Introduction
Human immunodeficiency virus (HIV) infection leads
to a progressive decline of immunity and eventually results in the
initiation and progression of acquired immune deficiency syndrome
(AIDS). The progressively weakened immune system of patients with
AIDS increases the incidence of opportunistic infections and leads
to higher mortality of patients with AIDS. CD4+ T cells
are the targets of invading HIV-1; therefore, the counts of
CD4+ T cells are used to evaluate the disease stage for
patients with AIDS (1).
Additionally, gut-associated lymphoid tissue (GALT) is the biggest
lymphoid tissue of the human body, which provides the specific
anatomical and physiological environment required by
CD4+ T cells and other immune cells (2). GALT is also the primary target of
HIV-1 infection; therefore, GALT was previously identified as the
potential reservoir of HIV-1 due to the depletion of
CD4+ T cells observed (3–5).
This was followed by damage in intestinal mucosa and disorders of
gut micro-ecology, which accelerated disease progression in
patients with AIDS (6–9). The intestinal immune dysfunction
following HIV infection may injure the gut barrier, leading to gut
bacterial translocation into the blood (10,11)
and chronic immune activation (11–13).
This may also increase the risks of other complications, such as
cardiovascular disease, osteoporosis, neurodegeneration and
metabolic disease (13). In view
of the fact that the exhaustion of CD4+ T cells from
GALT was much earlier than that of peripheral blood for AIDS
patients, CD4+ T cell counts in GALT may be used as
potential biomarkers for predicting the mortality rates in patients
with AIDS (14). Highly active
anti-retroviral therapy may improve CD4+ T cell counts
of patients with AIDS; however, the restoration of the immune
system is challenging, particularly the counts of CD4+ T
cells in gut intestinal mucosa remain at low levels (5,15).
Concurrently, this chronic inflammation may provide additional
activated CD4+ T cells as targets for HIV infection,
subsequently accelerating AIDS progression and exacerbating the
symptoms of the patient (7,14,16).
Therefore, the treatment of gut barrier damage in patients with
AIDS would improve the reconstructing of the immune system and
reduce the diffusion of HIV-1 in patients with AIDS.
MicroRNAs (miRNAs) are a type of endogenous
non-coding single-stranded RNAs with a length of 21 to 23
nucleotides. miRNAs regulate the gene expression by employing the
mechanism termed RNA interference at the post-transcription level,
which may lead to the degradation of the target mRNA or suppression
of its translation (17). A
previous study has predicted that miRNAs may regulate >50% of
human protein-coding genes (18).
miRNAs have important regulatory roles in different diseases by
participating in various processes, such as cell proliferation and
development (19,20). Dysregulation of miRNAs was also
detected in the disturbance of cellular processes of diabetes and
cancer progression (21). The
function of miRNAs in the pathogenesis of AIDS has been previously
illustrated in terms of the host cells aspect and the virus aspect
(22–24). In the host miRNAs, miR-132 was
identified to be upregulated following CD4+ T cell
activation, which increased HIV-1 replication in CD4+ T
cells (22). Conversely, miR-198
inhibited HIV-1 gene expression and replication (23). The HIV-1 tat protein also regulates
miR-217 for the regulation of sirtuin 1, IκB kinase and
phosphorylated nuclear factor-κB, subsequently inducing various
effects on multiple pathways and cellular responses, such as
p65-NFκB and AMPK signaling (24).
In addition to the aforementioned endogenous miRNAs of host cells,
HIV-1 also encodes two miRNAs. One is miR-N367, which suppresses
the HIV-1 nef protein and inhibits the transcription of HIV-1, thus
maintaining the virus at a latent stage in patients with HIV
infection where they are termed long-term non-progressors (LTNPs)
(25). The other miRNA is
HIV1-miR-H1, which suppresses the expression of the apoptosis
antagonizing transcription factor and the downstream B cell
leukemia/lymphoma 2 and MYC proto-oncogene and downregulated the
expression of cellular miR-149, which targets Viral Protein R of
HIV-1, facilitating HIV-1 replication and impairing cellular
responses to infection (26).
Therefore, previous studies have identified the complicated
networks of miRNAs in HIV-1 and host cells that influenced the
pathogenesis and progression of AIDS. However, the interactions
between the miRNAs that have been identified, remain to be
elucidated. Therefore, identifying the miRNAs active in the
intestinal mucosa of patients with AIDS may elucidate the networks
of miRNAs for HIV-infected patients and identify potential novel
mechanisms of miRNA regulation in patients with HIV.
The present study isolated RNA from colon biopsy
samples of HIV+ antiretroviral therapy-naïve (without
therapeutic intervention) patients and HIV− healthy
individuals. The samples were subjected to miRNA array
hybridization. MiRNAs with significantly different expression
levels were identified between the HIV/AIDS and control groups with
a threshold of P<0.05. The target genes of the significantly
different miRNAs were predicted and subsequent GO and KEGG pathway
analyses were used to predict the potential signal pathways that
may be associated with gut barrier dysfunction of patients with HIV
infection and the potential disease progression. RT-qPCR was
performed in order to verify the expression of the significantly
expressed genes, which modulated the gut barrier dysfunction of
patients with HIV infection.
Materials and methods
Patients
Colon biopsy samples from a total of 26 male
participants aged 27–53 years were collected by electron endoscopy
from the First Affiliated Hospital of Kunming Medical University
(Kunming, China) between January 2013 and January 2014 and informed
consent was obtained. The HIV-infected antiretroviral therapy-naïve
group contained 3 participants and other 3 healthy individuals were
assigned to control group. Additional colon biopsies from 10
HIV-infected patients and 10 healthy control individuals were
performed. The samples were immediately cryopreserved for
transcriptional analysis. Peripheral blood samples were also
collected at the time of the endoscopy. Detailed information on the
clinical characteristics of the participants are presented in
Table I. HIV-infected individuals
enrolled in the current study if they adhered to the following
criteria: Antiretroviral therapy naïve; viral load of >10,000
HIV-1 RNA copies/ml of plasma; and period for infection with HIV-1
>1 year. The HIV-1 patients with current opportunistic or other
infections were excluded from the present study. The procedures in
the current study were approved by the Ethics Committee Review
Board of Kunming Medical University.
 | Table I.Patient data and clinical parameters
of male Chinese HIV+ patients and normal
individuals. |
Table I.
Patient data and clinical parameters
of male Chinese HIV+ patients and normal
individuals.
| ID | HIV status | CD4+
cell count, cells/µl) | Viral load,
copies/ml | Age, years | Duration of
infection, years |
|---|
| N78 | HIV- | 836 | NA | 29 | NA |
| N121 | HIV- | 921 | NA | 47 | NA |
| N324 | HIV- | 865 | NA | 31 | NA |
| Y130 | HIV+ | 217 | 1790 | 36 | 4 |
| G45 | HIV+ | 414 | 1326 | 30 | 1 |
| W72 | HIV+ | 205 | 2204 | 30 | 2 |
| N-1 | HIV- | 664 | NA | 27 | NA |
| N-2 | HIV- | 567 | NA | 27 | NA |
| N-3 | HIV- | 681 | NA | 29 | NA |
| N-4 | HIV- | 688 | NA | 32 | NA |
| N-5 | HIV- | 696 | NA | 28 | NA |
| N-6 | HIV- | 718 | NA | 34 | NA |
| N-7 | HIV- | 643 | NA | 33 | NA |
| N-8 | HIV- | 1089 | NA | 30 | NA |
| N-9 | HIV- | 810 | NA | 31 | NA |
| N-10 | HIV- | 769 | NA | 38 | NA |
| P-1 | HIV+ | 106 | 2088 | 33 | 1 |
| P-2 | HIV+ | 201 | 1557 | 41 | 5 |
| P-3 | HIV+ | 206 | 1432 | 38 | 3 |
| P-4 | HIV+ | 368 | 2421 | 33 | 3 |
| P-5 | HIV+ | 140 | 1805 | 38 | 2 |
| P-6 | HIV+ | 222 | 1717 | 37 | 2 |
| P-7 | HIV+ | 231 | 1314 | 43 | 5 |
| P-8 | HIV+ | 440 | 1022 | 51 | 4 |
| P-9 | HIV+ | 117 | 1840 | 53 | 3 |
| P-10 | HIV+ | 225 | 1950 | 45 | 3 |
Total RNA extraction and quality
inspection
Total RNA from each colon tissue sample was isolated
with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and purified with miRNeasy Mini kit (Qiagen,
Inc., Valencia, CA, USA) according to the manufacturer's protocols.
RNA quality and quantity were quantified using a NanoDrop
spectrophotometer at a wavelength of 280 nm (ND-1000; Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). RNA integrity was
determined by gel electrophoresis (1% agarose). Subsequently, the
isolated RNA was purified using RNeasy Mini kit (Qiagen, Inc.) and
the purity was examined with NanoDrop ND-1000.
RNA labeling and array
hybridization
Following quality control, the miRCURY Hy3/Hy5 Power
labeling kit (Exiqon, Vedbæk, Denmark) was used for miRNA labeling
according to the manufacturer's protocol. Subsequently, Hy3-labeled
samples were hybridized on the miRCURY LNA Array (v.18.0; Exiqon)
according to the array manual. The total mixture with hybridization
buffer was hybridized to the microarray in a 12-Bay Hybridization
system (hybridization system; NimbleGen Systems, Inc., Madison, WI,
USA), which provided an active mixing action and a constant
incubation temperature to improve hybridization uniformity and
enhanced the signal. Following hybridization, the slides were
washed three times using Wash buffer kit (Exiqon) and scanned using
the Axon GenePix 4000B microarray scanner (Molecular Devices, LLC,
Sunnyvale, CA, USA). GeneChip microarray experiments were conducted
by KangChen Biotech (Shanghai, China).
Microarray data analysis
Scanned images were imported into GenePix Pro
version 6.0 software package (Molecular Devices, LLC) for grid
alignment and data extraction. Replicated miRNAs were averaged and
miRNAs with intensity ≥30 in all samples were selected for the
calculation of the normalization factor. Expressed data were
normalized using the median normalization. Significantly
differentially-expressed miRNAs between the two groups were
identified through fold-change and P<0.05. A cut-off of >2.0
fold-change of gene expression was used to identify miRNAs for
analysis of hierarchical clustering patterns and downstream
biofunctional assessment. The difference between two groups,
evaluated by P-value was calculated based on a Student's t-test.
Hierarchical clustering was performed to identify distinguishable
miRNA expression profiling among samples. A >2.0 fold-change and
P<0.05 cut-offs were selected to analyze gene expression data
(27), obtain sufficient
information and generate a large data set to be used for downstream
pathway analysis.
Bioinformatics analysis
To predict the target genes of the
differentially-expressed miRNAs, the 3 most popular databases,
TargetScan (28), miRanda
(29) and miRDB (30) were fully referenced. In order to
reduce false positive results, genes which were predicted by all 3
databases were selected as differential miRNA targets for further
analysis. The miRNA expression changes were arranged from high to
low according to the standardized intensity and fold-changes. The
biological functions, including biological processes (BP, pathways
and larger processes made up of the activities of multiple gene
products), cellular compounds (CC, where gene products are active)
and molecular function (MF, molecular activities of gene products)
for the potential target genes, were analyzed using Gene Ontology
(GO; www.geneontology.org) terms. GO defines
concepts/classes used to describe gene function and the
relationships between these concepts. Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analysis was also performed with the
target genes, which identified those most likely to be involved in
intracellular signal transduction pathways. The major GO terms
associated with BP and CC were manually summarized based on
gene-term enrichment buttons provided for each functional group at
P<0.05.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
In order to confirm the expression profiles of key
miRNAs, total miRNA was isolated from colon biopsies of 10
additional HIV+ patients and 10 normal individuals that
were used as negative control. Poly-A tails were added to the
miRNAs and reverse transcribed into cDNA with miRcute miRNA cDNA
First-Strand Synthesis kit (Tiangen Biotech Co., Ltd., Beijing,
China). For polyA tail addition, 1 µg RNA was mixed with 0.4 µl
E. coli poly(A) polymerase (5 U/µl), 2 µl 10X poly(A)
polymerase buffer and 4 µl 5X rATP solution for incubation at 37°C
for 60 min. For reverse transcription, the above polyA reaction mix
was further mixed with 0.5 µl Quant RTase and other additives for
incubation at 37°C for 60 min. The quantity of miRNA was determined
using a miRcute miRNA quantification kit (Tiangen Biotech Co.,
Ltd.). The forward primers for the miRNAs are as follows (U6 was
used as miRNA reference): hsa-miR-199a-3 pF,
5′-CAGACAGTAGTCTGCACATTGGTTA-3′; hsa-miR-20b-5 pF,
5′-GCAAAGTGCTCATAGTGCAGGTAG-3′; hsa-miR-32-5 pF,
5′-CGCAGTATTGCACATTACTAAGTTG-3′; U6-F,
5′-CGATACAGAGAAGATTAGCATGGC-3′.
The primers for the mRNAs are as follows: (Actin was
used as mRNA reference): ITGA5-F, 5′-ACCCAGACCCTGCTCATCCA-3′ and
ITGA5-R, 5′-TGTGAATCGGCGAGAGTTTGTC-3′; FBN1-F,
5′-CGTGCACCCTATGCCAAGTT-3′ and FBN1-R, 5′-GCATTCCTCAGTACCCCAGG-3′.
The thermocycling conditions are as follows: 95°C for 10 min; 45
cycles of 95°C for 15 sec and 60°C for 60 sec. The experiments were
performed in triplicate with SYBR Green dye (Tiangen Biotech Co.,
Ltd.). Expression levels were calculated using the
2−ΔΔCq method (31) and
data were presented as the mean ± standard error. P<0.05 was
considered to indicate a statistically significant difference.
Results
Differentially-expressed miRNA
profiles in AIDS and healthy groups
In order to identify the miRNAs from the colon
biopsy samples of AIDS patients and healthy controls, a 7th
generation miRNA array containing 3,100 capture probes, covering
all human, mouse and rat miRNAs annotated in miRBase 18.0, all
viral miRNAs associated with these species and 25 miRPlus human
miRNAs were used. The latter were proprietary miRNAs that were not
found in miRBase. RNA quality was inspected prior to performing
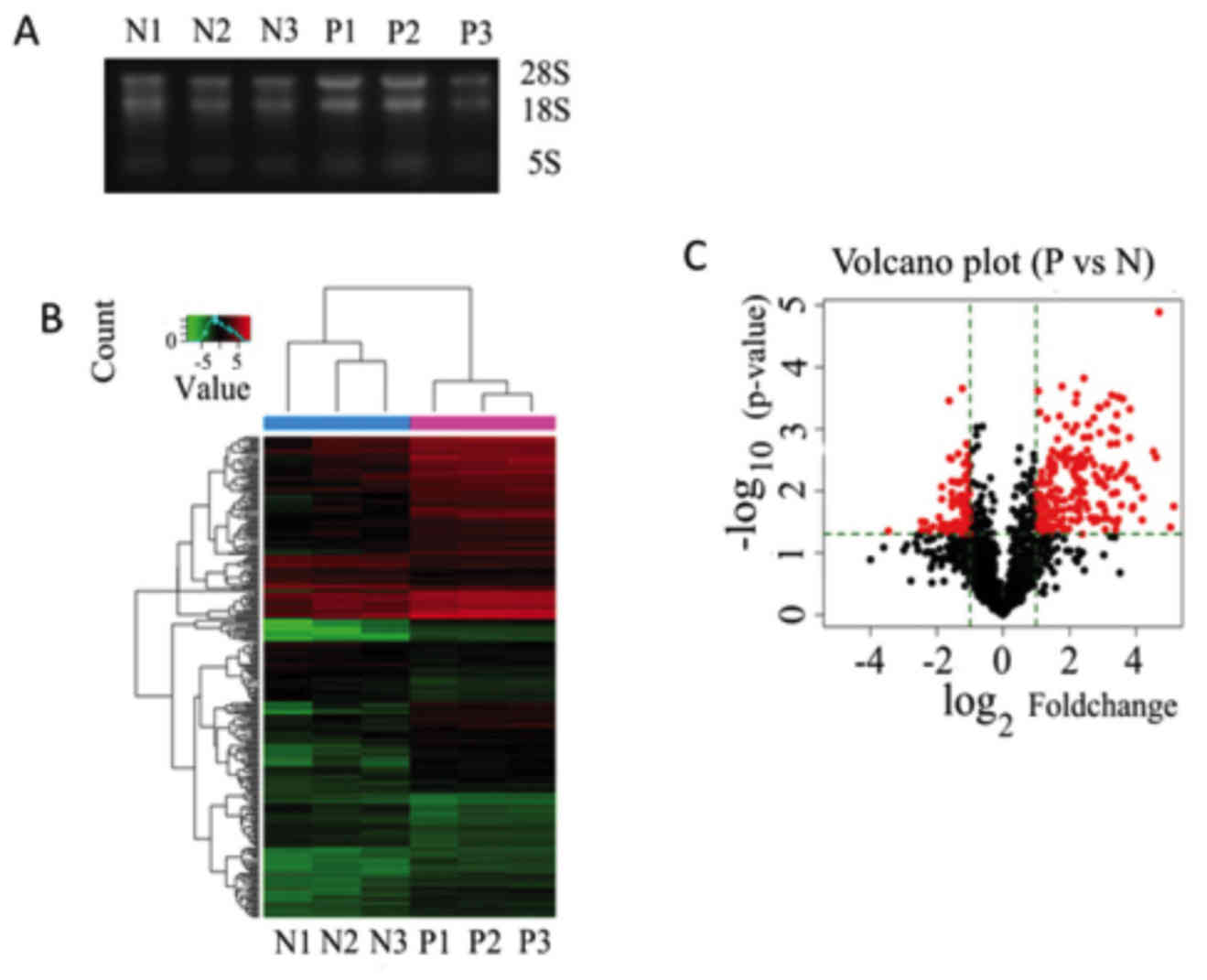
microarray hybridization. As presented in Fig. 1A, three bands were evident after
RNA electrophoresis, denoting 28S, 18S and 5S RNA. Filtering was
performed with microarray detection flags (presence or absence of
signals), using thresholds of fold-change (≥2) and P<0.05 to
identify differentially-expressed miRNAs, 257 miRNAs were
upregulated and 219 miRNAs were downregulated (data not shown) in
the HIV-infected patients compared with the HIV-negative normal
individuals. A heatmap was also generated to display a two-way
hierarchical clustering of miRNAs and samples (Fig. 1B). The red color represents
upregulated miRNAs, whereas green bars represent downregulated
miRNAs. The three patients and the three control samples were
grouped as different clusters, due to their similarities in miRNA
expression profiles. A Volcano plot was also composed to visually
identify the significantly differentially-expressed miRNAs between
the two groups, based on the aforementioned thresholds (Fig. 1C). The red dots represent the
miRNAs that had significantly altered expression level (P<0.05),
either upregulated (the red dots to the right) or downregulated
(the red dots to the left).
Prediction of target genes from
differentially-expressed miRNAs
Functions of miRNAs were predicted by their target
mRNAs; therefore, the putative target genes of
differentially-expressed miRNAs between AIDS patients and healthy
control individuals were predicted using the 3 aforementioned
databases and the associated target genes were retrieved from all 3
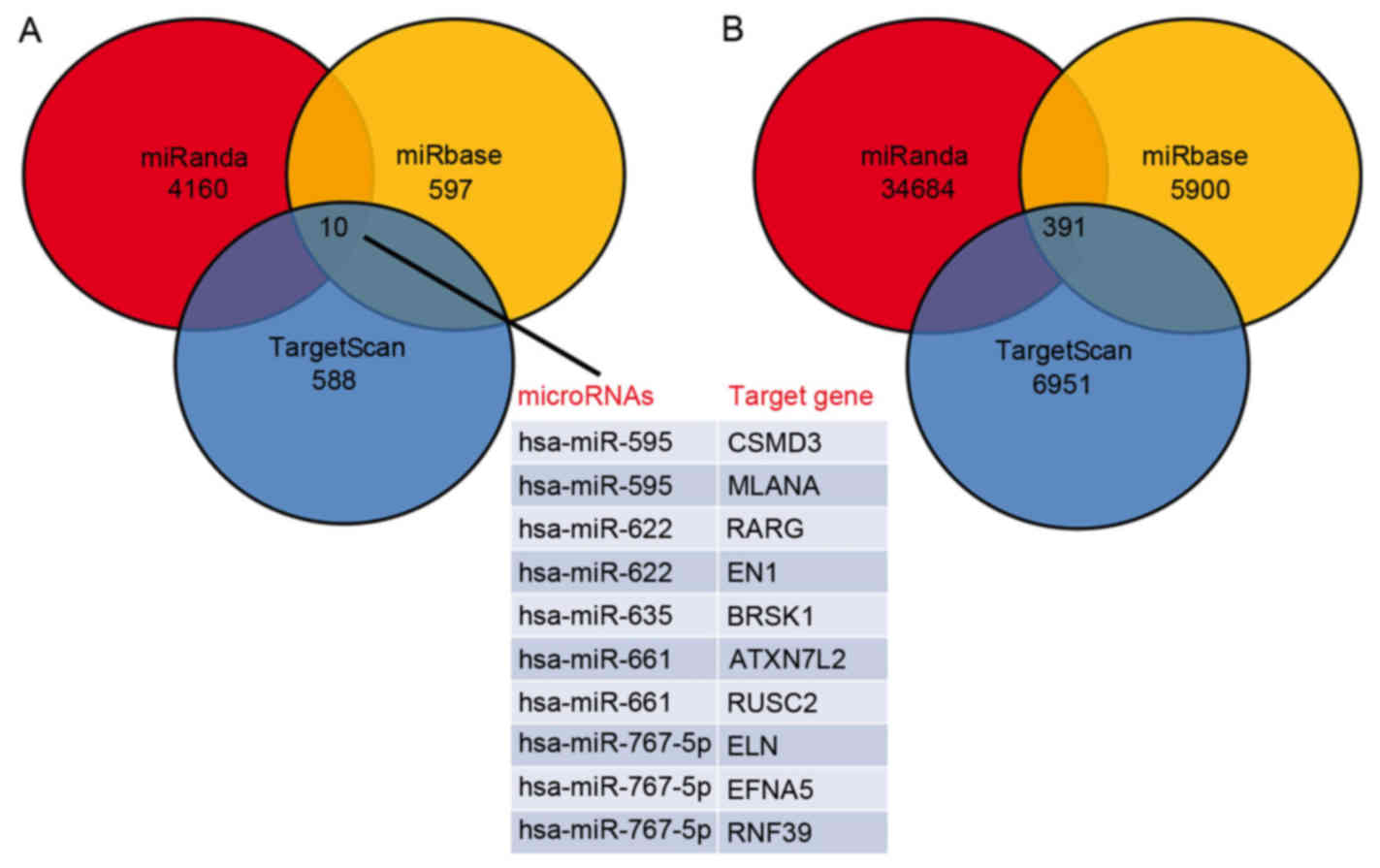
databases. A total of 10 target genes of significantly
downregulated miRNAs were predicted, including CSMD3, MLANA, RARG,
EN1, BRSK1, ATXN7L2, RUSC2, ELN, EFNA5 and RNF39 (Fig. 2A). Additionally, 391 target genes
of significantly upregulated miRNAs were also predicted (Fig. 2B).
GO analysis for potential target genes
of upregulated miRNAs
GO analysis of these target genes, which were
predicted according to the differentially-expressed miRNAs, was
performed, including BP, CC and MF. As only 10 overlapping target
genes of downregulated miRNAs were predicted through the 3
databases, GO analysis for these 10 genes was not performed and the
present study focused on GO and KEGG pathway analysis based on the
target genes of the upregulated miRNAs.
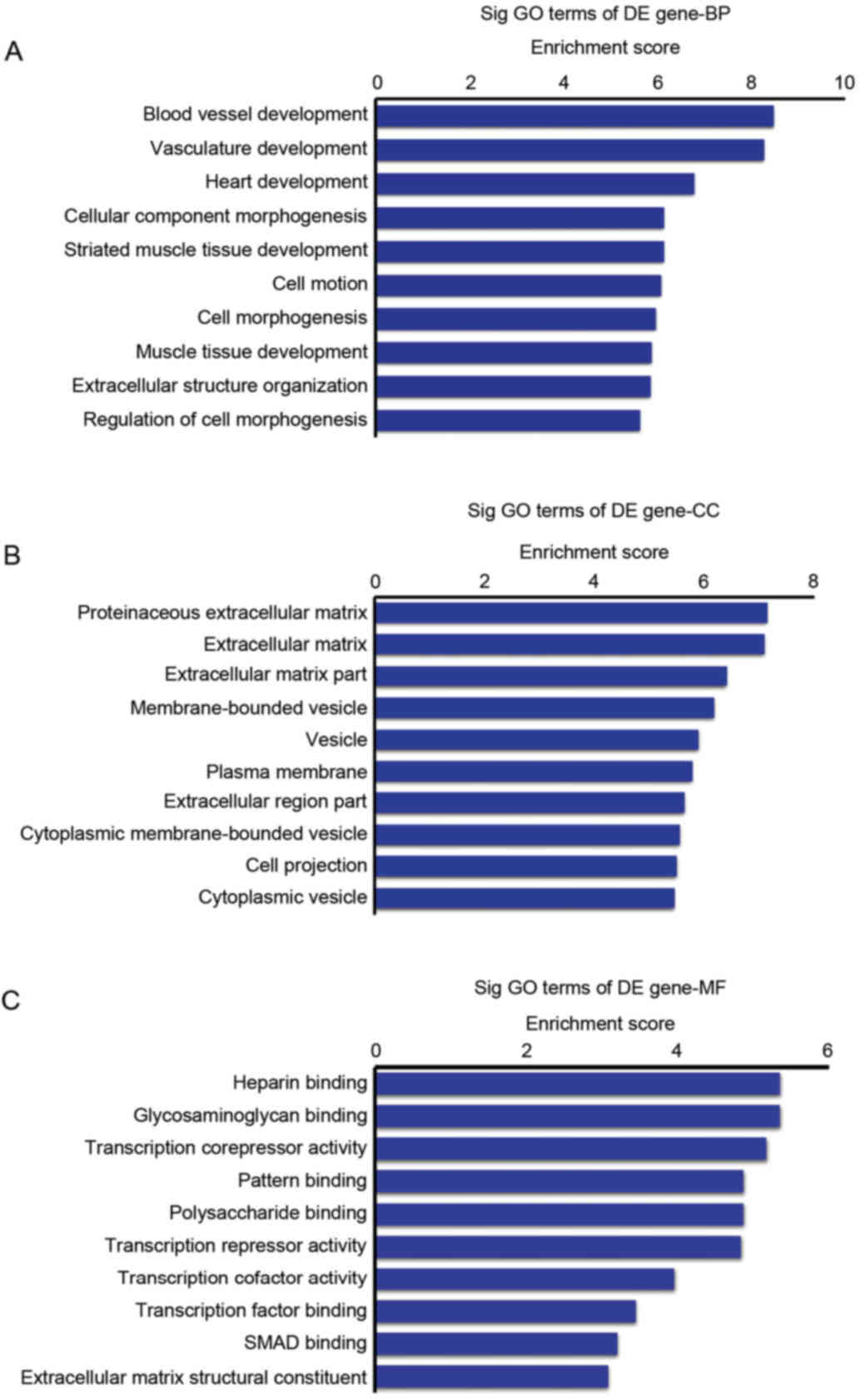
The target genes were sorted into various BP
according to their enrichment scores. The top 10 BP were identified
as follows: Blood vessel development, vasculature development,
heart development, cellular component morphogenesis, striated
muscle tissue development, cell motion, cell morphogenesis, muscle
tissue development, extracellular structure organization and
regulation of cell morphogenesis (Fig.
3A).
The predicted target genes of upregulated miRNAs
were also sorted into various categories of CC according to their
enrichment scores, with the top 10 CC categories being listed as
follows: Proteinaceous extracellular matrix, extracellular matrix,
extracellular matrix part, membrane-bounded vesicle, vesicle,
plasma membrane, extracellular region part, cytoplasmic
membrane-bounded vesicle, cell projection and cytoplasmic vesicle
(Fig. 3B).
The GO analysis was used to sort the predicted
target genes of upregulated miRNAs were into various categories of
MF (Fig. 3C). The top 10
categories of MF were: Heparin binding, glycosaminoglycan binding,
transcription corepressor activity, pattern binding, polysaccharide
binding, transcription repressor activity, transcription cofactor
activity, transcription factor binding, SMAD binding and
extracellular matrix structural constituent.
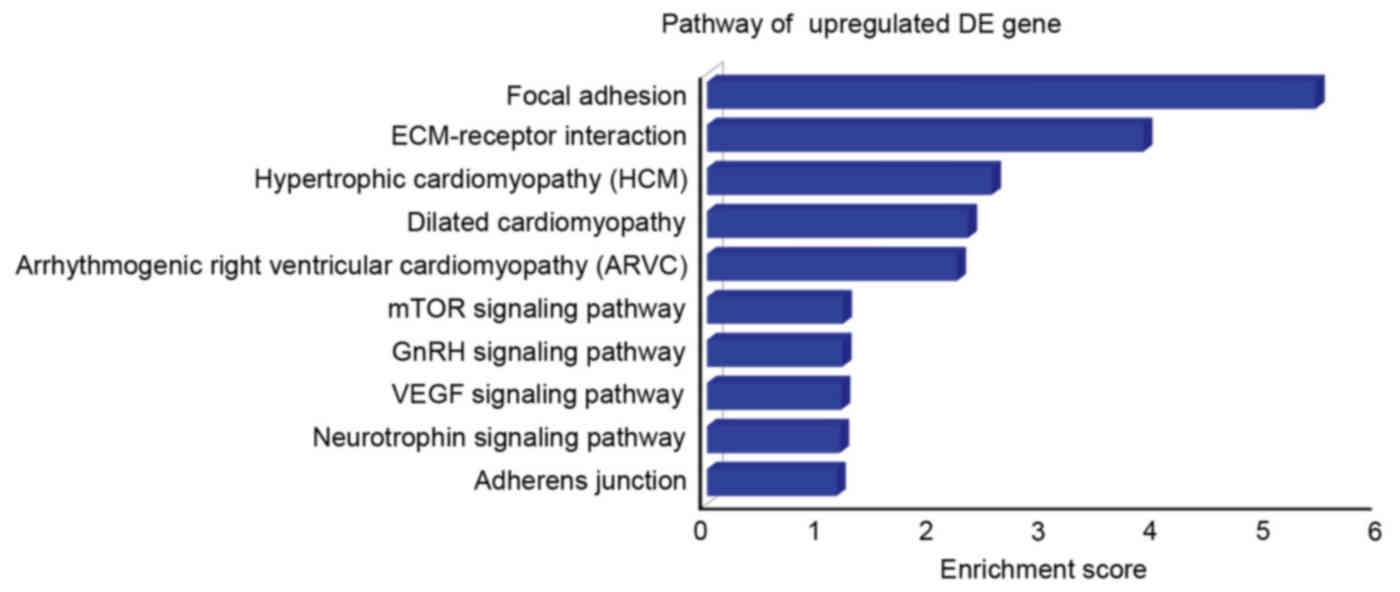
KEGG pathway analysis of predicted
target genes of upregulated miRNAs
KEGG pathway analysis was used to analyze the
pathways, which were most likely to be altered by upregulated
miRNAs (Fig. 4). The top 10
signaling pathways identified for the predicted target genes of
upregulated miRNAs according to their enrichment scores were focal
adhesion, ECM-receptor interaction, hypertrophic cardiomyopathy,
dilated cardiomyopathy, arrhythmogenic right ventricular
cardiomyopathy, mechanistic target of rapamycin signaling pathway,
gonadotropin releasing hormone signaling pathway, vascular
endothelial growth factor signaling pathway, neutrophin signaling
pathway and adherens junction.
Confirmation of expression levels of
key upregulated miRNAs
To confirm the expression levels of key miRNAs,
RT-qPCR was performed with colon biopsies from 10 additional
HIV-infected patients and 10 healthy control individuals.
Normalized expression levels of AIDS patients, and the fold-changes
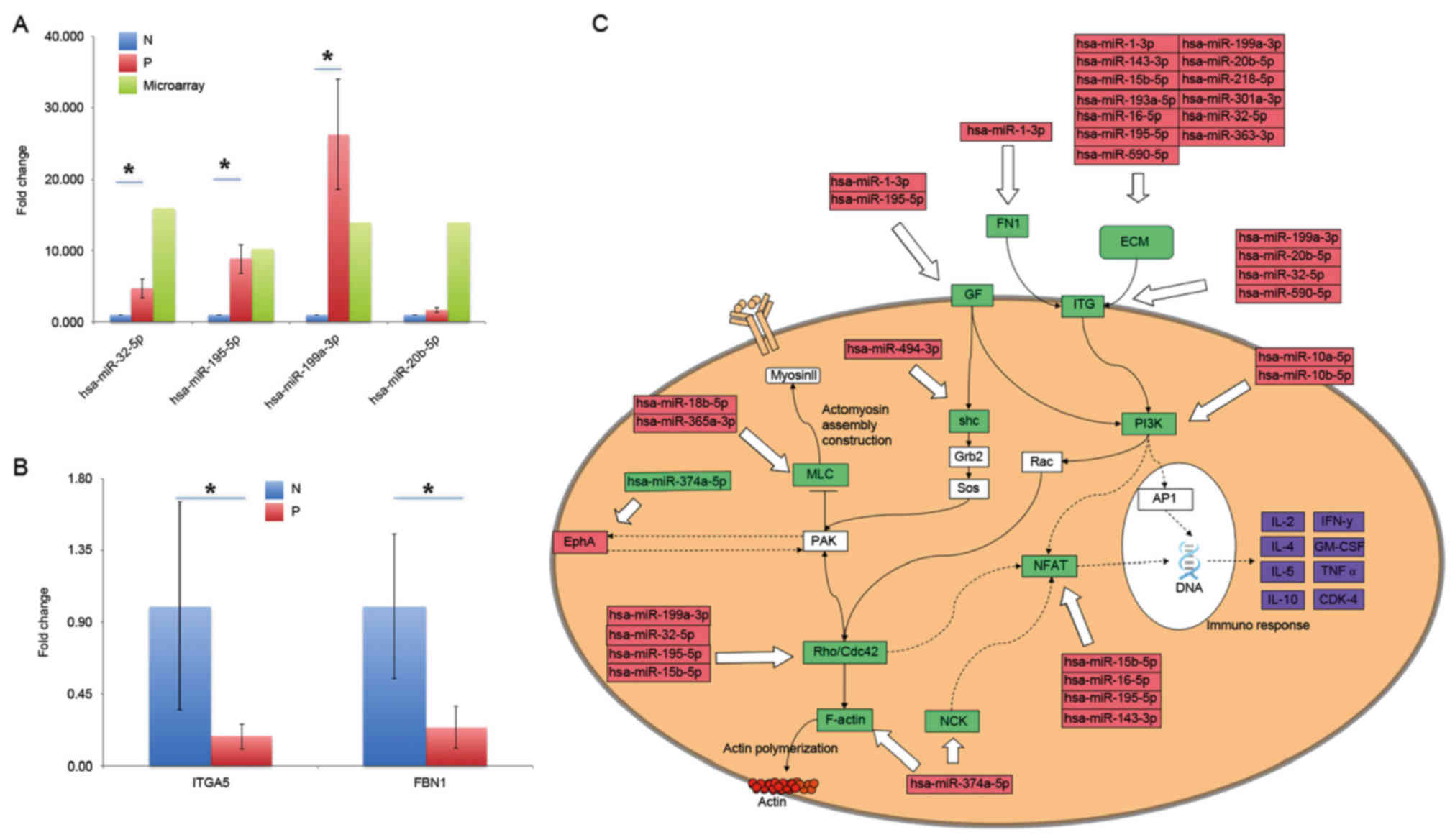
from microarray data are presented in Fig. 5A. A total of 5 miRNAs were tested,
4 of which produced the consistent results with the microarray
data. These miRNAs were hsa-miR-32-5p, hsa-miR-195-5p,
hsa-miR-199a-3p and hsa-miR-20b-5p. The other miRNA,
hsa-miR-590-5p, was upregulated in the samples of HIV-infected
individuals from microarray data, but was demonstrated to be
decreased in qPCR data. These miRNAs were selected as they were
predicted to regulate some important genes that are implicated in
intestinal mucosa dysfunction. For example, hsa-miR-32-5p was
predicted to modulate the expression of integrin α5 (ITGA5), which
has an important role in colon epithelium functions. All of them
had high fold-changes of >10 in the P group, and the expression
of some miRNAs, including hsa-miR-32-5p, hsa-miR-195-5p and
hsa-miR-20b-5p was confirmed by RT-qPCR, had fold-changes of 4.764,
8.914, 1.712 and 26.290, respectively. Additionally, expression of
some putative target genes was also confirmed. As presented in
Fig. 5B, expression of ITGA5 and
fibrillin 1 (FBN1), the two putative target genes of upregulated
hsa-miR-32-5p, were downregulated. Although the error bars are
large for some groups due to the variability between the samples,
the difference between the groups was significant (P<0.05).
These findings partially confirmed the accuracy of the microarray
data and validated the further analysis performed.
Discussion
The present study identified a large number of
miRNAs with altered expression levels in HIV-infected individuals
compared with healthy individuals. The data in the current study
revealed that the regulation of some miRNAs was consistent with
previous studies, such as miR-132, which was reported to be
upregulated in activated CD4+ T cells and enhance HIV-1
replication (22). Some miRNAs
activate CD4+ T cells, thereby creating a favorable
environment for HIV-1 replication (32,33),
such as miR-21, miR-142-3p/5p, miR-155, miR-181a and miR-27b
(34–40). Among them, miR-181a was capable of
activating CD4+ T cells by enhancing various associated
pathways, including ERK and calcium flux (40). The importance of miR-181a was also
confirmed by the present study with a 3-fold increase in expression
in patients with AIDS, in association with CD4+ T cell
activation. Conversely, some inconsistencies were also detected
between the current study and previous literature. For example,
miR-142-3p/5p were downregulated in CD4+ T cells from
patients with systemic lupus erythematosus, which may also lead to
the over activation of CD4+ T cells via their targets,
such as SAP, CD84, interleukin (IL)-10 (37). However, the higher levels of
miR-142-3p in the present study suggested a possible deactivation
of CD4+ T cells. Additionally, lower levels of
hiv1-miR-TAR-3p, an HIV-1-encoded miRNA, in HIV-infected
individuals were observed, which may reveal that HIV-1 infection
may trigger apoptosis. This was consistent previous studies of
HIV-1 entry, facilitated by CD4 and CCR5 chemokine receptor. This
elevated Fas levels in the cells and rendered the cells susceptible
to apoptosis induced by the Fas/Fas ligand interaction (41).
Using criterions of fold change ≥2 and P<0.05 a
total of 476 human and virus-derived miRNAs were identified as
significantly altered in HIV/AIDS when compared with the control
group (P<0.05). Among them, a lower number of target genes of
downregulated miRNAs were predicted compared with the target genes
of upregulated miRNAs (10 vs. 391). These 10 genes include retinoic
acid receptor γ (RARG), an antagonist of RAR signaling. Deficiency
in retinoic acid receptors α (RARα) may lead to defects in
CD4+ T cell activation (42), which implied that the retinoic acid
pathway through RARα was essential for CD4+ T cell
effector responses. The reduced expression of some miRNAs, such as
miR-622, may predict an increase in RARG. As a nuclear receptor,
RARG shares functional similarities with other isoforms, such as
RARα and RARβ (43). Therefore,
the transcription of associated genes would be altered. Therefore,
it is possible that RARG may also contribute to the promotion of
CD4+ T cell effector responses and acceleration of
disease progression may occur, although definitive evidence is
still required.
Although the expression patterns of some miRNAs were
confirmed, their roles remain to be fully elucidated. As gene
expression may be modulated by a broad range of biological factors,
including other gene products, miRNAs, long non-coding RNA,
prediction of changes in the expression of target genes only based
on their regulator miRNAs may not be entirely reliable. By
considering the aforementioned factors, GO and KEGG pathways
analyses were performed, where the direction of the expression
alterations (upregulation or downregulation) was not required.
Therefore, the present study demonstrated pathways that are
modulated by the differentially-expressed miRNAs. The GO and KEGG
pathway analyses identified a number of pathways involved in heart
development and cardiomyopathy development. Previous studies have
determined that AIDS patients are more prone to be dilated
cardiomyopathy and other heart diseases (44,45).
The actin, a, cardiac muscle 1 (ACTC1) gene, which encodes cardiac
muscle α actin and is responsible for heart muscle contraction, was
regulated by miR-30a-5p, miR-32-5p and miR-363-3p. Some of the
validated miRNAs had important functions in AIDS and associated
diseases. For example, nuclear factor of activated T-cells 3
(NFATC3) was a predicted target of hsa-miR-195-5p. Upregulation of
hsa-miR-195-5p reduced the NFATC3 mRNA level, which confirmed by
the mRNA microarray data (not shown). This protein is a member of
the nuclear factors of activated T cells and regulates the
expression of various cytokines, including IL-2 and tumor necrosis
factor-a (TNF-α; Fig. 5C.
Therefore, NFATC3 and its regulator hsa-miR-195-5p, are important
for the immune response. Another important molecule is
hsa-miR-32-5p, whose putative targets include RAB23, member RAS
oncogene family, FBN1, Kruppel like factor 2 and ITGA5. These genes
modulate multiple biological processes and components, such as the
extracellular matrix. The expression for parts of these putative
target genes were confirmed by RT-qPCR as presented in Fig. 5B.
Using the significantly changed miRNAs, the
potential genes that may have crucial roles on regulating gut
barrier dysfunction for HIV-infected patients were predicated using
the DAVID comprehensive functional classification. Three pathways
and an important immune response mediator, NFAT (Fig. 5C) were identified. A number of
significantly upregulated miRNAs that may alter NFAT expression
were identified, including miR-10a-5p (46). NFAT, present in the cytosol, was
translocated to the nucleus by T cell receptor stimulation and
became a member of the nuclear factors for activating T cells
transcription complex (47). Due
to the reduced NFAT expression, its translocation and regulatory
functions may be severely impaired, leading to reduced levels of
target cytokine expression, including IL-2, IL-4, IL-5, IL-10,
interferon-γ, TNF-α. Their reduced expression may lead to
autoimmune diseases and immune deficiency (48).
It is of note that various altered miRNAs were
identified to affect components of the extracellular matrix (ECM).
These ECM molecules, including fibronectin, bind to integrin
subtypes and affect multiple cellular processes, including
cell-cell and cell-matrix interaction, cell motility and signaling
pathways (49). Actin
polymerization was a prerequisite of highly concentrated CD4 and
CXCR4 membrane receptors, which was determined to be essential for
HIV propagation (50,51). Therefore, actin polymerization and
cytoskeleton remodeling was hijacked by infected cells to
facilitate HIV attack and AIDS progression. Conditions may be
worsened with weakened adherens junctions regulated by some
upregulated miRNAs, such as miR-10b-5p (52).
In conclusion, the present study provided a complete
network of miRNAs with altered expression in HIV-infected
individuals. Some of the alterations were consistent with those
previously observed in literature (22,40).
Target genes of the significantly different miRNAs were predicted
using the TargetScan, mirBase and miRanda databases, and the shared
target genes from the three databases were selected. GO and KEGG
pathway analyses focused on the 391 downregulated target genes and
predicted important pathways including cell-EMC interaction and
chemokine regulation. Alterations in these pathways are closely
associated with CD4+T cell activation and reduction in
chemokine levels. They also lead to gut barrier dysfunction of
patients with HIV infection. miRNAs that potentially influence
these target genes and pathways included hsa-miRNA-32-5p,
hsa-miRNA-195-5p, hsa-miRNA-20b-5p and hsa-miRNA-590-5p. Expression
of some miRNAs and target genes was confirmed with RT-qPCR. The
present study identified the crucial roles of miRNAs in the
regulation of the gut barrier dysfunction via multiple regulatory
molecules and signaling pathways, which elucidated the underlying
molecular mechanism for gut barrier dysfunction of HIV infection.
The specific roles of these miRNAs in disease progression remain to
be further investigated.
Acknowledgements
The present study was financially supported by the
Key Science and Technology planning Project of Yunnan Provincial
Science and Technology Department (grant no. 2016FC005). Foundation
of Medical Leading Talent of Yunnan Province (grant no. L-201205),
the National Natural Science Foundation of China (grant no.
81360069) and Department of Science and Technology of Yunnan
Province-Kunming Medical University (grant no. 2013FB105). The Key
Science and Technology planning Project of Kunming Science and
Technology Bureau, the China Guanghua Foundation and the Major
project of Yunnan Provincial Bureau of Education (grant no.
ZD2015010).
References
|
1
|
Kelleher AD and Zaunders JJ: Decimated or
missing in action: CD4+ T cells as targets and effectors
in the pathogenesis of primary HIV infection. Curr HIV/AIDS Rep.
3:5–12. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vergnon-Miszczycha D, Lucht F, Roblin X,
Pozzetto B, Paul S and Bourlet T: Key role played by the gut
associated lymphoid tissue during human immunodeficiency virus
infection. Med Sci (Paris). 31:1092–1101. 2015.(In French).
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Estes JD, Harris LD, Klatt NR, Tabb B,
Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira
KM, et al: Damaged intestinal epithelial integrity linked to
microbial translocation in pathogenic simian immunodeficiency virus
infections. PLoS Pathog. 6:e10010522010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Andrew AL, Mahesh M and Ronald SV: The
gastrointestinal tract and AIDS pathogenesis. Gastroenterology.
136:1966–1978. 2009. View Article : Google Scholar
|
|
5
|
Chun TW, Nickle DC, Justement JS, Meyers
JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI,
et al: Persistence of HIV in gut-associated lymphoid tissue despite
long-term antiretroviral therapy. J Infect Dis. 197:714–720. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heise C, Miller CJ, Lackner A and Dandekar
S: Primary acute simian immunodeficiency virus infection of
intestinal lymphoid tissue is associated with gastrointestinal
dysfunction. J Infect Dis. 169:1116–1120. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vyboh K, Jenabian MA, Mehraj V and Routy
JP: HIV and the gut microbiota, partners in crime: Breaking the
vicious cycle to unearth new therapeutic targets. J Immunol Res.
2015:6141272015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vujkovic-Cvijin I, Dunham RM, Iwai S,
Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM,
Huang Y, Somsouk M, et al: Dysbiosis of the gut microbiota is
associated with HIV disease progression and tryptophan catabolism.
Sci Transl Med. 5:193ra912013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lozupone CA, Li M, Campbell TB, Flores SC,
Linderman D, Gebert MJ, Knight R, Fontenot AP and Palmer BE:
Alterations in the gut microbiota associated with HIV-1 infection.
Cell Host Microbe. 14:329–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Novati S, Sacchi P, Cima S, Zuccaro V,
Columpsi P, Pagani L, Filice G and Bruno R: General issues on
microbial translocation in HIV-infected patients. Eur Rev Med
Pharmacol Sci. 19:866–878. 2015.PubMed/NCBI
|
|
11
|
Vassallo M, Mercié P, Cottalorda J,
Ticchioni M and Dellamonica P: The role of lipopolysaccharide as a
marker of immune activation in HIV-1 infected patients: A
systematic literature review. Virol J. 9:1742012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Steele AK, Lee EJ, Vestal B, Hecht D, Dong
Z, Rapaport E, Koeppe J, Campbell TB and Wilson CC: Contribution of
intestinal barrier damage, microbial translocation and HIV-1
infection status to an inflammaging signature. PLoS One.
9:e971712014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hunt PW: Role of immune activation in HIV
pathogenesis. Curr HIV/AIDS Rep. 4:42–47. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hunt PW, Sinclair E, Rodriguez B, Shive C,
Clagett B, Funderburg N, Robinson J, Huang Y, Epling L, Martin JN,
et al: Gut epithelial barrier dysfunction and innate immune
activation predict mortality in treated HIV infection. J Infect
Dis. 210:1228–1238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Asmuth DM, Pinchuk IV, Wu J, Vargas G,
Chen X, Mann S, Albanese A, Ma ZM, Saroufeem R, Melcher GP, et al:
Role of intestinal myofibroblasts in HIV-associated intestinal
collagen deposition and immune reconstitution following combination
antiretroviral therapy. AIDS. 29:877–888. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nwosu FC, Avershina E, Wilson R and Rudi
K: Gut microbiota in HIV infection: Implication for disease
progression and management. Gastroenterol Res Pract.
2014:8031852014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rice AP: Roles of microRNAs and
long-noncoding RNAs in human immunodeficiency virus replication.
Wiley Interdiscip Rev RNA. 6:661–670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012.PubMed/NCBI
|
|
19
|
Li LM, Wang D and Zen K: MicroRNAs in
drug-induced liver injury. J Clin Transl Hepatol. 2:162–169.
2014.PubMed/NCBI
|
|
20
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:pp. 13944–13949. 2005; View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiang K, Liu H and Rice AP: miR-132
enhances HIV-1 replication. Virology. 438:1–4. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sung TL and Rice AP: miR-198 inhibits
HIV-1 gene expression and replication in monocytes and its
mechanism of action appears to involve repression of cyclin T1.
PLoS Pathog. 5:e10002632009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang HS, Wu TC, Sang WW and Ruan Z:
MiR-217 is involved in Tat-induced HIV-1 long terminal repeat (LTR)
transactivation by down-regulation of SIRT1. Biochim Biophys Acta.
1823:1017–1023. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Omoto S, Ito M, Tsutsumi Y, Ichikawa Y,
Okuyama H, Brisibe EA, Saksena NK and Fujii YR: HIV-1 nef
suppression by virally encoded microRNA. Retrovirology. 1:442004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kaul CD, Ahlawat A and Gupta SD: HIV-1
genome-encoded hiv1-mir-H1 impairs cellular responses to infection.
Mol Cell Biochem. 323:143–148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM,
Wu JY, Chen HY, Wang ZW, Qiu GQ and Peng ZH: Identification of
recurrence-related microRNAs in hepatocellular carcinoma following
liver transplantation. Mol Oncol. 6:445–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective miRNA target sites in mammalian mRNAs. Elife.
4:e050052015. View Article : Google Scholar :
|
|
29
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X: miRDB: A microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Siliciano RF and Greene WC: HIV latency.
Cold Spring Harb Perspect Med. 1:a0070962011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang J, Wang F, Argyris E, Chen K, Liang
Z, Tian H, Huang W, Squires K, Verlinghieri G and Zhang H: Cellular
microRNAs contribute to HIV-1 latency in resting primary
CD4+ T lymphocytes. Nat Med. 13:1241–1247. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nathans R, Chu CY, Serquina AK, Lu CC, Cao
H and Rana TM: Cellular microRNA and P bodies modulate host-HIV-1
interactions. Mol Cell. 34:696–709. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Triboulet R, Mari B, Lin YL, Chable-Bessia
C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P,
Baillat V, et al: Suppression of microRNA-silencing pathway by
HIV-1 during virus replication. Science. 315:1579–1582. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chiang K, Sung TL and Rice AP: Regulation
of cyclin T1 and HIV-1 Replication by microRNAs in resting
CD4+ T lymphocytes. J Virol. 86:3244–3252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ding S, Liang Y, Zhao M, Liang G, Long H,
Zhao S, Wang Y, Yin H, Zhang P, Zhang Q and Lu Q: Decreased
microRNA-142-3p/5p expression causes CD4+ T cell
activation and B cell hyperstimulation in systemic lupus
erythematosus. Arthritis Rheum. 64:2953–2963. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stahl HF, Fauti T, Ullrich N, Bopp T,
Kubach J, Rust W, Labhart P, Alexiadis V, Becker C, Hafner M, et
al: miR-155 inhibition sensitizes CD4+ Th cells for TREG
mediated suppression. PLoS One. 4:e71582009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fenoglio C, Cantoni C, De Riz M, Ridolfi
E, Cortini F, Serpente M, Villa C, Comi C, Monaco F, Mellesi L, et
al: Expression and genetic analysis of miRNAs involved in
CD4+ cell activation in patients with multiple
sclerosis. Neurosci Lett. 504:9–12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Palin AC, Ramachandran V, Acharya S and
Lewis DB: Human neonatal naive CD4+ T cells have
enhanced activation-dependent signaling regulated by the microRNA
miR-181a. J Immunol. 190:2682–2691. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pan Z, Radding W, Zhou T, Hunter E, Mountz
J and McDonald JM: Role of calmodulin in HIV-potentiated
Fas-mediated apoptosis. Am J Pathol. 149:903–910. 1996.PubMed/NCBI
|
|
42
|
Hall JA, Cannons JL, Grainger JR, Dos
Santos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G,
Robinson M, et al: Essential role for retinoic acid in the
promotion of CD4(+) T cell effector responses via retinoic acid
receptor alpha. Immunity. 34:435–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Safi R, Muramoto GG, Salter AB, Meadows S,
Himburg H, Russell L, Daher P, Doan P, Leibowitz MD, Chao NJ, et
al: Pharmacological manipulation of the RAR/RXR signaling pathway
maintains the repopulating capacity of hematopoietic stem cells in
culture. Mol Endocrinol. 23:188–201. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Barbaro G, Di Lorenzo G, Grisorio B and
Barbarini G: Incidence of dilated cardiomyopathy and detection of
HIV in myocardial cells of HIV-positive patients. Gruppo italiano
per lo studio cardiologico dei pazienti affetti da AIDS. N Engl J
Med. 339:1093–1099. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lewis W: Cardiomyopathy in AIDS: A
pathophysiological perspective. Prog Cardiovasc Dis. 43:151–170.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou X, Wang H, Burg MB and Ferraris JD:
Inhibitory phosphorylation of GSK-3β by AKT, PKA, and PI3K
contributes to high NaCl-induced activation of the transcription
factor NFAT5 (TonEBP/OREBP). Am J Physiol Renal Physiol.
304:F908–F917. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Macián F, López-Rodríguez C and Rao A:
Partners in transcription: NFAT and AP-1. Oncogene. 20:2476–2489.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Brocker C, Thompson D, Matsumoto A, Nebert
DW and Vasiliou V: Evolutionary divergence and functions of the
human interleukin (IL) gene family. Hum Genomics. 5:30–55. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Leyme A, Marivin A, Perez-Gutierrez L,
Nguyen LT and Garcia-Marcos M: Integrins activate trimeric G
proteins via the nonreceptor protein GIV/Girdin. J Cell Biol.
210:1165–1184. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu Y, Belkina NV and Shaw S: HIV
infection of T cells: Actin-in and actin-out. Sci Signal.
2:pe232009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Blanco J, Bosch B, Fernández-Figueras MT,
Barretina J, Clotet B and Esté JA: High level of
coreceptor-independent HIV transfer induced by contacts between
primary CD4 T cells. J Biol Chem. 279:51305–51314. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sufiawati I and Tugizov SM: HIV-associated
disruption of tight and adherens junctions of oral epithelial cells
facilitates HSV-1 infection and spread. PLoS One. 9:e888032014.
View Article : Google Scholar : PubMed/NCBI
|