Introduction
Renal cell carcinoma (RCC) is one of the third most
common types of urological cancer worldwide (1). According to previous report, the
incidence of RCC has increased in the last 20 years (2% per year),
and patients with advanced RCC experience reduced survival rates
(2,3). Chemotherapy, radiotherapy and surgery
are the most common treatment methods for RCC, however, response to
treatment remains poor (4). In
addition, there are few effective and specific biomarkers for RCC,
which is the reason for failed early detection and treatment of
RCC. As a result, it is necessary to identify sensitive biomarkers
for RCC and to develop novel targets for the treatment of RCC.
Previous studies have found that long non-coding
RNAs (lncRNAs) are important in the development and progression of
cancer (5,6). lncRNAs are a type of non-coding RNA,
which are >200 nucleotides in length. Certain lncRNAs have been
shown to regulate the proliferation, migration and drug resistance
of human cancer. Hu et al reported that lncRNA GAPLINC
regulates the malignant activity of gastric cancer cells by
regulating the expression of CD44 (7). Sun et al found that lncRNA
HOXA11-AS promotes the proliferation and invasion of gastric cancer
by regulating the histone methylation state and sponging for
microRNA (miR)-1297 (8). Yuan
et al found that lncRNA DANCR increases the stemness of
hepatocellular carcinoma by regulating the expression of CTNNB1 via
inhibiting miR-214, miR-320a and miR-199a (9). These findings indicate that lncRNAs
may be novel potential targets for treating cancer. However, the
function of lncRNAs in RCC remains to be fully elucidated.
lncRNA ROR has been reported as an oncogene in
several types of cancer. Studies have shown that lncRNA ROR can
promote the proliferation and invasion of breast cancer via
targeting miR-145 and miR-205, regulating the targets of these
miRNAs (10,11). In addition, previous studies have
shown that lncRNA ROR can regulate the sensitivity to radiotherapy
and chemotherapy in cancer treatment via the p53/miR-145 signaling
pathway, indicating that lncRNA ROR can be used as a novel target
in cancer treatment (12). Wang
et al also found that lncRNA ROR can predict outcome in
patients with gallbladder cancer, and promotes the proliferation,
migration and invasion of gallbladder cancer (13). Huang et al found that lncRNA
ROR can promote cell proliferation and tumorigenesis by promoting
the stability of c-Myc mRNA via binding hnRNP I and AU-rich element
RNA-binding protein 1 (AUF1) (14). These studies show that lncRNA ROR
is involved in regulating the malignant activities of different
types of cancer, indicating that lncRNA ROR may be a potential
treatment target. However, the role of lncRNA ROR in RCC remains to
be elucidated.
The present study aimed to investigate the
expression of lncRNA ROR in RCC tissues and adjacent tissues, and
to examine the clinical characteristics of lncRNA ROR in RCC. The
possible mechanism underlying the effect of lncRNA ROR on RCC was
also investigated. The results indicated that lncRNA ROR may offer
potential as a novel biomarker for the diagnosis of RCC and a
target for its treatment.
Materials and methods
Clinical sample collection and
preparation
A total of 36 RCC tissues and matched adjacent
non-tumor tissues were collected from patients who underwent
radical nephrectomy in the Department of Urology, The first
Affiliated Hospital of Liaoning Medical University (Jinzhou,
China), between June 2014 and July 2015. None of these patients had
received chemotherapy or radiotherapy prior to collection. The
samples were cut into small sections following washing with the
RNase-free phosphate-buffered saline (PBS), and then collected and
stored in the liquid nitrogen. The clinical stages of the RCC
tissue samples were confirmed according to the TNM classification
(4). All patients were informed of
the use of samples and written consent was provided. The entire
protocol was approved by the Institutional Review Board of The
First Affiliated Hospital of Liaoning Medical University.
Cell culture
The immortalized normal human proximal tubule
epithelial cell line (HK-2) and renal cancer cell lines (Caki-1 and
Caki-2) were the cells used for investigation in the present study,
which were purchased from the Cell Bank of Type Culture Collection
of Chinese Academy of Sciences (Shanghai, China). The HK-2 cell
line was cultured in KSFM medium (Hyclone; GE Healthcare Life
Sciences, Logan, UT, USA), and Caki-1 and Caki-2 were cultured in
RPMI 1640 medium (Hyclone, GE Healthcare Life Sciences). These
media were supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and 1% penicillin and
streptomycin (Beyotime Institute of Biotechnology, Haimen, China).
The cells were maintained at 37°C with 5% CO2.
Plasmid transfection
The cells were plated in a six-well plate at 70–90%
confluence (~1×105 target cells), to which fresh
serum-free culture medium was added 2 h prior to transfection. The
plasmids used to knockdown the expression of lncRNA ROR in renal
cancer cell lines (shRNA:
GATCCCCCCTGAGAGTTGGCATGAATTTCAAGAGAATTCATGCCAACTCTCAGGTTTTTC) were
provided by Sangon Biotech (Shanghai, China). The plasmids were
transfected into cells using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) for 12 h according to the manufacturer's
protocol. The fluorescence was observed after 24 h.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA from the cells and clinical samples were
extracted using RNAiso Plus (Takara Bio Inc., Otsu, Japan),
according to the manufacturer's protocol. The RNA was then reverse
transcribed into cDNA using a PrimeScript™ RT reagent kit with gDNA
Eraser, according to the manufacturer's protocol (Takara Bio,
Inc.). The RT-qPCR procedure (2 µl cDNA, 1 µl forward primer, 1 µl
reverse primer, 0.4 µl ROX, 5.6 µl RNase-free Water and 10 µl SYBR)
was performed on a Real time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.), as follows: 10 min at 95°C, 40 cycles of
10 sec at 95°C, 30 sec at 60°C, and 20 sec at 72°C. GAPDH was used
as a control. The relative expression of lncRNA ROR (forward:
5′-CCAGGACAATGAAACCAC-3′; reverse: 5′-AGGAGCCCAAAGTAACAG-3′) was
calculated using the comparative cycle threshold 2−ΔΔCq
method, with GAPDH (forward: 5′-GGTGAAGGTCGGAGTCAACG-3′; reverse:
5′-CAAAGTTGTCATGGATGHACC-3′) as the endogenous control for data
normalization.
Cell Counting Kit-8 (CCK-8)
assays
The target cells were seeded into a 96-well
flat-bottomed plate, each well containing 3,000 cells in 200 µl
cell suspension. A 20-µl volume of CCK8 (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was added to each well and
cultured for 2 h at 37°C. The absorbance value of each well at 450
nm was measured and recorded, which represented the proliferation
of the target cells. Each experiment included five replicates and
was repeated three times.
Analysis of apoptosis
The activity of Caspase 3 was measured to determine
the level of apoptosis. A Caspase-3 Colorimetric Activity Assay kit
(EMD Millipore, Billerica, MA, USA) was used to detect the level of
apoptosis of the target cells, according to the standard assay
protocol.
Western blot analysis
The cells were washed three times with pre-cooled
PBS. Total cell proteins were extracted using RIPA buffer
containing the protein inhibitor, PMSF. Protein concentrations were
assessed using the standard curve established with bovine serum
albumin (Beyotime Institute of Biotechnology, Beijing, China). The
total proteins (30 µg) were subjected to sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% denaturing
gel. Following SDS-PAGE, the proteins were transferred onto a PVDF
membrane (EMD Millipore). The membrane was inhibited with 5%
non-fat milk for 2 h at room temperature. This was followed by
incubation of the membrane with primary antibodies at 4°C
overnight. The source and dilution of these antibodies were as
follows: p53 (ab32049; 1:1,000; Abcam, Cambridge, MA, USA); c-Myc
(ab39688; 1:1,000; Abcam); GAPDH (ab8245; 1:1,000; Abcam). The
membrane was then washed in TBS-T (Wuhan Boster Biological
Technology, Ltd., Wuhan, China) for 5 min, and he membrane was
incubated with secondary antibody (A16072; 1:1,000; and A16104;
1:1,000; Thermo Fisher Scientific, Inc.). The blots were visualized
using an ECL chemiluminescence detection system, and protein bands
were quantified using densitometric analysis with Quantity One
software version 4.62 (Bio-Rad Laboratories, Inc., Hercules,
USA).
Statistical analysis
All experiments were performed at least three times
and data are presented as the mean ± standard deviation.
Statistical analyses were performed using Student's t-test or
one-way analysis of variance using GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA). Survival analysis was performed
using the log-rank test in GraphPad. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression levels of lncRNA ROR are
high in RCC tissues and RCC cell lines
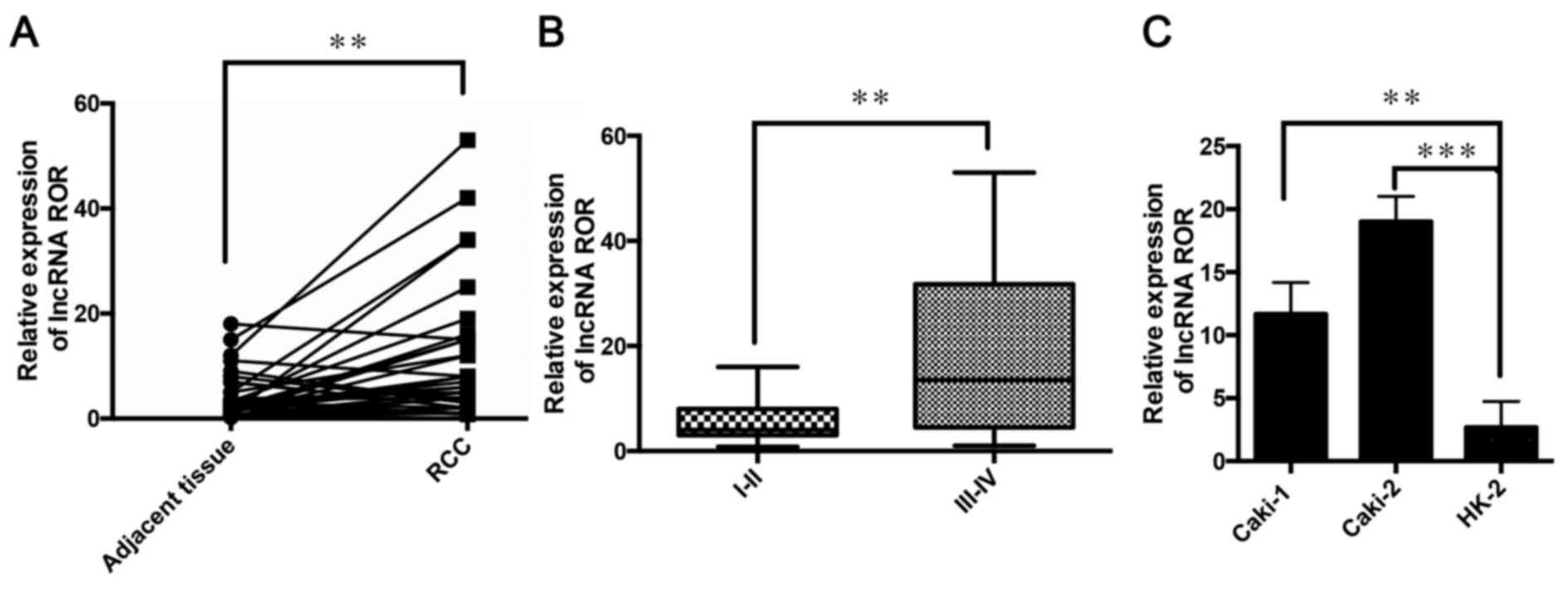
The relative expression of lncRNA ROR was determined
in 36 patients with RCC using RT-qPCR analysis. It was found that
the expression of lncRNA ROR was high in the RCC tissues, compared
with the adjacent non-tumor tissues (Fig. 1A). It was also found that the
expression of lncRNA ROR was higher at clinical stages III and IV,
compared with that at clinical stages I and II (Fig. 1B). The expression of lncRNA ROR was
also detected in Caki-1 and Caki-2 RCC cell lines and the HK-2
normal human proximal tubule epithelial cell line. It was found
that the expression levels of lncRNA ROR in the RCC lines were
higher, compared with that in the HK-2 cell line (Fig. 1C). These results indicated that
lncRNA ROR was expressed at high levels in the RCC tissues and cell
lines, and that lncRNA ROR may be involved in the progression and
development of RCC.
High expression of lncRNA ROR predicts
poor patient prognosis
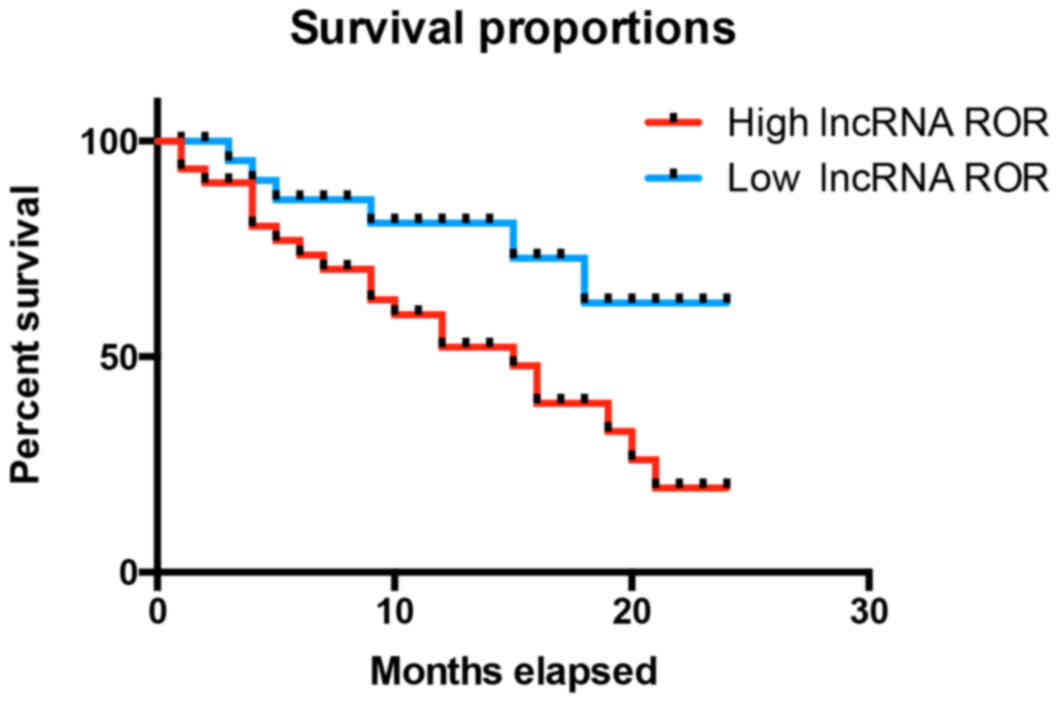
To further examine the effect of lncRNA ROR in RCC,
the present study examined the association between the expression
of lncRNA ROR and the survival rates of patients with RCC. It was
found that the patients with RCC and high expression levels of
lncRNA ROR had reduced survival rates, compared with patients with
low expression levels of lncRNA ROR (Fig. 2). As shown in Table I, it was found that the expression
of lncRNA ROR was closely associated with tumor size and the
clinical stage of RCC, however, no significant associations were
found between the expression of lncRNA ROR and age or gender. These
results indicated that lncRNA ROR was closely associated with tumor
size in RCC. Taken together, these results suggested that the
expression of lncRNA ROR was closely associated with the prognosis
of patients with RCC and may be associated with proliferation.
 | Table I.Associations between the expression
level of lncRNA ROR and clinical characteristics of patients. |
Table I.
Associations between the expression
level of lncRNA ROR and clinical characteristics of patients.
|
|
| lncRNA ROR |
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | High (n) | Low (n) | P-value |
|---|
| Age (years) |
|
|
| 0.532 |
|
<60 | 16 | 7 | 9 |
|
|
>60 | 20 | 11 | 9 |
|
| Gender |
|
|
| 0.376 |
| Male | 21 | 12 | 9 |
|
|
Female | 15 | 7 | 8 |
|
| Clinical stage |
|
|
| 0.0043 |
| I–II | 19 | 6 | 13 |
|
|
III–IV | 17 | 13 | 4 |
|
| Tumor size |
|
|
| 0.0032 |
| <3
cm | 14 | 5 | 9 |
|
| >3
cm | 22 | 17 | 5 |
|
Knockdown of lncRNA ROR inhibits
proliferation and promotes apoptosis of RCC cells
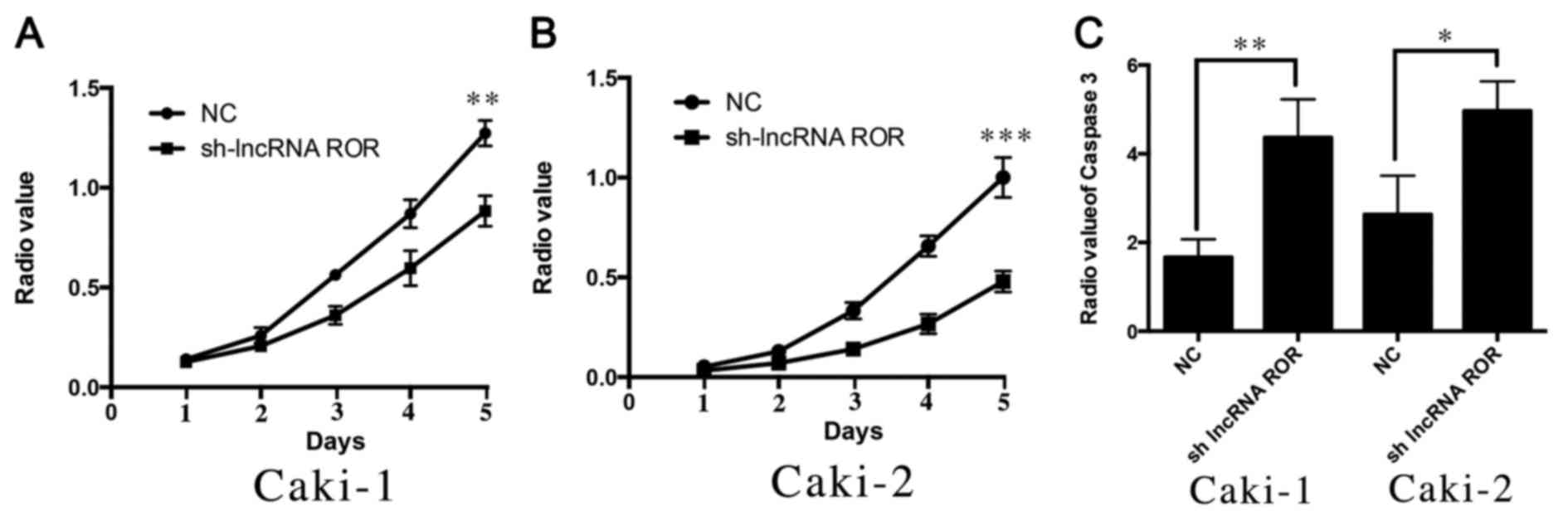
As described in the results above, it was found that
lncRNA ROR may be associated with the proliferation of RCC. To
further investigate the possible effect of lncRNA ROR on the
proliferation of RCC cells, the present study used a CCK8 assay to
detect changes in the proliferation ability of Caki-1 and Caki-2
cells following the suppression of lncRNA ROR. It was found that
the suppression of lncRNA ROR led to a decrease in the
proliferation ability of the Caki-1 and Caki-2 cells (Fig. 3A and B). The apoptosis of Caki-1
and Caki-2 was then detected following suppression of the
expression of lncRNA ROR. It was also found that apoptosis was
increased following transfection with short hairpin (sh)-lncRNA
ROR, compared with that in the control group (Fig. 3C). These findings indicated that
knockdown of the expression of lncRNA ROR inhibited the
proliferation and promoted the apoptosis of RCC cells.
lncRNA ROR promotes the proliferation
of RCC cells via suppressing the expression of p53 and increasing
the expression of c-Myc
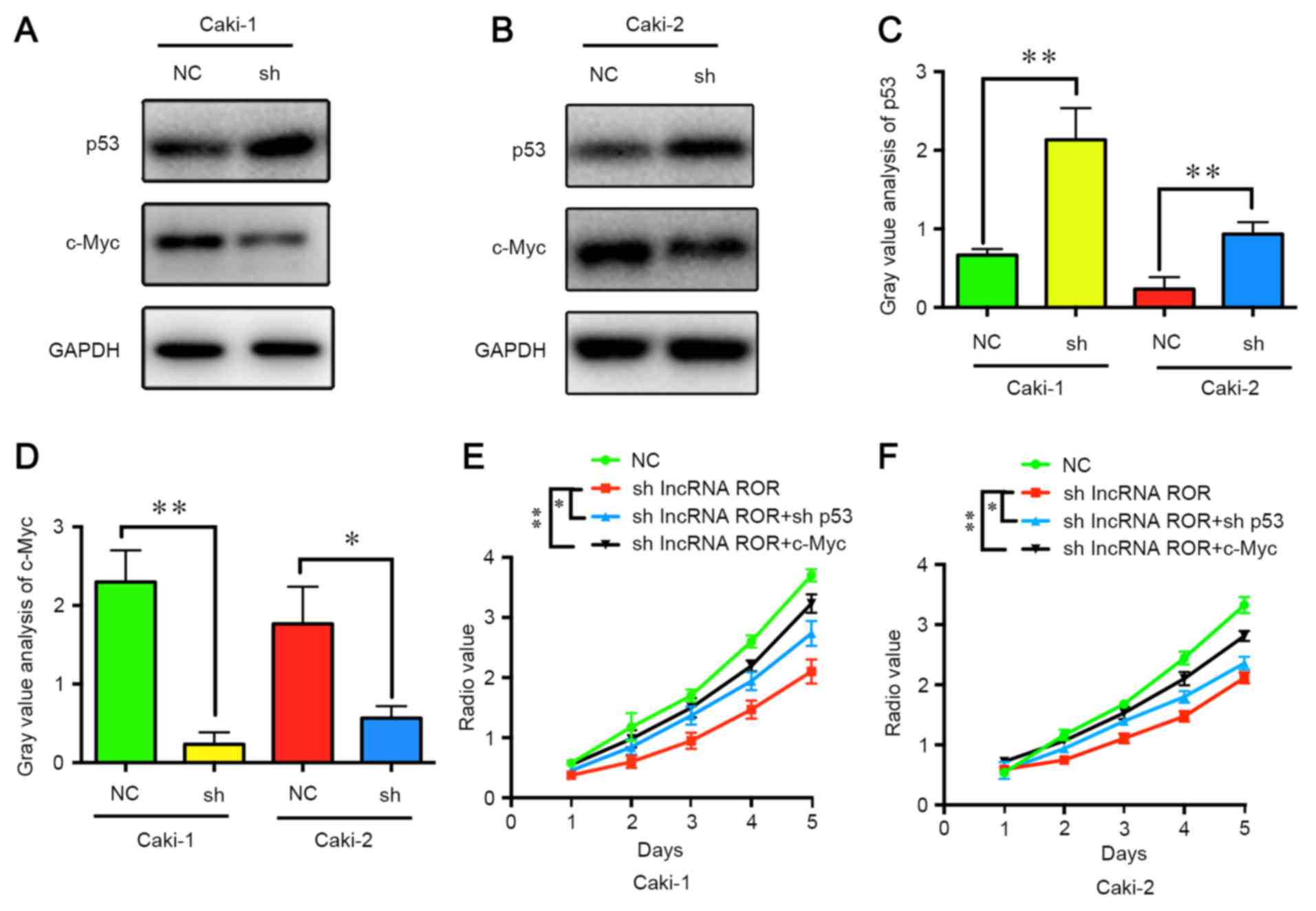
Although the present study demonstrated that lncRNA
ROR promoted the proliferation of RCC, the detailed mechanism
remains to be fully elucidated. According to a previous study,
lncRNA ROR promotes the resistance of human colorectal cancer cells
to radiotherapy via suppressing the expression of p53. In addition,
Huang et al (14)
demonstrated that lncRNA ROR promotes the expression of c-Myc to
increase cell proliferation and tumorigenesis. In the present
study, the expression levels of c-Myc and p53 were detected
following the suppression of lncRNA ROR. It was found that the
expression of p53 was increased, whereas the expression of c-Myc
was decreased (Fig. 4A-D). It was
also found that the suppression of p53 and the overexpression of
c-Myc restored the proliferation ability of the RCC cells (Fig. 4E and F). These results indicated
that lncRNA ROR promoted the expression of c-Myc and suppressed the
expression of p53 in the RCC cells, leading to cell
proliferation.
Discussion
RCC remains one of the leading causes of
cancer-associated mortality worldwide; therefore, novel targets for
its treatment, prognosis and diagnosis are required to improve
clinical treatment. An increasing number of studies have found that
lncRNA is involved in the development and progression of different
types of tumor, which may be developed as novel targets and
biomarkers for cancer treatment. Wang et al (15) has found that a panel of four
lncRNAs (BANCR, NR_026817, NR_029373 and NR_034119) can be used to
detect colorectal cancer. In addition, certain lncRNAs, including
lncRNA MALAT1, HOTAIR, lncRNA CCAT2 and H19, have shown potential
in predicting prognosis, survival rates and the effect of treatment
(16). Among the lncRNAs, lncRNA
ROR has been investigated extensively, as its function is important
in the development of different types of cancer. However, the
effect of lncRNA ROR in the development of RCC has note been fully
elucidated, requiring further investigation.
In the present study, the expression of lncRNA ROR
in patients with RCC was first investigated. It was found that the
expression of lncRNA ROR was high in RCC tissues, compared with
that in adjacent non-tumor tissues, determined using RT-qPCR
analysis. The results also indicated that the expression of lncRNA
ROR was closely associated with patient survival rates. Patients
with RCC and higher expression levels of lncRNA ROR experienced
reduced survival rates, compared to those with lower expression
levels of lncRNA ROR. The expression of lncRNA was also closely
associated with tumor size, suggesting that lncRNA ROR affected the
proliferation of RCC cells. Taken together, these results indicated
that lncRNA ROR is an independent prognostic factor for patients
with RCC, and is important in the proliferation of RCC.
To further confirm the above findings, the effect of
lncRNA ROR on the proliferation of RCC cells was determined.
Changes in the proliferation ability of the Caki-1 and Caki-2 RCC
cells were detected following the suppression of lncRNA ROR. The
proliferation of the cells was suppressed, indicating that lncRNA
ROR promoted the proliferation of the Caki-1 and Caki-2 cells. This
result was in agreement with the clinical findings.
Previous studies have found that lncRNA ROR promotes
the proliferation of different types of cancer, including the
proliferation ability of gastric cancer, gallbladder cancer and
nasopharyngeal carcinoma, which indicates that lncRNA ROR is
important in controlling the proliferation of cancer (13,17,18).
However, the detailed mechanism underlying the effect of lncRNA ROR
in regulating proliferation remains to be elucidated. lncRNA ROR
was first identified as a regulator for the reprogramming of
differentiated cells into induced pluripotent stem cells; the
suppression of lncRNA ROR increases apoptosis and the activation of
p53 (19). In addition, a previous
study demonstrated that lncRNA ROR can act as a competing
endogenous RNA to sponge miR-145, and miR-145 is closely linked
with p53 (20). Another previous
study found that lncRNA ROR also increased the mRNA level of c-Myc
in colon cancer and breast cancer cell lines (14). It has also been shown that lncRNA
is unable to bind with AUF1, which binds c-Myc mRNA, enhancing the
stability of c-Myc mRNA (14).
These previous findings suggest that p53 and c-Myc are regulated by
lncRNA ROR. According to these findings, the present study aimed to
detect whether lncRNA ROR promoted the proliferation ability of RCC
via p53 and c-Myc. The results revealed that the suppression of
lncRNA ROR led to an increase in the expression of p53 and a
decrease in the expression of c-Myc. It was also found that the
overexpression of c-Myc and suppression of p53 restored the
proliferation ability of the RCC cells, suggesting that lncRNA ROR
may regulate the proliferation ability of RCC cell lines via c-Myc
and p53.
In conclusion, the present study found that lncRNA
ROR was expressed at high levels in RCC tissues, and its expression
was negatively correlated with the survival rates of patients with
RCC. This indicated that lncRNA ROR may be used as a potential
target to predict the prognosis of RCC. It was also found that
lncRNA ROR regulated the proliferation of RCC cell lines, which may
occur via regulating the expression of p53 and c-Myc.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Olshan AF, Kuo TM, Meyer AM, Nielsen ME,
Purdue MP and Rathmell WK: Racial difference in histologic subtype
of renal cell carcinoma. Cancer Med. 2:744–749. 2013.PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Russo P: Renal cell carcinoma:
Presentation, staging, and surgical treatment. Semin Oncol.
27:160–176. 2000.PubMed/NCBI
|
|
5
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang
Y, Chen H, Hong J, Zou W, Chen Y, et al: Long noncoding RNA GAPLINC
regulates CD44-dependent cell invasiveness and associates with poor
prognosis of gastric cancer. Cancer Res. 74:6890–6902. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: LncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1 and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yuan SX, Wang J, Yang F, Tao QF, Zhang J,
Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, et al: Long noncoding RNA
DANCR increases stemness features of hepatocellular carcinoma by
derepression of CTNNB1. Hepatology. 63:499–511. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y,
Zhao L, Zhang Y, Huang B and Lu J: LincRNA-ROR induces
epithelial-to-mesenchymal transition and contributes to breast
cancer tumorigenesis and metastasis. Cell Death Dis. 5:e12872014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
13:330–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang P, Yang Y, An W, Xu J, Zhang G, Jie J
and Zhang Q: The long noncoding RNA-ROR promotes the resistance of
radiotherapy for human colorectal cancer cells by targeting the
P53/miR-145 pathway. J Gastroenterol Hepatol. 32:837–845. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang SH, Zhang MD, Wu XC, Weng MZ, Zhou D
and Quan ZW: Overexpression of LncRNA-ROR predicts a poor outcome
in gallbladder cancer patients and promotes the tumor cells
proliferation, migration, and invasion. Tumour Biol.
37:12867–12875. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang J, Zhang A, Ho TT, Zhang Z, Zhou N,
Ding X, Zhang X, Xu M and Mo YY: Linc-RoR promotes c-Myc expression
through hnRNP I and AUF1. Nucleic Acids Res. 44:3059–3069. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang R, Du L, Yang X, Jiang X, Duan W, Yan
S, Xie Y, Zhu Y, Wang Q, Wang L, et al: Identification of long
noncoding RNAs as potential novel diagnosis and prognosis
biomarkers in colorectal cancer. J Cancer Res Clin Oncol.
142:2291–2301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang
Q, Ren M, Chen L, Yuan D, Zhang Y, et al: The lncRNA MALAT1 is a
novel biomarker for gastric cancer metastasis. Oncotarget.
7:56209–56218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang S, Liu F, Deng J, Cai X, Han J and
Liu Q: Long noncoding RNA ROR regulates proliferation, invasion,
and stemness of gastric cancer stem cell. Cell Reprogram.
18:319–326. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Gu M, You B, Shi S, Shan Y, Bao L
and You Y: Long non-coding RNA ROR promotes proliferation,
migration and chemoresistance of nasopharyngeal carcinoma. Cancer
Sci. 107:1215–1222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loewer S, Cabili MN, Guttman M, Loh YH,
Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al:
Large intergenic non-coding RNA-RoR modulates reprogramming of
human induced pluripotent stem cells. Nat Genet. 42:1113–1117.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang P, Yang Y, An W, Xu J, Zhang G, Jie J
and Zhang Q: The long non-coding RNA-ROR promotes the resistance of
radiotherapy for human colorectal cancer cells by targeting the
P53/miR-145 pathway. J Gastroenterol Hepatol. 32:837–845. 2017.
View Article : Google Scholar : PubMed/NCBI
|