Introduction
Salivary adenoid cystic carcinoma (SACC) is a common
malignant tumor and is characterized by distinctive clinical
features and behaviors, including neural and blood invasiveness,
aggressive growth, distant metastasis and poor long-term survival
(1). SACC occurs in the major and
minor salivary glands and spreads to the oral and oropharyngeal
mucosa, tracheobronchial tree and the esophagus (2). Previous studies have indicated that
40–60% of patients with SACC develop distant metastases in the soft
tissues, lungs and bone (1,3).
Typically, distant metastasis is associated with poor patient
survival (3). It could be
beneficial for patients with SACC to be able to identify potential
molecular targets for the early diagnosis, therapy and prognostic
analysis (4–9).
Inhibitor of DNA-binding (ID) proteins, including
ID1, ID2, ID3 and ID4, belong to the helix-loop-helix (HLH) protein
family (10). ID proteins inhibit
DNA binding and the transcriptional activity of basic HLH proteins
(10). Previous studies suggest
that ID1 is overexpressed in various types of cancer, including
melanoma, breast and gastric cancer, endometrial carcinoma,
osteosarcoma, oral squamous cell carcinoma and lung cancer
(11–18). In addition, a correlation between
ID1 protein and tumor angiogenesis in SACC has been determined
(19). ID1 serves a key role in
cell growth, senescence and differentiation. Furthermore, ID1 may
mediate tumor progression and metastasis by promoting tumor
angiogenesis in SACC (20).
In the present study, the role of ID1 in SACC was
investigated. The expression of ID1 in clinical SACC samples and
normal salivary tissues was determined using immunohistochemistry.
In addition, the correlation between ID1 expression and clinical
pathological characteristics, including age, gender, tumor stage,
tumor invasion and metastasis was examined. ID1 expression was
silenced using small interfering RNA (siRNA), or overexpressed
using plasmids to investigate its effects on the growth, invasion
and migration of SACC cells in vitro. To explore the
potential mechanisms of ID1 in SACC, the expression of known ID1
target genes, including S100A9, CDKN2A and matrix
metalloproteinase-1 (MMP1), was analyzed using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis following transfection of SACC cells with ID1 siRNA.
Materials and methods
Cell culture and clinical samples
The SACC-83 cell line was provided by the Peking
University School of Stomatology (Beijing, China). The cells were
maintained in RPMI 1640 (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) and incubated in humidified atmosphere
containing 5% CO2 at 37°C. Surgically resected salivary
tissue samples were obtained from the First Affiliated Hospital of
the Fujian Medical University (Fuzhou, China) and the Fujian
Medical University Union Hospital (Fuzhou, China) between May 2004
and September 2014 and all the samples were reviewed and diagnosed
by two independent pathologists. A total of 50 normal
tumor-adjacent salivary tissues (24 females and 26 males, age range
23–75, mean age 48.57±14.22) and 68 SACC samples (40 females and 28
males, age range 18–79, mean age 50.13±14.92) were included. The
Institutional Review Board of Fujian Medical University (Fuzhou,
China) approved the present study, and written informed consent was
obtained from each participant.
Immunohistochemistry
Salivary tissues were fixed in 10% neutral buffered
formalin at 4°C for 24 h, embedded in paraffin and sections (5-µm
in thickness) were mounted on slides coated with poly-L-lysine.
Following deparaffinization in xylene at 37°C, the sections were
rehydrated in a decreasing ethanol series, washed for 10 min in
phosphate-buffered saline (PBS; pH 7.2) and incubated in methanol
containing 3% H2O2 for 10 min at room
temperature. Following several washes in PBS, the sections were
blocked with a universal 5% blocking reagent (Fuzhou Maixin
Biotech. Co., Ltd., Fuzhou, China) for 10 min at room temperature,
and subsequently incubated with a primary antibody against ID1
(ab66495; 1:30; Abcam, Cambridge, MA, USA) for 1 h at room
temperature. Following several washes with PBS, the sections were
incubated with a biotin-conjugated secondary antibody
(ready-to-use; Rabbit Anti-Mouse lgG H&L EliVision™plus kit;
KIT-9902-B; Fuzhou Maixin Biotech. Co., Ltd.) for 10 min at room
temperature. The sections were then washed several times with PBS
and incubated with streptavidin-peroxidase (ready-to-use, ABD-0030;
Fuzhou Maixin Biotech. Co., Ltd.) for 10 min at room temperature.
The sections were subsequently rinsed with PBS and the antibody
complexes were visualized following incubation with
diaminobenzidine chromogen (DAB chromogenic kit; DAB-0031; Fuzhou
Maixin Biotech. Co., Ltd.) for 2 min at 37°C. Sections were
counterstained with hematoxylin (Dako; Agilent Technologies, Inc.,
Santa Clara, CA, USA) for 2 min at room temperature, dehydrated and
examined by light microscopy. Two pathologists reviewed all slides
independently and in a blinded fashion. The staining results were
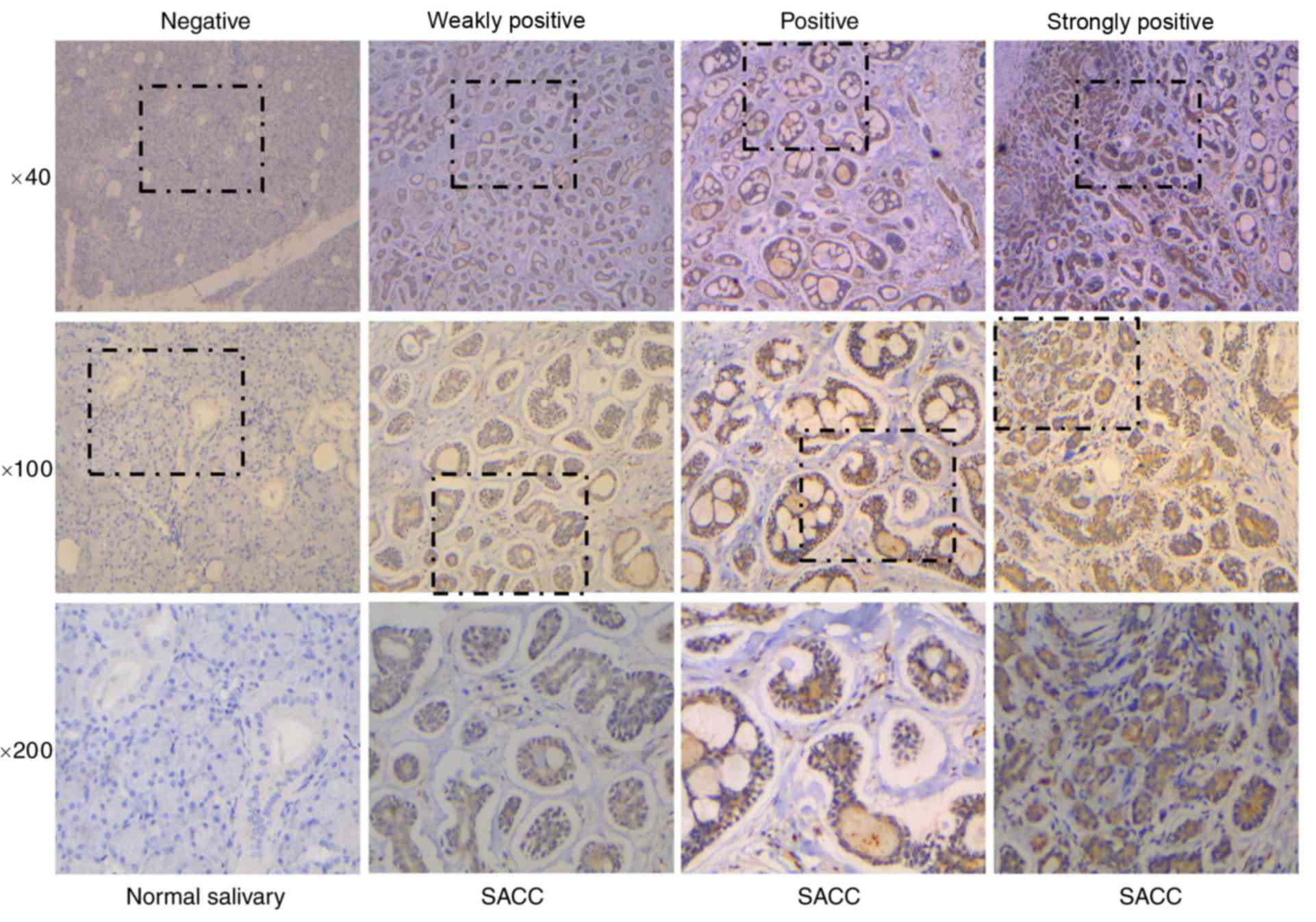
quantified using the following 4-tier scale: Negative, no staining;
1+, weakly-positive staining; 2+, positive staining; and 3+,
strongly-positive staining. Cell staining was also assessed using
the following four-tier scale: Negative, no staining in the cells;
1+, staining was present in <30% of cells; 2+, indicated
staining in 30–50% of cells; and 3+, staining was present in
>50% of cells. The immunohistochemical results were graded with
4 different scores (negative, weakly-positive, positive and
strongly-positive).
RNA interference (RNAi) and plasmid
transfection
Negative control (NC) siRNA and two siRNAs against
ID1 (siRNA800 and siRNA858) were synthesized by Shanghai GenePharma
Co., Ltd. (Shanghai, China). The siRNA sequences are listed in
Table I. At 24 h prior to
transfection, SACC-83 cells in the exponential phase of growth were
trypsinized by Trypsin-EDTA solution (Gibco; Thermo Fisher
Scientific, Inc.), counted and plated in 6-well plates at a density
of 3×105 cells/well. Cells were transfected with 50 nM
siRNAs using Lipofectamine RNAiMAX reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), or the ID1 recombinant plasmid pEX-2 (2
µg/well; Shanghai GenePharma, Co., Ltd.) using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. SACC-83 cells were transfected with NC
siRNA or empty vector as a negative control for siRNA and plasmid
transfection, respectively.
 | Table I.Sequences of siRNAs employed in the
present study. |
Table I.
Sequences of siRNAs employed in the
present study.
| siRNA | Sense (5′-3′) | Antisense
(5′-3′) |
|---|
| ID1-800 |
GGGAUUCCACUCGUGUGUUTT |
AACACACGAGUGGAAUCCCTT |
| ID1-858 |
GUUUGGUGCUUCUCAGAUUTT |
AAUCUGAGAAGCACCAAACTT |
| Negative control |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
RT-qPCR
At 48 h following transfection with siRNA, total RNA
was extracted from SACC-83 cells using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and 2.5 µg total RNA were reverse
transcribed into cDNA using the PrimeScript RT reagent kit (Takara
Bio, Inc., Otsu, Japan, Japan) according to the manufacturer's
protocols. The cDNA was used as a template to detect gene
expression levels of ID1 and known ID1 target genes, S100A9, CDKN2A
and MMP1, by RT-qPCR analysis using SYBR Premix Ex Taq™ (Takara
Bio, Inc.) The 20 µl PCR mixture contained 10 µl SYBR Premix Ex
Taq™ (2X), 0.4 µl PCR Forward Primer (10 µM), 0.4 µl PCR Reverse
Primer (10 µM), 0.4 µl ROX Reference Dye I (50X), 2 µl cDNA
template and 6.8 µl ddH2O. GAPDH was used as an internal
control. The primers used in the present study are listed in
Table II. The reaction was as
follows: Denaturation at 95°C for 2 min, followed by 40 cycles at
95°C for 15 sec, 60°C for 30 sec. The fluorescent signal was
measured at the end of the annealing phase of every cycle. Target
gene expression was quantified using the 2−ΔΔCq method
(21).
 | Table II.Reverse transcription-quantitative
polymerase chain reaction primers. |
Table II.
Reverse transcription-quantitative
polymerase chain reaction primers.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| ID1 |
ATTACGTGCTCTGTGGGTCTCC |
TAGTAGGTGTGCAGAGAGGAGC |
| S100A9 |
GCTGGAACGCAACATAGAGACC |
TCTGCATTTGTGTCCAGGTCCT |
| CDKN2A |
GGCACCAGAGGCAGTAACCAT |
GAAAGCGGGGTGGGTTGTG |
| MMP1 |
GGGGAGATCATCGGGACAACTC |
AGAATGGCCGAGTTCATGAGCT |
| GAPDH |
TGCACCACCAACTGCTTAGC |
AGCTCAGGGATGACCTTGCC |
Western blotting
At 48 h following transfection with siRNA and
recombinant ID1 plasmids, total proteins were extracted from
SACC-83 cells with IP lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China). The protein concentration was
determined by BCA method and 25 µg of protein was loaded per lane
for SDS-PAGE separation. The proteins were separated by 8% SDS-PAGE
and transferred onto polyvinylidene difluoride membranes (GE
Healthcare; Chicago, IL, USA). Membranes were blocked in 5% bovine
serum albumin (VWR International, Radnor, PA, USA) at 4°C for 60
min. Subsequently, the membranes were immunoblotted overnight at
4°C with primary antibodies against ID1 (ab66495; 1:1,000; Abcam)
or GAPDH (M20006; 1:2,000; Abmart, Inc., Berkeley Heights, NJ,
USA). Membranes were then washed three times in Tris-buffered
saline with 0.1% Tween 20 and subsequently incubated with a
secondary antibody (Goat Anti-Mouse IgG H&L (HRP); ab205719;
1:2,000; Abcam) at room temperature for 60 min. Protein bands were
visualized using CDP-Star reagent (Roche Diagnostics, Indianapolis,
IN, USA). The signals were detected by exposure to X-Ray film for
different times (1–10 min).
Cell proliferation assay
The number of viable SACC-83 cells in the
logarithmic phase of growth following siRNA or plasmid transfection
was measured at 24 h intervals using the Cell Counting Kit-8 assay
(CCK-8; CK04; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) according to the manufacturer's protocols. SACC-83 cells
were transfected with siRNAs or plasmids and 2×103
cells/well were seeded in a 96-well plate. CCK8 reagent (10
µl/well) was added to the cells at the same time over 5 consecutive
days and incubated at 37°C for 1 h and the absorbance of each well
was measured at 450 nm using a microplate reader (Pharmacia
Biotech, Inc.; GE Healthcare). This procedure was performed in
triplicate for statistical analysis and to construct the
corresponding proliferation curves.
Colony formation assay
At 24 h following siRNA or plasmid transfection,
cells were plated in 6-well plates at a density of 500 cells/well,
and cultured for 2 weeks. Colonies were fixed with cold 100%
methanol for 10 min and stained with 1% crystal violet for 30 min
at room temperature and the number of colonies in each well was
counted. The experiments were repeated three times.
Wound healing assay
Then, 24 h after seeded in a 6-well plate at a
density of 7×105 cells/well, SACC-83 cells were
transfected with siRNAs or plasmids. The cells formed a monolayer
covering the bottom of the plate, and a 20-µl pipette tip was used
to generate a scratch-wound. The medium was replaced with RPMI-1640
supplemented with 0.1% FBS at 0 and 24 h following generation of
the scratch-wound. The cells were visualized under a light
microscope at both time points, and images were captured. The width
of scratch-wound was measured by the ruler to evaluate the cell
invasion.
In vitro cell invasion assay
Cell invasion was determined using 24-well
Matrigel-coated Transwell chambers (8-µm pore size; 354480; BD
Bioscience, Bedford, MA, USA). 24 h after transfection with siRNA
or plasmids, SACC-83 cells were serum-starved for 24 h and then
harvested and resuspended in RPMI 1640 containing 1% FBS. Cells
were subsequently plated in the upper chamber of the Transwell
plate at a density of 1×105 cells, and 500 µl RPMI 1640
containing 10% FBS was added to the lower chamber. Following
incubation at 37°C for 48 h, the Matrigel and cells in the upper
chamber were removed using a cotton swab and the chamber was
stained with 1% crystal violet for 10 min at room temperature. The
cells were counted under a light microscope in at least five random
fields of view (magnification, ×200) and the images were
captured.
In vitro cell migration assay
Cell migration assays were performed using 24-well
Transwell chambers (8 µm pore size; 353097; BD Biosciences,
Bedford, MA, USA). The same procedure for the cell invasion assay
was followed with the exception that the plate was not
Matrigel-coated.
Statistical analysis
The results were analyzed using the SPSS 22.0
statistics software package (IBM Corp., Armonk, NY, USA). Data are
presented as the mean ± standard deviation. Rank-sum tests were
used to compare the two groups of immunohistochemistry data, the
Mann-Whitney U test was used to correlate ID1 expression with
clinicopathological features and multi-sample mean one-way analysis
of variance tests with Dunnett's test were used for group
comparisons. P<0.05 was considered to indicate a statistically
significant difference.
Results
ID1 is upregulated in SACC
The salivary tissue samples consisted of 68 SACC and
50 normal tumor-adjacent tissue samples. As shown in Fig. 1 and Table III, tissue samples were divided
into four groups depending on ID1 staining intensity. The results
demonstrated that ID1 protein expression was upregulated in SACC
tissue samples when compared with normal tissues (P<0.001;
Table III). Subsequently, the
association of ID1 protein expression levels with clinical features
of SACC was determined. The results shown in Table IV demonstrate that the expression
of ID1 was significantly increased in the late-stage tumors when
compared with early-stage tumors (P=0.001), and ID1 expression
levels were significantly correlated with tumor invasion (P=0.002)
and tumor metastasis (P=0.019) in patients with SACC. These results
indicate that ID1 may serve an important role in SACC.
 | Table III.Expression of ID1 in normal salivary
and SACC samples. |
Table III.
Expression of ID1 in normal salivary
and SACC samples.
| Sample | Case | Negative | Weakly
positive | Positive | Strongly
positive | P-value |
|---|
| Normal
salivary | 50 | 46 | 1 | 1 | 2 | <0.001 |
| SACC | 68 | 24 | 16 | 23 | 5 |
|
 | Table IV.Association between the inhibitor of
DNA binding 1 expression and the clinical and pathological
characteristics of patients with salivary adenoid cystic
carcinoma. |
Table IV.
Association between the inhibitor of
DNA binding 1 expression and the clinical and pathological
characteristics of patients with salivary adenoid cystic
carcinoma.
|
| Staining
intensity |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Negative | Weakly
positive | Positive | Strongly
positive | Total | P-value |
|---|
| Gender |
|
|
|
|
|
|
|
Female | 13 | 10 | 14 | 3 | 40 | 0.661 |
|
Male | 11 | 6 | 9 | 2 | 28 |
|
| Age (years) |
|
|
|
|
|
|
|
≤55 | 15 | 9 | 13 | 4 | 41 | 0.942 |
|
>55 | 9 | 7 | 10 | 1 | 27 |
|
| Stage |
|
|
|
|
|
|
|
Early | 18 | 5 | 7 | 1 | 31 | 0.001 |
|
Late | 6 | 11 | 16 | 4 | 37 |
|
| Invasion |
|
|
|
|
|
|
| No | 15 | 7 | 6 | 0 | 28 | 0.002 |
|
Yes | 9 | 9 | 17 | 5 | 40 |
|
| Metastasis (lymph
node and distant) |
|
|
|
|
|
|
| No | 20 | 14 | 14 | 2 | 50 | 0.019 |
|
Yes | 4 | 2 | 9 | 3 | 18 |
|
ID1 promotes SACC cell proliferation
in vitro
To investigate the role of ID1 in the proliferation
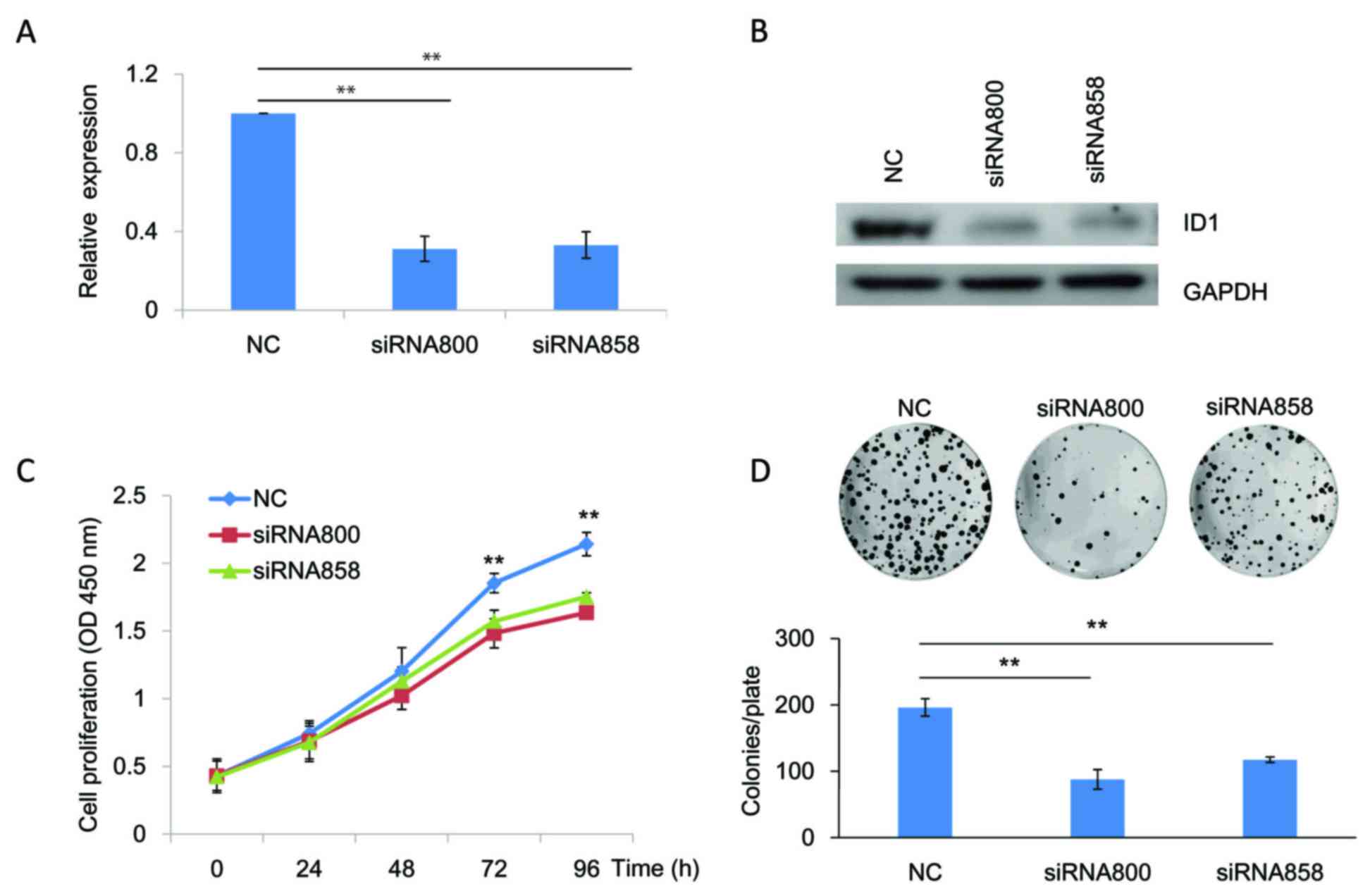
of SACC cells, siRNAs targeting ID1 (siRNA800 and siRNA858) were
transfected into SACC-83 cells to knockdown ID1 expression. As
indicated by RT-qPCR (Fig. 2A;
P<0.01) and western blotting (Fig.
2B) analyses, both siRNAs targeting ID1 efficiently reduced ID1
expression levels in SACC-83 cells when compared with the NC. The
results of the CCK-8 assay demonstrated that the growth of SACC-83
cells at 72 and 96 h following knockdown of ID1 expression was
significantly reduced when compared with the NC (P<0.01;
Fig. 2C). These results were
consistent with the colony formation assay, which demonstrated that
knockdown of ID1 was associated with a significant reduction in the
number of colonies (P<0.01; Fig.
2D). To further verify these results, SACC-83 cells were
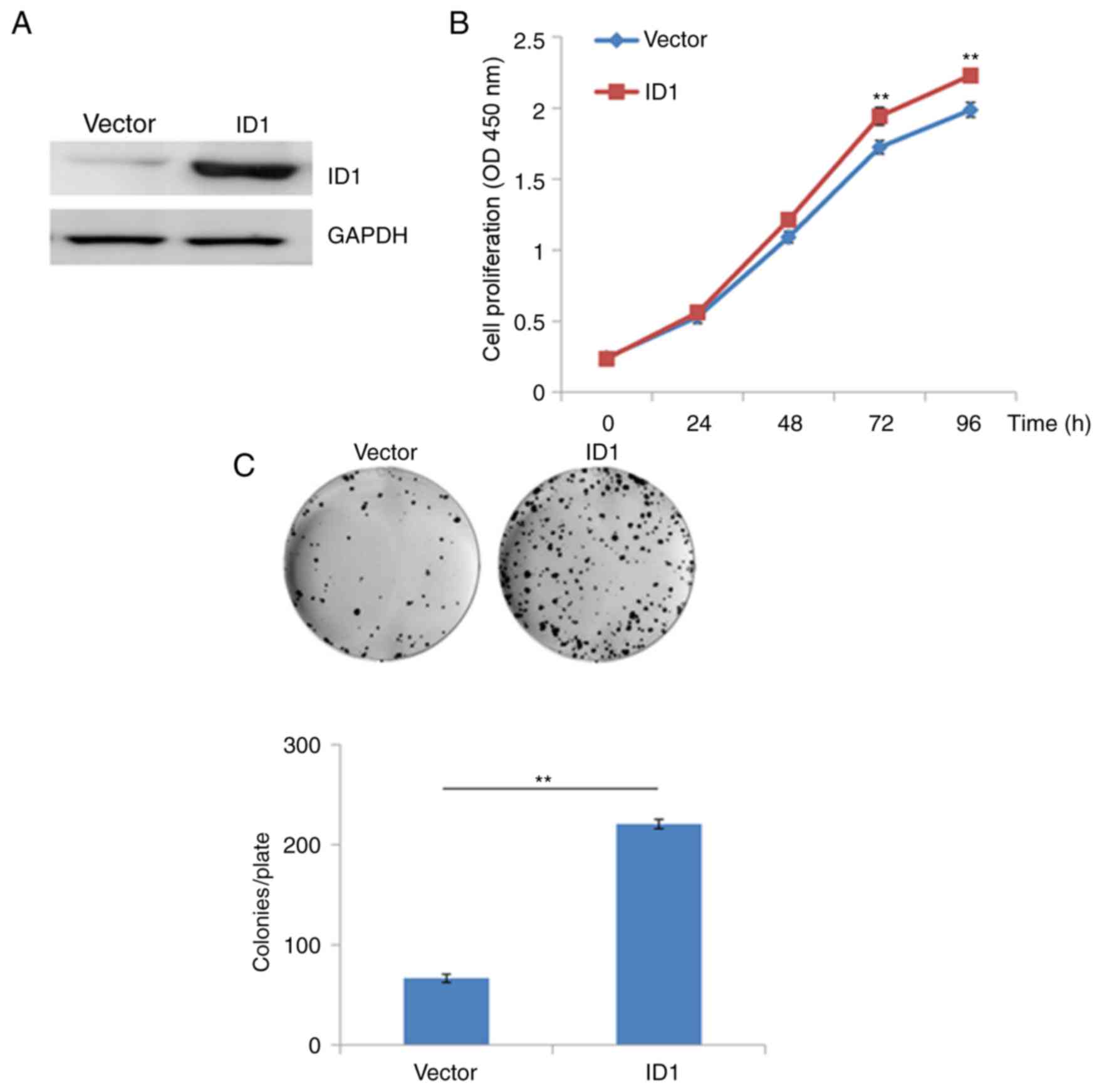
transfected with a recombinant plasmid (pEX-2) containing the
coding sequence of ID1. As demonstrated in Fig. 3A, transfection recombinant
expression plasmids containing the ID1 sequence was associated with
a marked increase in the protein expression levels of ID1 compared
with empty vector controls (Fig.
3A). Overexpression of ID1 in SACC-83 cells significantly
promoted cell growth at 48, 72 and 96 h following transfection when
compared with vector-only transfected cells (P<0.01; Fig. 3B). Similarly, the results of the
colony formation assay demonstrated that overexpression of ID-1
significantly increased the growth of SACC-83 cells when compared
with vector-only transfected cells (P<0.01; Fig. 3C). These results suggest that
overexpression of ID1 may promote the proliferation of SACC cells
in vitro, and ID1 may therefore exert an oncogenic role in
SACC.
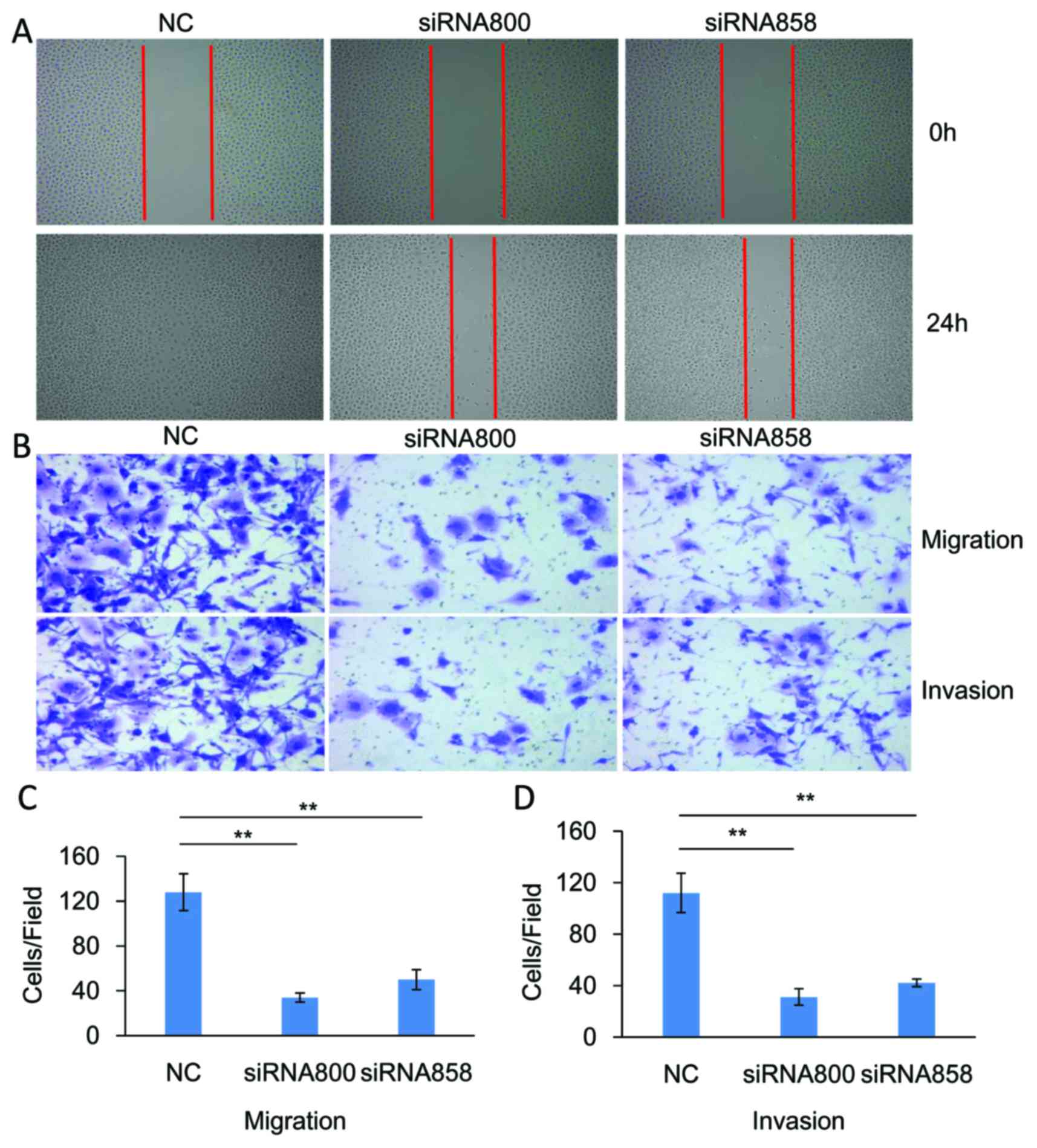
ID1 promotes cell migration and
invasion in vitro
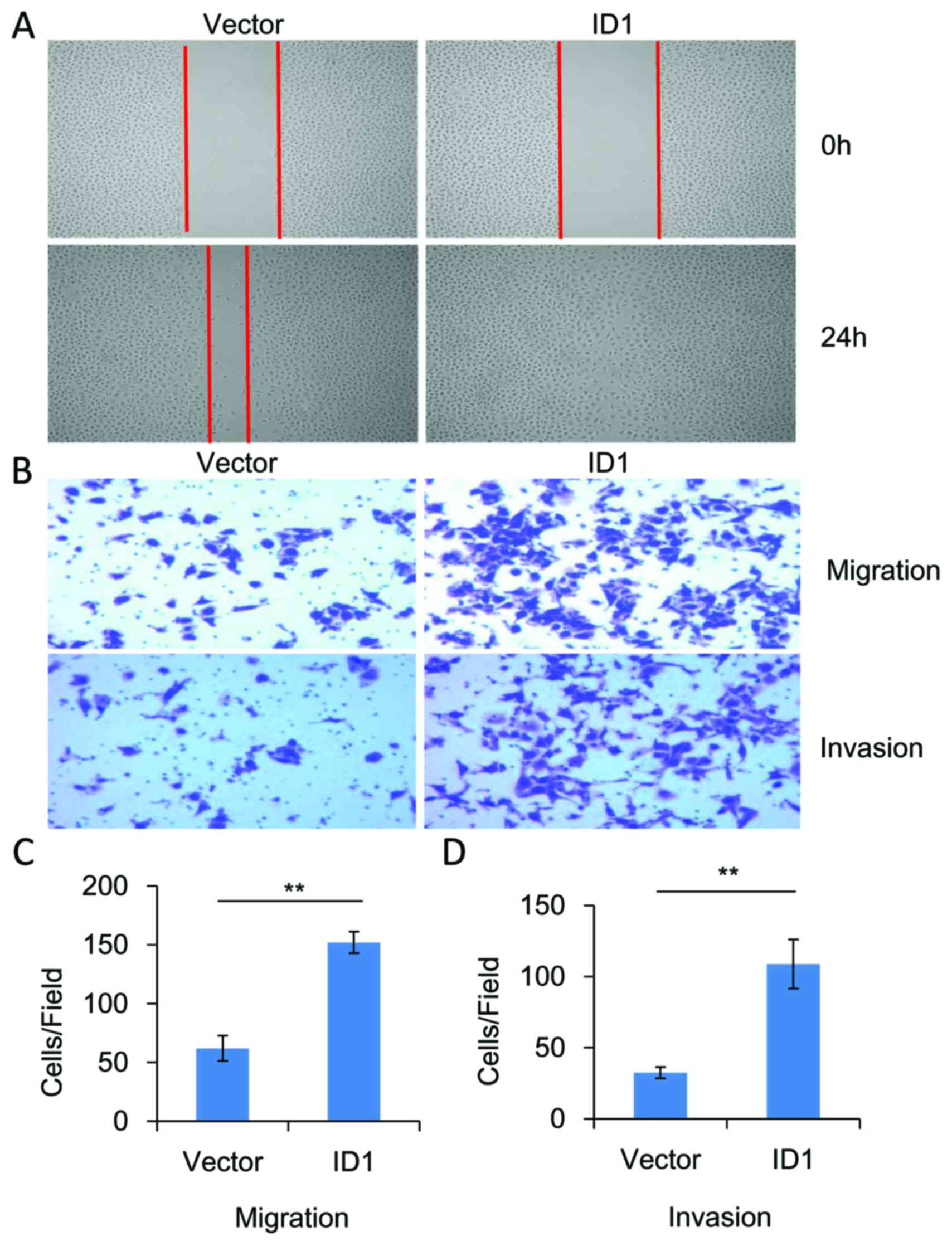
The functional role of ID1 in the migration and
invasion of SACC cells was investigated. As demonstrated in
Fig. 4, knockdown of significantly
inhibited the migration (P<0.01, n=3) and invasion (P<0.01,
n=3) of SACC-83 cells when compared with controls. By contrast,
overexpression of ID1 in SACC-83 cells significantly promoted cell
motility when compared with controls, as indicated by the wound
healing (Fig. 5A) and Transwell
assays (P<0.01, n=3; Fig. 5B and
C). As demonstrated in Fig. 5B and
D, ID1 overexpression was associated with a significant
increase in cell invasiveness (P<0.01, n=3). These results
provide further evidence to suggest that ID1 may function as an
oncogene in SACC, and contribute to the migration and invasion of
SACC cells.
Knockdown of ID1 reduces the
expression of MMP1 and promotes the expression of S100A9 and
CDKN2A
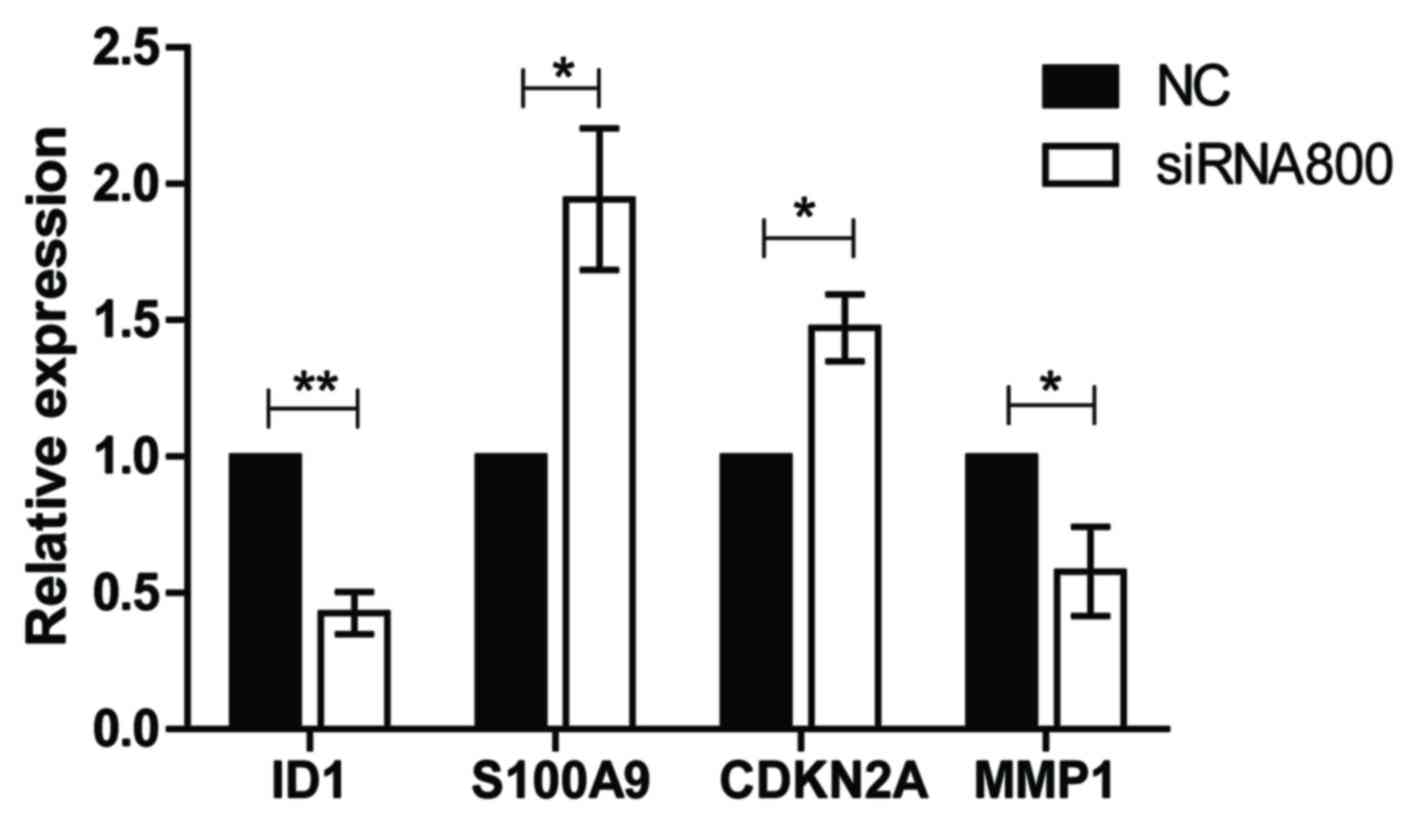
To explore the potential mechanisms of ID1 in SACC
further, the gene expression levels of known ID1 target genes
(S100A9, CDKN2A and MMP1) were determined following the
transfection of SACC-83 cells with ID1 siRNA800 or NC controls
using RT-qPCR. Compared with NC, the expression level of MMP1 was
significantly downregulated following knockdown of ID1 (P<0.05),
whereas the S100A9 and CDKN2A gene expression levels were
significantly upregulated in SACC-83 cells (both P<0.05;
Fig. 6). These results indicate
that MMP1, S100A9 and CDKN2A may be involved in ID1-mediated
alterations in SACC cell proliferation, migration and invasion.
Discussion
ID1 serves an important role in the regulation of a
number of biological behaviors in various tumors. Silencing of ID1
was demonstrated to induce apoptosis and inhibit the growth of
osteosarcoma cells, and the level of phosphorylated AKT was
downregulated by ID1 RNAi (17).
Previous studies have indicated that ID1 serves a role in promoting
cell survival and proliferation and it is involved in tumor
differentiation (21–25). In addition, ID1 contributes to cell
invasion in thyroid tumor cells by inducing mesenchymal features
(26). Pillai et al
(27) revealed that ID1
facilitates the growth and metastasis of non-small cell lung
cancer.
In a previous study, the expression of ID1 in SACC
was increased by 65.2% compared with normal salivary tissues
(20). In the present study, the
expression of ID1 in 68 cases of SACC and 50 tumor-adjacent normal
tissue samples was compared using immunohistochemical staining. The
results suggested that the expression of ID1 was significantly
higher in SACC tissues compared with that in the normal salivary
tissues. In addition, the results demonstrated a positive
correlation between ID1 expression and tumor stage, invasion and
metastasis in patients with SACC. This suggests that ID1 may serve
an oncogenic role in SACC.
In the present study, knockdown and overexpression
of ID1 in SACC cells by siRNA or plasmid transfection,
respectively, was performed to investigate the effect of ID1 on
cell growth using CCK-8 and colony formation assays. The results
demonstrated that ID1 significantly promoted the proliferation of
SACC cells in vitro, thereby supporting the role of ID1 as
an oncogene in SACC. Metastasis and invasion are two important
additional factors that affect the prognosis and recurrence of
patients with SACC. In the current study, knockdown of ID1
significantly inhibited the migration and invasion of SACC-83
cells, whereas overexpression of ID1 significantly promoted the
migration and invasion of the SACC-83 cells. This suggests that ID1
may regulate the migration and invasion of SACC cells; however, the
molecular mechanism of ID1 in SACC remains to be fully elucidated.
In breast cancer, ID1 promotes tumor metastasis by regulating
S100A9 (28). The results of the
current study are consistent with those of previous studies
(28–30) indicating that MMP1, S100A9 and
CDKN2A are ID1 target genes. Knockdown of ID1 significantly
inhibited the expression level of MMP1 mRNA, and significantly
promoted the expression of S100A9 and CDKN2A mRNA. These results
demonstrated that the expression of these genes was altered in
response to ID1 silencing, indicating that MMP1, S100A9 and CDKN2A
may serve a role in mediating the effects of ID1 in SACC cells.
However, additional studies are required to investigate the
potential mechanisms by which ID1 functions to regulate the
proliferation and metastasis of SACC cells. In prostate cancer,
overexpression of ID1 promotes angiogenesis through the activation
of vascular endothelial growth factor (31). Li et al (32) suggested that the extracellular
signal-related-dependent downregulation of S-phase
kinase-associated protein 2 reduced myc activity with hepatocyte
growth factor, which lead to the inhibition of hepatocyte
proliferation via a decrease in ID1 expression. In addition, Cheng
et al (33) revealed that
ID1 promotes the proliferation of lung cancer cells and lung tumor
growth via the Akt signaling pathway. Similarly, Yang et al
(34) demonstrated that
downregulation of ID1 in gastric cancer inhibits cell growth via
the Akt signaling pathway. In the present study, ID1 expression was
associated with the proliferation, invasion and migration of SACC
cells. The observed inhibition of SACC cell growth, invasion and
migration following knockdown of ID1 expression in the present
study, may have been due to restoration of the balance between
oncogenic and tumor-suppressive effects resulting from changes in
the expression of downstream genes or associated proteins. Further
studies are required to determine the molecular mechanisms of ID1
in SACC. The results of the current study suggest that ID1 may
present a novel therapeutic target for the treatment of patients
with SACC.
Acknowledgements
The present study was supported by the National
Natural Sciences Foundation of China (grant no. 81172583), the
Natural Sciences Foundation of Fujian (grant no. 2011J01167), and
the Key Project of Science and Technology Foundation of Fujian
Province of China (grant no. 2011Y0025).
References
|
1
|
Rapidis AD, Givalos N, Gakiopoulou H,
Faratzis G, Stavrianos SD, Vilos GA, Douzinas EE and Patsouris E:
Adenoid cystic carcinoma of the head and neck. Clinicopathological
analysis of 23 patients and review of the literature. Oral Oncol.
41:328–335. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Su BH, Qu J, Song M, Huang XY, Hu XM, Xie
J, Zhao Y, Ding LC, She L, Chen J, et al: NOTCH1 signaling
contributes to cell growth, anti-apoptosis and metastasis in
salivary adenoid cystic carcinoma. Oncotarget. 5:6885–6895. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ding LC, Huang XY, Zheng FF, Xie J, She L,
Feng Y, Su BH, Zheng DL and Lu YG: FZD2 inhibits the cell growth
and migration of salivary adenoid cystic carcinomas. Oncol Rep.
35:1006–1012. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang CY, Zhao YX, Xia RH, Han J, Wang BS,
Tian Z, Wang LZ, Hu YH and Li J: RASSF1A promoter hypermethylation
is a strong biomarker of poor survival in patients with salivary
adenoid cystic carcinoma in a Chinese population. PLoS One.
9:e1101592014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Zhang Y, Ren W and Rao G: erbB2
gene silencing and its effect on PTEN in SACC-83salivary adenoid
cystic carcinoma cells. Oncol Rep. 24:1291–1296. 2010.PubMed/NCBI
|
|
6
|
Shimoda M, Sugiura T, Imajyo I, Ishii K,
Chigita S, Seki K, Kobayashi Y and Shirasuna K: The T-box
transcription factor Brachyury regulates epithelial-mesenchymal
transition in association with cancer stem-like cells in adenoid
cystic carcinoma cells. BMC Cancer. 12:3772012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J, Shao C, Tan ML, Mu D, Ferris RL and
Ha PK: Molecular biology of adenoid cystic carcinoma. Head Neck.
34:1665–1677. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Laurie SA, Ho AL, Fury MG, Sherman E and
Pfister DG: Systemic therapy in the management of metastatic or
locally recurrent adenoid cystic carcinoma of the salivary glands:
A systematic review. Lancet Oncol. 12:815–824. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bell D, Bell A, Roberts D, Weber RS and
El-Naggar AK: Developmental transcripti-on factor EN1-a novel
biomarker in human salivary gland adenoid cystic carcinoma. Cancer.
118:1282–1292. 2012. View Article : Google Scholar
|
|
10
|
O'Toole PJ, Inoue T, Emerson L, Morrison
IE, Mackie AR, Cherry RJ and Norton JD: Id proteins negatively
regulate basic helix-loop-helix transcription factor function by
disrupting subnuclear compartmentalization. J Biol Chem.
278:45770–45776. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Healey MA, Deaton SL, Alder JK,
Winnepenninckx V, Casero RA Jr and Herman JG: Id1 overexpression is
independent of repression and epigenetic silencing of tumor
suppressor genes in melanoma. Epigenetics. 5:410–421. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yap WN, Zaiden N, Tan YL, Ngoh CP, Zhang
XW, Wong YC, Ling MT and Yap YL: Id1, inhibitor of differentiation,
is a key protein mediating anti-tumor responses of
gamma-tocotrienol in breast cancer cells. Cancer Lett. 291:187–199.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong Z, Liu S, Zhou C, Sumida T, Hamakawa
H, Chen Z, Liu P and Wei F: Overexpression of Id-1 is associated
with tumor angiogenesis and poor clinical outcome in oral squamous
cell carcinoma. Oral Oncol. 46:154–157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu B, Song C and Wang P: Expression and
clinical significance of ID1 in gastric cancer. J Shanghai Jiao
tong Med Univ(Med Sci). 33:298–302. 2013.
|
|
15
|
Sun L, Li X and Liu G: Expression of Id1
and Id3 in endometrial carcinoma and their roles in regulating
biological behaviors of endometrial carcinoma cells in vitro. J
South Med Univ. 33:812–818. 2013.
|
|
16
|
Chen YY, Zhang S and Jiang ZQ: Expression
and clinical significance of Id-1, −3 in cervical intraepithelial
neoplasia and cervical sqamous cell cancer. Chin J Obstet Gynecol
Pediatr. 10:177–180. 2014.
|
|
17
|
Chen Y, Yang G and Yang ZH: Effect of DNA
binding protein inhibitor Id1 on the proliferation of osteosarcoma
cells. J Trop Med. 14:71–74. 2014.
|
|
18
|
Rothschild SI, Kappeler A, Ratschiller D,
Betticher DC, Tschan MP, Gugger M and Gautschi O: The stem cell
gene ‘inhibitor of differentiation 1’ (ID1) is frequently expressed
in non-small cell lung cancer. Lung Cancer. 71:306–311. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Liu Y, Zhou J, Dong SZ and Jin Y:
Expression of Id1 and its correlation with tumor angiogenesis in
salivary adenoid cystic carcinoma. J Clin Stomatol. 27:343–345.
2011.
|
|
20
|
Xei W, Li X, Ren G and Guo W: Expression
and importance of inhibitor of DNA binding helix-loop-helix protein
in salivary adenoid cystic carcinoma. Br J Oral Maxillofac Surg.
48:434–437. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mellick AS, Plummer PN, Nolan DJ, Gao D,
Bambino K, Hahn M, Catena R, Turner V, McDonnell K, Benezra R, et
al: Using the transcription factor inhibitor of DNA binding 1 to
selectively target endothelial progenitor cells offers novel
strategies to inhibit tumor angiogenesis and growth. Cancer Res.
70:7273–7282. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Langenfeld E, Deen M, Zachariah E and
Langenfeld J: Small molecule antagonist of the bone morphogenetic
protein type I receptors suppresses growth and expression of Id1
and Id3 in lung cancer cells expressing Oct4 or nestin. Mol Cancer.
12:1292013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu X, Xu X, Han B and Zhou R: Inhibitor of
DNA Binding-1 overexpression in prostate cancer: Relevance to tumor
differentiation. Pathol Oncol Res. 15:91–96. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ling YX, Tao J, Fang SF, Hui Z and Fang
QR: Down regulation of Id1 by small interfering RNA in prostate
cancer PC3 cells in vivo and in vitro. Eur J Cancer Prev. 20:9–17.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ciarrocchi A, Piana S, Valcavi R, Gardini
G and Casali B: Inhibitor of DNA binding-1 induces mesenchymal
features and promotes invasiveness in thyroid tumor cells. Eur J
Cancer. 47:934–945. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pillai S, Rizwani W, Li X, Rawal B, Nair
S, Schell MJ, Bepler G, Haura E, Coppola D and Chellappan S: ID1
facilitates the growth and metastasis of non-small cell lung cancer
in response to nicotinic acetylcholine receptor and epidermal
growth factor receptor signaling. Mol Cell Biol. 31:3052–3067.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gumireddy K, Li A, Kossenkov AV, Cai KQ,
Liu Q, Yan J, Xu H, Showe L, Zhang L and Huang Q: ID1 promotes
breast cancer metastasis by S100A9 regulation. Mol Cancer Res.
12:1334–1343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Asp J, Brantsing C, Lövstedt K, Benassi
MS, Inerot S, Gamberi G, Picci P and Lindahl A: Evaluation of p16
and Id1 status and endogenous reference genes in human
chondrosarcoma by real-time PCR. Int J Oncol. 27:1577–1582.
2005.PubMed/NCBI
|
|
30
|
Zheng W, Wang H, Xue L, Zhang Z and Tong
T: Regulation of cellular senescence and p16 (INK4a) expression by
Id1 and E47 proteins in human diploid fibroblast. J Biol Chem.
279:31524–31532. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ling MT, Lau TC, Zhou C, Chua CW, Kwok WK,
Wang Q, Wang X and Wong YC: Overexpression of Id1 in prostate
cancer cells promotes angiogenesis through the activation of
vascular endothelial growth factor (VEGF). Carcinogenesis.
26:1668–1676. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X, Bian Y, Takizawa Y, Hashimoto T,
Ikoma T, Tanaka J, Kitamura N, Inagaki Y, Komada M and Tanaka T:
ERK-dependent downregulation of Skp2 reduces Myc activity with HGF,
leading to inhibition of cell proliferation through a decrease in
Id1 expression. Mol Cancer Res. 11:1437–1447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheng YJ, Tsai JW, Hsieh KC, Yang YC, Chen
YJ, Huang MS and Yuan SS: Id1 promotes lung cancer cell
proliferation and tumor growth through Akt-related pathway. Cancer
Lett. 307:191–199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang G, Zhang Y, Xiong J, Wu J, Yang C,
Huang H and Zhu Z: Downregulation of Id1 by small interfering RNA
in gastric cancer inhibits cell growth via the Akt pathway. Mol Med
Rep. 5:1075–1079. 2012. View Article : Google Scholar : PubMed/NCBI
|