ollier WE, Silman AJ and Woo P; Southwood T; British
Paediatric Rheumatology Study Group, : Juvenile idiopathic
arthritis classified by the ILAR criteria: HLA associations in UK
patients. Rheumatology (Oxford). 41:1183–1189. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hollenbach JA, Thompson SD, Bugawan TL,
Ryan M, Sudman M, Marion M, Langefeld CD, Thomson G, Erlich HA and
Glass DN: Juvenile idiopathic arthritis and HLA class I and class
II interactions and age-at-onset effects. Arthritis Rheum.
62:1781–1791. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Prahalad S, Shear ES, Thompson SD,
Giannini EH and Glass DN: Increased prevalence of familial
autoimmunity in simplex and multiplex families with juvenile
rheumatoid arthritis. Arthritis Rheum. 46:1851–1856. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hinks A, Eyre S, Ke X, Barton A, Martin P,
Flynn E, Packham J, Worthington J and Thomson W; Childhood
Arthritis Prospective Study (CAPS), ; UKRAG Consortium, ; BSPAR
Study Group, : Overlap of disease susceptibility loci for
rheumatoid arthritis and juvenile idiopathic arthritis. Ann Rheum
Dis. 69:1049–1053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hinks A, Eyre S, Ke X, Barton A, Martin P,
Flynn E, Packham J, Worthington J and Thomson W; Childhood
Arthritis Prospective Study, ; UKRAG Consortium, ; BSPAR Study
Group, : Association of the AFF3 gene and IL2/IL21 gene region with
juvenile idiopathic arthritis. Genes Immun. 11:194–198. 2010b.
View Article : Google Scholar
|
|
14
|
Dimopoulou DG, Zervou MI, Trachana M,
Myrthianou E, Pratsidou-Gertsi P, Kardassis D, Garyfallos A and
Goulielmos GN: Investigation of juvenile idiopathic arthritis
susceptibility loci: Results from a Greek population. Hum Immunol.
74:1194–1198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ellis JA, Chavez RA, Pezic A, Ponsonby AL,
Akikusa JD, Allen RC and Munro JE: Independent replication analysis
of genetic loci with previous evidence of association with juvenile
idiopathic arthritis. Pediatr Rheumatol Online J. 11:122013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Herlin MP, Petersen MB and Herlin T:
Update on genetic susceptibility and pathogenesis in juvenile
idiopathic arthritis. EMJ Rheumatol. 1:73–83. 2014.
|
|
17
|
Singh S, Bhattad S and Danda D: Genetics
of juvenile idiopathic arthritis. Int J Rheum Dis. 17:233–236.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Finkel TH, Li J, Wei Z, Wang W, Zhang H,
Behrens EM, Reuschel EL, Limou S, Wise C, Punaro M, et al: Variants
in CXCR4 associate with juvenile idiopathic arthritis
susceptibility. BMC Med Genet. 17:242016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McIntosh LA, Marion MC, Sudman M, Comeau
ME, Becker ML, Bohnsack JF, Fingerlin TE, Griffin TA, Haas JP,
Lovell DJ, et al Boston Children's JIA Registry, ; German Society
for Pediatric Rheumatology (GKJR), ; JIA Gene Expression Studies, ;
NIAMS JIA Genetic Registry, ; TREAT Study, : Understanding TNF
Therapy in JIA Project: Genome-wide association meta-analysis
reveals novel juvenile idiopathic arthritis susceptibility loci.
Arthritis Rheumatol. July 18–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ombrello MJ, Arthur VL, Remmers EF, Hinks
A, Tachmazidou I, Grom AA, Foell D, Martini A, Gattorno M, Özen S,
et al British Society of Pediatric and Adolescent Rheumatology
(BSPAR) Study Group, ; Inception Cohort of Newly Diagnosed Patients
with Juvenile Idiopathic Arthritis (ICON-JIA) Study Group, ;
Childhood Arthritis Prospective Study (CAPS) Group, ; Randomized
Placebo Phase Study of Rilonacept in sJIA (RAPPORT) Investigators,
Sparks-Childhood Arthritis Response to Medication Study (CHARMS)
Group, ; Biologically Based Outcome Predictors in JIA (BBOP) Group,
: Genetic architecture distinguishes systemic juvenile idiopathic
arthritis from other forms of juvenile idiopathic arthritis:
Clinical and therapeutic implications. Ann Rheum Dis. 76:906–913.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Consolaro A, Varnier GC, Martini A and
Ravelli A: Advances in biomarkers for paediatric rheumatic
diseases. Nat Rev Rheumatol. 11:265–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tao JH, Zou YF, Feng XL, Li J, Wang F, Pan
FM and Ye DQ: Meta-analysis of TYK2 gene polymorphisms association
with susceptibility to autoimmune and inflammatory diseases. Mol
Biol Rep. 38:4663–4672. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vazgiourakis VI, Sidiropoulos P, Bertsias
G, Koutsounaki E, Fragouli E, Raptopoulou A, Kritikos H, Boumpas DT
and Goulielmos GN: Association of the nitric oxide synthase (eNOS)
gene polymorphism with increased risk for both lupus
glomerulonephritis and rheumatoid arthritis in a single genetically
homogeneous population. Lupus. 16:867–874. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miyahara K, Kawamoto T, Sase K, Yui Y,
Toda K, Yang LX, Hattori R, Aoyama T, Yamamoto Y, Doi Y, et al:
Cloning and structural characterization of the human endothelial
nitric-oxide-synthase gene. Eur J Biochem. 223:719–726. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang J, Tsui V, Van Abbema A, Bao L,
Barrett K, Beresini M, Berezhkovskiy L, Blair WS, Chang C, Driscoll
J, et al: Lead identification of novel and selective TYK2
inhibitors. Eur J Med Chem. 67:175–187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Myrthianou E, Zervou MI, Budu-Aggrey A,
Eliopoulos E, Kardassis D, Boumpas DT, Kougkas N, Barton A,
Sidiropoulos P and Goulielmos GN: Investigation of the genetic
overlap between rheumatoid arthritis and psoriatic arthritis in a
Greek population. Scand J Rheumatol. 46:180–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guzik TJ, Korbut R and Adamek-Guzik T:
Nitric oxide and superoxide in inflammation and immune regulation.
J Physiol Pharmacol. 54:469–487. 2003.PubMed/NCBI
|
|
28
|
Albrecht ED, Aberdeen GW, Niklaus AL,
Babischkin JS, Suresch DL and Pepe GJ: Acute temporal regulation of
vascular endothelial growth/permeability factor expression and
endothelial morphology in the baboon endometrium by ovarian
steroids. J Clin Endocrinol Metab. 88:2844–2852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Noiri E, Satoh H, Taguchi J, Brodsky SV,
Nakao A, Ogawa Y, Nishijima S, Yokomizo T, Tokunaga K and Fujita T:
Association of eNOS Glu298Asp polymorphism with end-stage renal
disease. Hypertension. 40:535–540. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Dudley D and Wang XL:
Haplotype-specific effects on endothelial NO synthase promoter
efficiency: Modifiable by cigarette smoking. Arterioscler Thromb
Vasc Biol. 22:e1–e4. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mantaka A, Goulielmos GN, Koulentaki M,
Tsagournis O, Voumvouraki A and Kouroumalis EA: Polymorphisms of
genes related to endothelial cells are associated with primary
biliary cirrhosis patients of Cretan origin. Hum Immunol.
73:829–835. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Galanakis EI, Kofteridis D, Stratigi K,
Petraki E, Vazgiourakis V, Fragouli E, Mamoulakis D, Boumpas DT and
Goulielmos GN: Intron 4 a/b polymorphism of the endothelial nitric
oxide synthase gene is associated with both type 1 and type 2
diabetes in a genetically homogeneous population. Hum Immunol.
69:279–283. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zervou MI, Vazgiourakis VM, Yilmaz N,
Kontaki E, Trouw LA, Toes RE, Bicakcigil M, Boumpas DT, Yavuz S and
Goulielmos GN: TRAF1/C5, eNOS, C1q, but not STAT4 and PTPN22 gene
polymorphisms are associated with genetic susceptibility to
systemic lupus erythematosus in Turkey. Hum Immunol. 72:1210–1213.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bunjevacki V, Maksimovic N, Jekic B, Milic
V, Lukovic L, Novakovic I, Damjanov N, Radunovic G and Damnjanovic
T: Polymorphisms of the eNOS gene are associated with disease
activity in rheumatoid arthritis. Rheumatol Int. 36:597–602. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goulielmos GN, Chiaroni-Clarke RC,
Dimopoulou DG, Zervou MI, Trachana M, Pratsidou-Gertsi P,
Garyfallos A and Ellis JA: Association of juvenile idiopathic
arthritis with PTPN22 rs2476601 is specific to females in a Greek
population. Pediatr Rheumatol Online J. 14:252016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Diogo D, Bastarache L, Liao KP, Graham RR,
Fulton RS, Greenberg JD, Eyre S, Bowes J, Cui J, Lee A, et al: TYK2
protein-coding variants protect against rheumatoid arthritis and
autoimmunity, with no evidence of major pleiotropic effects on
non-autoimmune complex traits. PLoS One. 10:e01222712015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsoi LC, Spain SL, Knight J, Ellinghaus E,
Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, et al
Collaborative Association Study of Psoriasis (CASP), ; Genetic
Analysis of Psoriasis Consortium, ; Psoriasis Association Genetics
Extension, ; Wellcome Trust Case Control Consortium 2, :
Identification of 15 new psoriasis susceptibility loci highlights
the role of innate immunity. Nat Genet. 44:1341–1348. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Neugebauer S, Baba T and Watanabe T:
Association of the nitric oxide synthase gene polymorphism with an
increased risk for progression to diabetic nephropathy in type 2
diabetes. Diabetes. 49:500–503. 2000. View Article : Google Scholar : PubMed/NCBI
|