Introduction
The decline in fertility in humans has been a
medical concern for ~25 years (1).
The development of in vitro fertilization (IVF) has emerged
as the most successful treatment for fertility problems. However,
despite the use of adjuvant therapies, the cumulative percentage of
live births remains at only ~40% worldwide (2). The major factor limiting the success
of IVF procedures is embryo implantation (3). Accumulating data has shown that low
pregnancy rates, despite high fertility rates, are primarily due to
implantation failure (4,5). Each failure of in vitro
fertilization has significant effects on the quality of life and
mental wellbeing of patients, in addition to being a financial
burden. Therefore, further investigations into the causes and
mechanisms of implantation failure are warranted.
Embryo implantation, which requires a viable
blastocyst and uterine receptivity, involves a series of steps,
including the interaction and invasion of the blastocyst to the
endometrium. The cross-talk between the embryo and the endometrium
is essential for successful implantation (2). As implantation is the result of
interactions between the blastocyst and endometrium, it is
considered that this process is initiated as soon as a chemical
interaction is established between the embryo and endometrium. The
interaction between the blastocyst and endometrium begins
immediately following entry of the embryo into the endometrial
cavity and prior to active penetration of the endometrium by the
developing blastocyst (6). In this
respect, it has been reported that embryo-specific signaling is
present prior to the embryo entering the endometrial cavity and
making intimate contact with the endometrium (7–10).
Preimplantation factor (PIF) is a unique peptide,
which is secreted only by viable embryos. PIF is detected first in
the early stages, and is present throughout the duration of
pregnancy in several species of mammal, where it is expressed in
the placenta (11,12). It has been noted that synthetic PIF
(sPIF), which replicates native PIF function, affects key processes
in early pregnancy implantation, including modulating peripheral
immune cells, contributing to the maternal adaptation to pregnancy,
and creating a favorable immune environment shortly following
fertilization (13). Of note, sPIF
can exert a positive autocrine effect on trophoblast invasion, and
the invasion of trophoblast cells is enhanced by exposure to sPIF
(14). Therefore, PIF is
considered to promote or rescue placental invasion and represents a
significant advance in reproductive technology.
In the present study, isobaric tags for relative and
absolute quantification (iTRAQ) technology, combined with liquid
chromatography-electrospray ionization-tandem mass spectrometry
(LCESI-MS/MS) analysis, were used to identify key proteins that
react with PIF, and western blot analysis was used for confirmation
of findings. Using these measurements, the present study aimed to
develop a profile of PIF-reactive proteins, and provide valuable
understanding and insight into the molecular mechanism underlying
the effect of PIF on the promotion of trophoblast invasion.
Materials and methods
Reagents and antibodies
Eight-plex iTRAQ reagent kits were acquired from
Applied Biosystems; Thermo Fisher Scientific, Inc. (Waltham, MA,
USA). Monoclonal antibodies against human PIF (18-802-392017) were
obtained from GenWay (San Diego, CA, USA) and polyclonal antibodies
against GAPDH (ab9485), MYH10 (ab684), tubulin, β5 class I (TUBB5;
ab15568) and heat shock protein family D1 (HSPD1; ab46798) were
purchased from Abcam (Cambridge, UK). MYH10-specific Stealth Select
RNAi™ small interfering (si)RNA (NM_005964.3), Stealth RNAi™
Negative Control siRNA (cat. no. 12935-400) and Lipofectamine 2000
transfection reagent were obtained from Thermo Fisher Scientific,
Inc. CytoSelect™ 24-Well Cell Migration and Invasion Assay kits (8
µm, colorimetric format) were purchased from Cell Biolabs (San
Diego, CA, USA).
Cell lines and tissues
The HTR-8/SVneo cell line was derived from an
explant culture of human first-trimester placenta and was used in
the present study (15).
HTR-8/SVneo cells, HEK293 cells (Sigma; Merck Millipore, Darmstadt,
Germany) HEC-1-B cells (Shanghai Institute of Cell Biology,
Shanghai, China) were cultured at 37°C, under 5.0% CO2,
in RPMI-1640 medium (Nanjing KeyGen Biotech. Co. Ltd., Nanjing,
China) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 100 IU/ml of penicillin. A total of 20 male and 20 female
Sprague-Dawley rats (7 weeks old, 200–250 g), obtained from the
Animals Experiment Center of Chongqing Medical University, were
housed under controlled conditions (25±2°C, 60±10% relative
humidity and 12-h light/dark cycle) with free access to tap water
and food throughout the experimental period. Following 1 week of
acclimation on an American Institute of Nutrition-93G diet, the
male rats were randomly allocated into two groups of 10 animals per
group: Healthy control group, and experimental group, in which rats
receiving sterilization by surgical ligation. The breeders were
paired every evening between 3:00 and 5:00 p.m., and were separated
every morning between 9:00 and 10:00 a.m., followed by vaginal plug
checks. After 4 days, all rats were euthanized with sodium
pentobarbital (1 mg/kg bodyweight i.p.), endometrial tissues were
extracted and fixed for 24 h in 4% paraformaldehyde in 1X PBS at
4°C. All tissues were frozen with liquid nitrogen and preserved at
−196°C. All rats received humane care in accordance with the
guidelines of the Animal Usage Committee of the Chongqing Medical
University (Chongqing, China), which approved the study prior to
commencement.
Peptide synthesis
The partial characterization and PIF assay
information were as described previously (16). Synthetic PIF, the purity of which
was documented using high-performance liquid chromatography and
mass spectrometry as 95%, was produced by Biosynthesis, Inc.
(Lewisville, TX, USA). Synthetic PIF (sPIF; MVRIKPGSANKPSDD),
intralipid and scrambled PIF (PIFscr; GRVDPSNKSMPKDIA) and
biotin-labeled ligands of >95% purity were also generated.
Invasion assays
Invasion assays were performed using a Cell Invasion
Assay kit (Cell Biolabs, Inc., Beijing, China), according to the
manufacturer's protocol. The HTR-8/SVneo (50,000) cells were
resuspended in medium with PIF or PIFscr (100 ng/ml). As determined
using trypan blue exclusion under a light micros'cope (CK40;
Olympus Corporation, Tokyo, Japan), ~1×105 viable
HTR-8/SVneo cells were seeded into the upper chamber of a 24-well
plate with polycarbonate membrane inserts. The numbers of cells to
invade through the ECM Matrix gel were determined using CyQuant GR
fluorescent dye (560 nm).
Co-immunoprecipitation (CoIP) and
iTRAQ labeling
The HEK293 cells were trypsinized and lysed in 500
µl lysis buffer (Western blot and immunoprecipitation; Beyotime
Institute of Biotechnology, Inc., Haimen, China) on ice for 20 min.
The cell lysates were centrifuged for 15 min at 17,949 × g and 4°C
to remove debris. Subsequently, 1 mg of lysates (1 µg/µl) was mixed
with biotin-labeled synthetic PIF or PIFscr overnight at 4°C, and
then precleared with 50 µl ulfo-NHS-SS-avidin protein A-Sepharose
beads (Pierce; Thermo Fisher Scientific, Inc.) for 2 h at 4 °C. The
beads were pelleted and washed three times with lysis buffer. The
bound proteins were eluted in SDS sample buffer and quantified by
2D Quantification kit assay (GE Healthcare Life Sciences, Little
Chalfont, UK) according to the manufacturer's protocol. Then 20 µg
protein per well was subjected to 10% SDS-PAGE and analyzed by
immunoblotting. The eluted protein (100 µg) was precipitated from
each pooled group, dissolved in dissolution buffer, denatured with
2% SDS, cysteine blocked, digested with 2 µg of sequencing grade
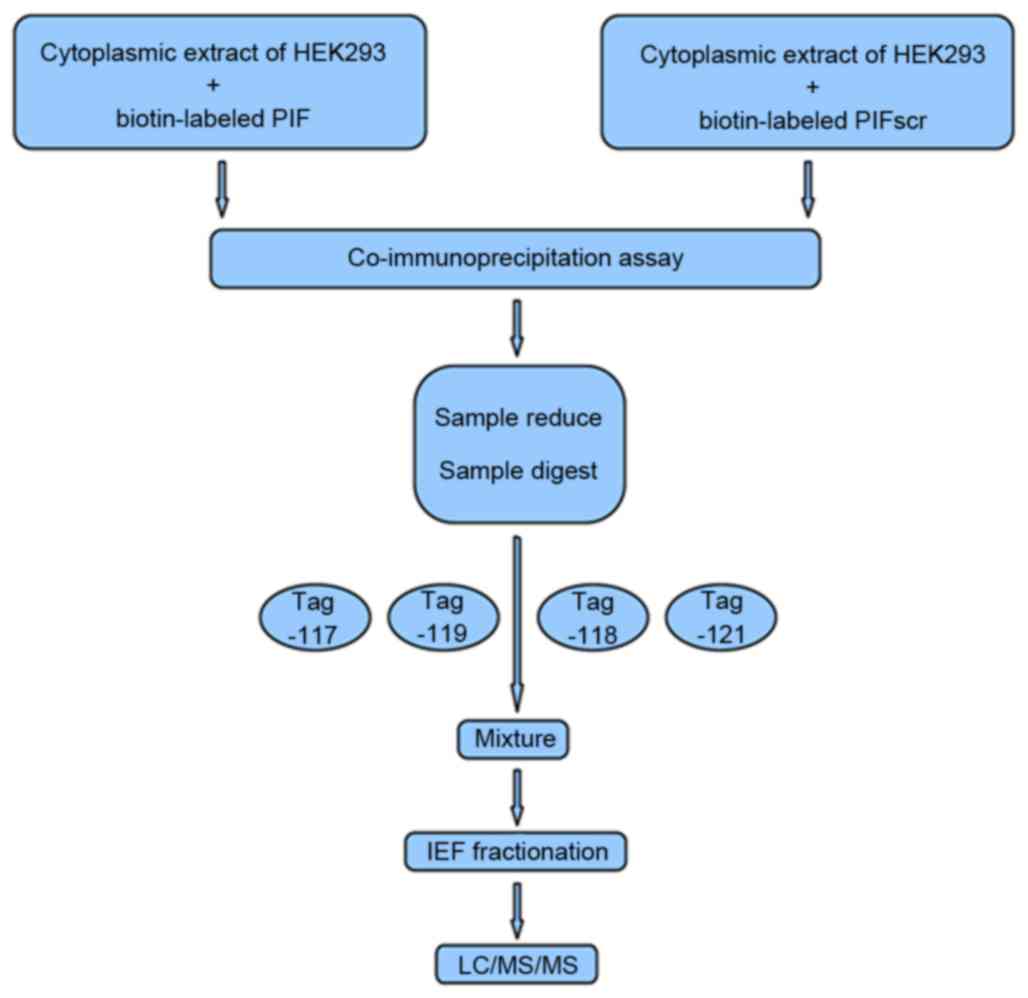
modified trypsin, and labeled using iTRAQ reagents (PIF, 117 tag;
PIFScr, 118 tag) provided with the iTRAQ kit (AB Sciex Analytical
Instrument Trading Co., Shanghai, China; Fig. 1). For the parallel experiment, the
same sample set was labeled with iTRAQ reagents 119 and 121,
respectively (Fig. 1). The
peptides from each sample set were mixed prior to subsequent
analysis.
Peptide fractionation
The labeled peptides were fractionated by
immobilized-pH-gradient isoelectric focusing (IPG-IEF), as
previously described (17,18). Briefly, the samples were dissolved
in a Pharmalyte (GE Healthcare Life Sciences, Chalfont, UK) and
urea solution, rehydrated on a pH 3–10 IPG strip, and subjected to
IEF at 68 kV/h using an IPG phor system (GE Healthcare Life
Sciences). The peptides were extracted from the gel using an
acetonitrile (ACN) and formic acid solution (19). The fractions were lyophilized, and
purified with SPE Discovery DSC-18 columns (Supelco, Inc.,
Bellefonte, PA, USA). The purified peptides were re-lyophilized and
stored at −20°C until use.
Mass spectrometry
The purified peptide fractions were reconstituted in
solvent A, comprising water/ACN (98:2 v/v) with 0.1% formic acid,
and separated using a C18-PepMap column (Thermo Fisher Scientific,
Inc.) with a solvent gradient of 2–100% Buffer B (0.1% formic acid
and 98% acetonitrile) in Buffer A at a flow rate of 0.3 µl/min. The
peptides were electrosprayed using a nanoelectrospray ionization
source at an ion spray voltage of 2,300 eV, and were analyzed using
the NanoLC-ESI-Triple TOF 5600 system (AB Sciex Analytical
Instrument Trading Co.). The mass spectrometer was set in the
positive ion mode at a mass range of 300–1,800 m/z. The two most
intensely charged peptides >20 counts were selected for tandem
MS/MS at a dynamic exclusion of 30 sec (19). Data were processed using
ProteinPilot v2.0 (AB Sciex Analytical Instrument Trading Co.) and
compared with the UniProt database (http://www.uniprot.org/). Cysteine modified by methane
thiosulfate was specified as a fixed modification. Protein
identification was based on a threshold protein score of >1.3.
For quantitation, at least two unique peptides with 95% confidence
and a P-value of <0.05 were required.
Bioinformatics analysis
The Gene Ontology was analyzed using the PANTHER
classification system (http://www.pantherdb.org/) to determine biological
processes, protein classes and molecular functions.
Western blot analysis
To validate the results of the proteomic analysis,
the same sets of protein samples were subjected to immunoblot
analyses. Each non-depleted sample was diluted 10-fold with 10 mM
PBS. Two equal volumes of diluted samples (20 µg) were separated by
10% SDS-PAGE, following which one volume was stained with Coomassie
blue for protein loading determination and the other volume was
transferred onto PVDF membranes for immunoblotting. The membranes
were blocked in 5% skim milk in Tris-buffered saline with Tween-20
at room temperature for 1 h, and incubated with primary antibodies
against GAPDH (1:2,500), PIF (1:200), MYH10 (1:2,000), TUBB5
(1:1,000) and HSPD1 (1:10,000) at 4°C overnight. The membranes were
then incubated with HRP-conjugated goat anti-mouse immunoglobulin
(Ig) G (sc-2005; 1:5,000) or goat anti-rabbit IgG (sc-2004;
1:5,000; both from Santa Cruz Biotechnology, Santa Cruz, CA, USA)
at room temperature for 1 h. Finally, the membranes were visualized
using the ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The expression levels of PIF, MYH10 and HSPD1
in the endometrial tissues of rats were detected using the same
immunoblotting procedure. The quantification of target proteins was
determined using densitometry and normalized against GAPDH (Multi
Sciences Biotech Co., Ltd., Hangzhou, China).
MYH10 siRNA transfection, Transwell
and wound healing assays
When the cultures of HEC-1-B cells grown in 10 cm
dishes with or without medium spiked with PIF or PIFscr (100 ng/ml)
reached 95% confluence, they were transfected with 100 nm of
MYH10-specific siRNA or a negative control siRNA (cat. no.
12935-400) using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Cell
viability was determined using a trypan blue exclusion assay, and
only cells with >95% viability were used for subsequent assays.
The Transwell assays were performed as described previously
(20). For the wound healing
assays, the cells were cultured in 6-well plates until they reached
100% confluence. A 200-µl pipette tip was used to scratch the cell
monolayer, followed by washing with growth medium to remove debris.
The resultant gap was monitored for up to 24 h under light
microscopy (Eclipse 80i; Nikon Corporation, Tokyo, Japan).
Statistical analysis
Statistical analyses were performed using SPSS
software v13.0 (SPSS Inc., Chicago, IL, USA). Quantitative
variables are presented as the mean ± standard deviation.
Comparisons between groups were analyzed using Student's t-test or
a Mann-Whitney U test. Qualitative variables are presented as
counts and percentages, which were analyzed using the χ2
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
PIF promotes HTR-8/Svneo cell
invasion
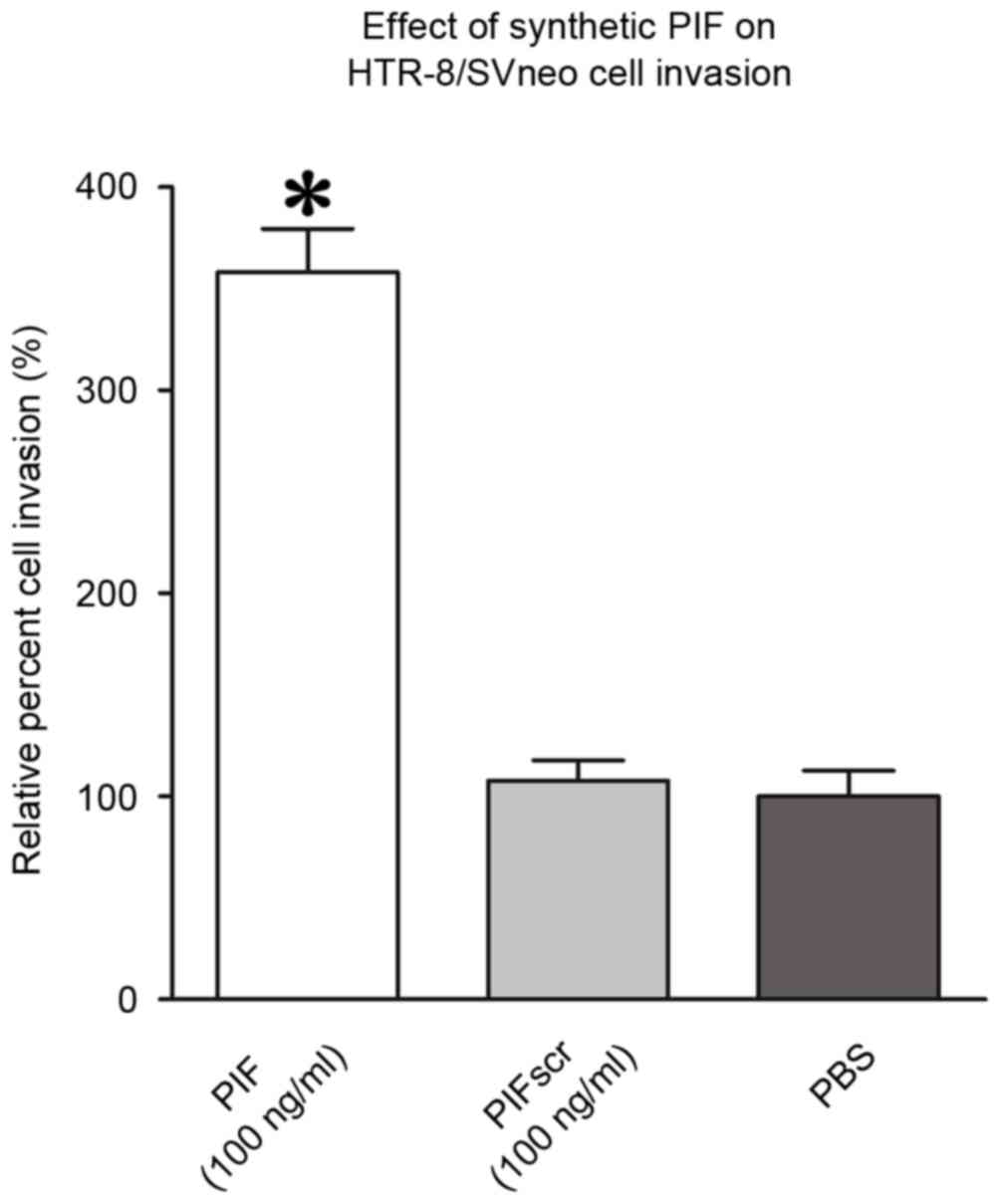
To confirm the bioactivity of synthetic PIF in tumor
cell motility, HTR-8/Svneo cells were resuspended in medium with
PIF or PIFscr (100 ng/ml). A Matrigel invasion assay revealed that
PIF increased the invasiveness of HTR-8/Svneo cells by 40–52%,
compared with the controls (P<0.05; Fig. 2).
MS identification and iTRAQ
quantification of aberrantly expressed proteins
To investigate the key proteins reacting with PIF,
quantitative proteomics analysis using iTRAQ tags was performed. At
least two peptides were used for quantification and protein
identification. For Protein Pilot-based database searching and
identification, the threshold [unused protscore (conf)] was set to
achieve 95% confidence at 5% false discovery rate. The protein
identification threshold, a ProtScore value >1.3, was used to
attain a confidence of 95%. When the proteins were classified as
significantly regulated or not, an additional >1.3 (1×1.3) or
<0.77 (1/1.3)-fold cutoff was applied to all iTRAQ ratios to
minimize false positives when determining whether proteins were
overexpressed or underexpressed. This cutoff value was used as
overall technical variation of data from the duplicate experiments
was estimated <30% (data not shown), and, in other
investigations using the iTRAQ approach, this value has been widely
used.
A total of 70 unique proteins were successfully
identified by using at least two peptides (data not shown). A total
of 21 proteins were differentially expressed in the PIF binding
proteins of HEK 293 (Table I), of
which 12 were upregulated and nine were downregulated in the PIF
binding proteins, compared with the PIFscr proteins.
 | Table I.List of differentially expressed
proteins, identified using iTRAQ analysis, between PIF and PIFscr
binding of cytoplasmic extracts in HEK293 cells. |
Table I.
List of differentially expressed
proteins, identified using iTRAQ analysis, between PIF and PIFscr
binding of cytoplasmic extracts in HEK293 cells.
| n | Accession | Gene symbol | Protein name | Peptides (95%) | Cytoplasmic extract
of HEK293+biotin-labeled PIF: cytoplasmic extract of
HEK293+biotin-labeled PIFscr 117:118a (fold) | Cytoplasmic extract
of HEK293+biotin-labeled PIF: cytoplasmic extract of
HEK293+biotin-labeled PIFscr 119:121a (fold) |
|---|
| 1 | P05067 | APP | Amyloid βA4 protein
isoform d |
5 | 67.0963 | 70.4693 |
| 2 | O94832 | MYO1D | Myosin-Id | 27 |
4.8306 |
4.4463 |
| 3 | P35580 | MYH10 | Isoform 1 of
Myosin-10 | 741 |
4.1281 |
4.1305 |
| 4 | Q12965 | MCM3AP | 80 kDa
MCM3-associated protein |
3 |
3.3729 |
2.6546 |
| 5 | P68371 | TUBB2C | Tubulin β-2C
chain | 14 |
1.8707 |
2.4434 |
| 6 | O00159 | MYO1C | Isoform 1 of
myosin-Ic |
2 |
1.7865 |
2.0137 |
| 7 | P67936 | TPM4 | Isoform 1 of
tropomyosin α-4 chain | 52 |
1.3062 |
1.6904 |
| 8 | P07437 | TUBB | Tubulin β chain | 13 |
2.1281 |
1.6444 |
| 9 | P68363 | TUBA1B | Tubulin α-1B
chain | 11 |
1.7219 |
1.6144 |
| 10 | Q9NYL9 | TMOD3 | Tropomodulin-3 | 13 |
1.5560 |
1.5704 |
| 11 | P07195 | LDHB | L-lactate
dehydrogenase B chain |
2 |
1.4450 |
1.4588 |
| 12 | P06753 | TPM3 | Isoform 2 of
tropomyosin α-3 chain | 75 |
1.3428 |
1.3428 |
| 13 | P02533 | KRT14 | Keratin, type I
cytoskeletal 14 |
7 |
0.5546 |
0.7112 |
| 14 | P13645 | KRT10 | Keratin, type I
cytoskeletal 10 | 35 |
0.1393 |
0.6918 |
| 15 | Q7Z406 | MYH14 | Isoform 1 of
myosin-14 | 297 |
0.5105 |
0.6252 |
| 16 | P08779 | KRT16 | Keratin, type I
cytoskeletal 16 |
7 |
0.4786 |
0.6026 |
| 17 | P04264 | KRT1 | Keratin, type II
cytoskeletal 1 | 49 |
0.2051 |
0.5754 |
| 18 | P35908 | KRT2 | Keratin, type II
cytoskeletal 2 epidermal | 18 |
0.4699 |
0.5200 |
| 19 | P08238 | HSP90AB1 | Heat shock protein
HSP 90-β |
2 |
0.2489 |
0.1459 |
| 20 | P02042 | HBD | Hemoglobin subunit
δ |
6 |
0.5754 |
0.0738 |
| 21 | P69905 | HBA2; HBA1 | Hemoglobin subunit
α |
9 |
0.7586 |
0.0136 |
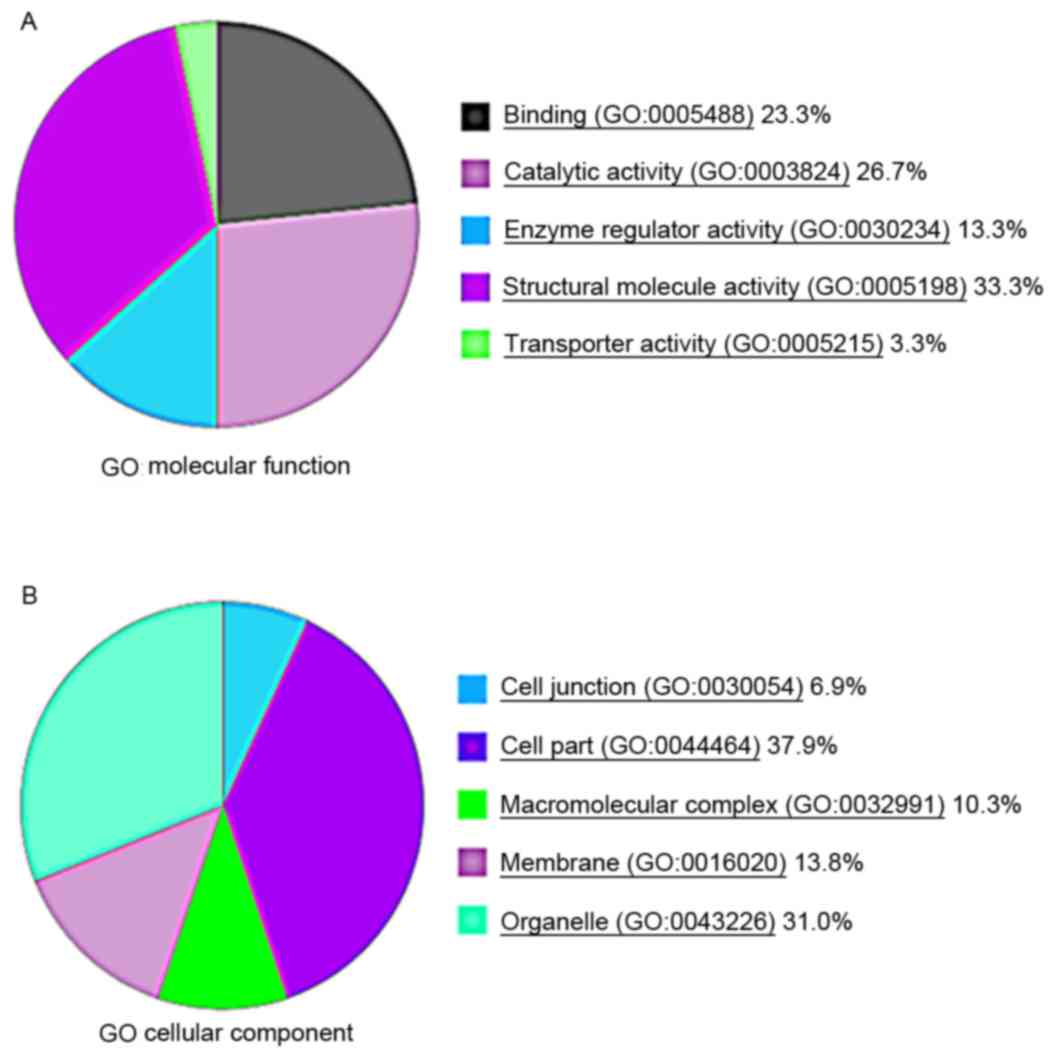
Gene Ontology analysis using PANTHER suggested that
the majority of the differentially expressed proteins were enzymes
or signaling molecules, followed by cell development regulators and
cellular structure-related proteins (Fig. 3A and B).
Verification of aberrant expression of
MYH10, TUBB5 and HSPD1
To determine the reliability of the iTRAQ analysis
data, western blot and CoIP analyses were performed to confirm
whether PIF interacts with MYH10, TUBB5 and HSPD1. As expected,
MYH10, TUBB5 and HSPD1 were captured when using PIF as the bait
protein (Fig. 4A-E).
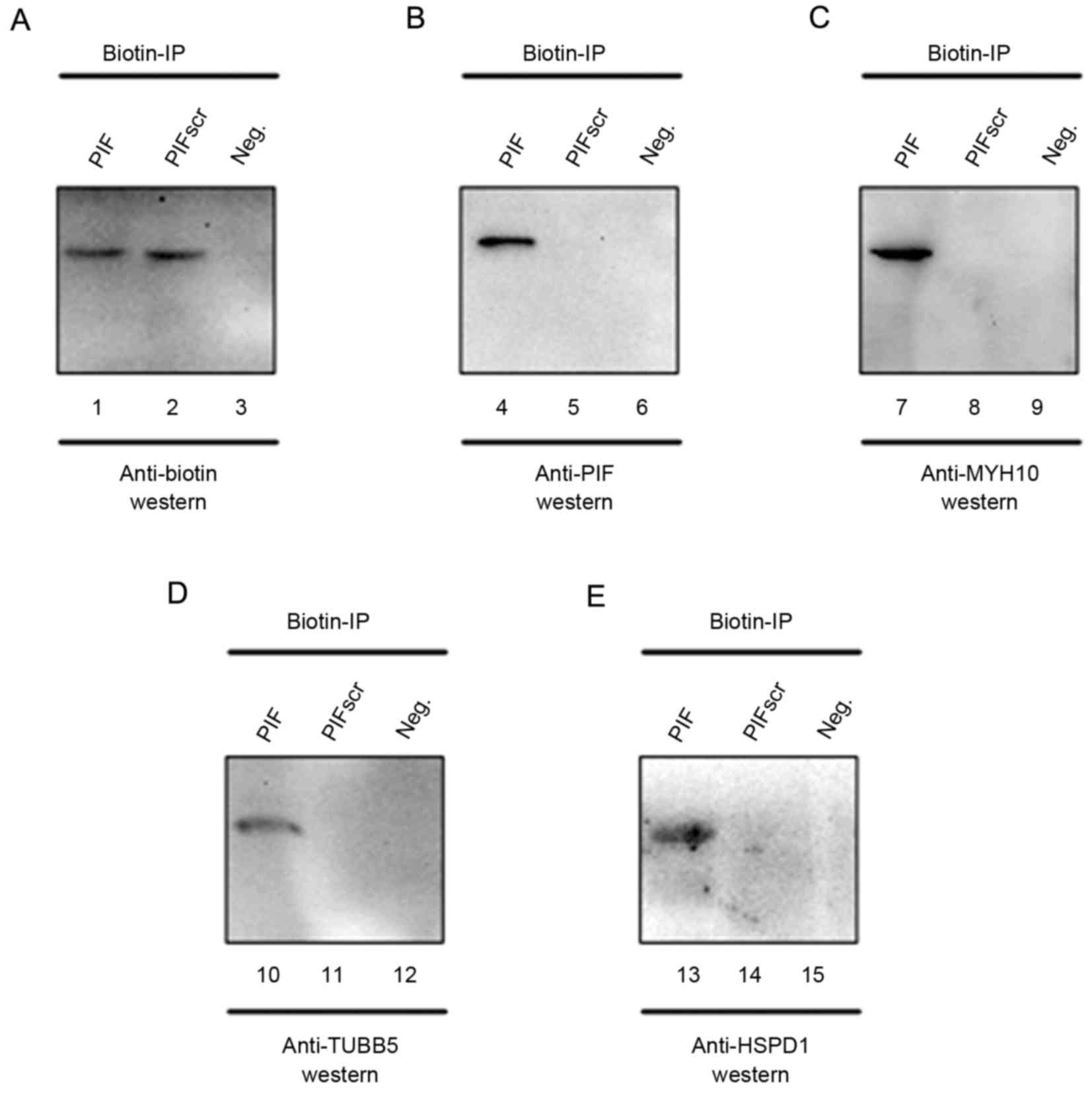 | Figure 4.Biochemical purification and
identification of PIF-associated protein factors. PIF protein
complexes from cytoplasmic extracts of HEK293 cells were mixed with
biotin-labeled PIF (lanes 1, 4, 7, 10 and 13) or PIFscr (lanes 2,
5, 8, 11 and 14). In lanes 3, 6, 9, 12 and 15, no biotin-labeled
PIF or PIFscr was added. Anti-biotin IPs were analyzed using
western blot analysis with (A) anti-biotin (lanes 1–3), (B)
anti-PIF (lanes 4–6), (C) anti-MYH10 (lanes 7–9), (D) anti-TUBB5
(lanes 10–12) and (E) anti-HSPD1 (lanes 13–15). PIF,
preimplantation factor; PIFScr, intralipid and scrambled PIF; IP,
immunoprecipitate; MYH10, and myosin heavy chain 10; TUBB5,
tubulin, β5 class I; HSPD1, heat shock protein family D1; Neg.,
negative control. |
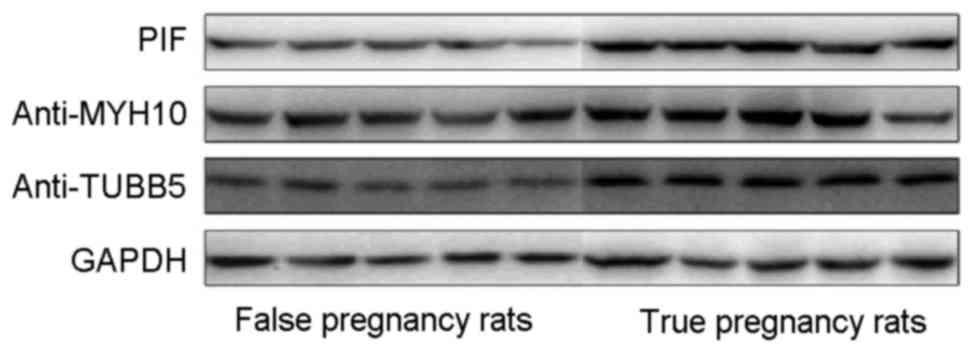
Expression of MYH10, TUBB5 and HSPD1
in tissues
Western blot analysis was used to assess the
expression of MYH10, TUBB5 and HSPD1 in the endometrial tissues of
the experimental rats. The results indicated that MYH10, TUBB5 and
HSPD1 were upregulated in the endometrial tissues of the pregnant
rats (Fig. 5).
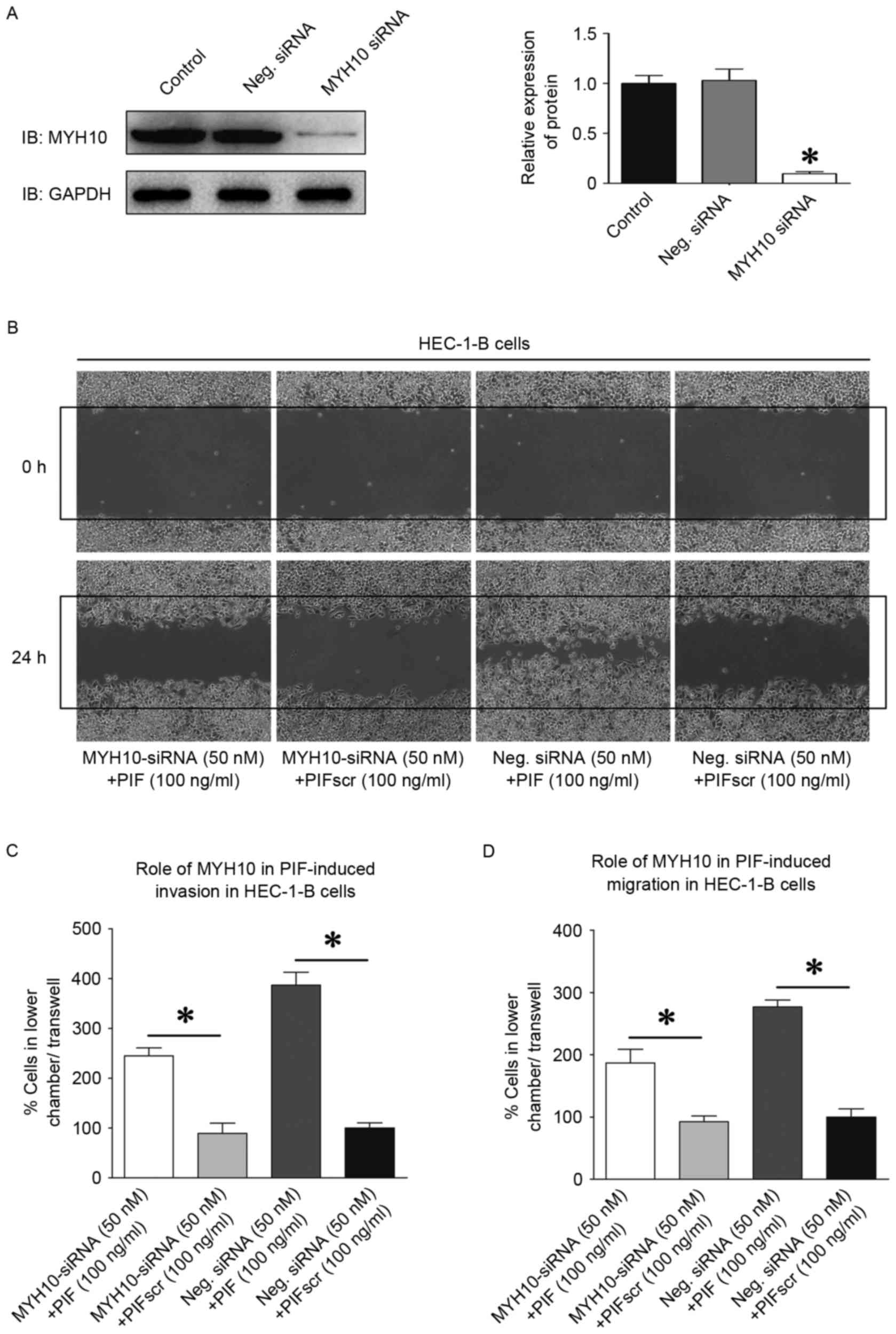
MYH10 knockdown attenuates HEC-1-B
cell migration and invasion
To examine the role of MYH10 in tumor cell motility,
protein expression was inhibited in HEC-1-B cell lines using RNA
interference. The effective silencing with MYH10-specific siRNA was
confirmed in the HEC-1-B cells using western blot analysis
(Fig. 6A). Transfection of the
HEC-1-B cells with MYH10-specific siRNA resulted in a 57–70%
decrease in the ability to close scratch wounds, compared with the
cells in the control siRNA group (P<0.05; Fig. 6B). The Matrigel invasion assay also
revealed that MYH10-specific siRNAs decreased the invasiveness of
HEC-1-B cells by 47–52%, and the migration capacity of the HEC-1-B
cells by 45–51%, compared with the controls (P<0.05 Fig. 6C and D).
Discussion
Placental development is dependent on adequate
invasion of the first-trimester trophoblast into the maternal
decidua to sufficiently remodel maternal spiral arteries (21). By contrast, incomplete invasion has
been implicated in a variety of adverse pregnancy outcomes,
including fetal loss, fetal growth restriction and preeclampsia
(22). The implantation of embryos
into the maternal endometrium and placental formation with
trophoblast invasion require a complex interplay of embryo-derived
cellular signaling for establishing maternal immune receptivity,
and for preparing the embryo itself to be suitable for invasion
(14,23). As an embryo-specific peptide, PIF
is produced as early as the two-cell stage (11), and is understood to facilitate
trophoblast invasion, eventually affecting placental development in
the peri-implantation period (14).
In the present study, a protein profile was used to
elucidate the molecular mechanism by which PIF promotes trophoblast
invasion. Utilizing the iTRAQ proteomics approach, proteins
interacting with PIF were identified. The use of
immunoprecipitation confirmed that MYH10, TUBB5 and HSPD1
potentially interacted with PIF. Functional investigations
indicated that MYH10 tended to act on the migration and invasion of
HTR-8 trophoblast cells. The results of the present study revealed
that the iTRAQ method for large-scale protein quantification was
credible and amenable to high throughput investigations, and that
the novel proteins uncovered may describe the PIF-relative
interaction network in promoting trophoblast invasion, thus
advancing the knowledge of its functional mechanism.
Certain aspects of the implantation process resemble
tumor invasion, whereas the implantation process consists of a
precisely controlled series of events. The initiation of
implantation requires that the trophoblast attaches, via its apical
plasma membrane, to the apical plasma membrane of the uterine
epithelium. As apical plasma membranes of epithelia are usually
non-adhesive, cells can express the mesenchymal/fibroblastoid
phenotype to allow the cells to move individually, or express the
epithelioid phenotype to migrate as sheets. The trophoblast of
blastocysts alters its motility apparatus to accommodate the
invasion process. Collective cell migration requires the dynamic
reorganization of cell-cell junction complexes and associated
cytoskeletal structures to allow cells to alter their positions
without losing cell-cell contacts. The invasion of trophoblast
cells occurs through the stroma in a regulated manner by the
remodeling of the extracellular matrix. A specialized submembrane
filamentous network supports stable cell-cell binding between these
cells (24). Myosin II is an
actin-binding protein, which is central in the control of cell
adhesion, cell migration and tissue architecture. Acting as an
actin cytoskeleton, MYH10 regulates cell polarity, adhesion and
migration. MYH10 can also affect cancer progression via the initial
acquisition of malignant properties by normal cells, invasion of
adjacent tissues and metastasis to distant sites (25,26).
These processes involve the dynamic remodeling of the actin
cytoskeleton and the interaction of the cell with its environment.
MYH10 facilitates protrusion formation via the generation of
retrograde flow of actin in the lamellum, which is connected to the
lamellipodium (27,28). Inhibition of the activity of MYH10
with blebbistatin, or the genetic deletion of MYH10, markedly
decreases the rate of actin retrograde flow in the lamellum
(27,28) and inhibits the coalescence of actin
into proto-bundles at the lamellipodium-lamellum interface
(29,30), which increases protrusiveness.
According to the role of MYH10 in regulating metastasis, actin
cytoskeleton-mediated cell migration and invasion can support the
PIF-induced promotion of trophoblast invasion.
The present study identified several potential
molecules to assist in explaining the mechanism of PIF-mediated
trophoblast invasion. Taken together, the interaction between PIF
and MYH10 significantly enhanced the invasion and migration
capabilities of HTR-8 trophoblast cells. These findings, coupled
with the known embryonic effects of sPIF, suggest that this peptide
requires investigation to promote or rescue placental invasion, and
are a significant advance in reproductive technology.
Acknowledgements
The present study was supported by the grant from
the National High Technology Research and Development Program of
China (863 Program; grant no. 2014AA022209), the Natural Science
Foundation Project of CQ CSTC (grant no. 2012jjA10064), the Natural
Science Foundation Project of CQ CSTC (grant no. 2013jcyj A 10060),
the class General Financial Grant from the China Postdoctoral
Science Foundation (grant no. 2012M511912), and the Chongqing
Postgraduate Research Innovation Project (grant no. CYB15091).
References
|
1
|
Carlsen E, Giwercman A, Keiding N and
Skakkebaek NE: Evidence for decreasing quality of semen during past
50 years. BMJ. 305:609–613. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeyneloglu HB and Onalan G: Remedies for
recurrent implantation failure. Semin Reprod Med. 32:297–305. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kulak D, Jindal SK, Oh C, Morelli SS,
Kratka S and McGovern PG: Reporting in vitro fertilization cycles
to the society for assisted reproductive technology database: Where
have all the cycles gone? Fertil Steril. 105:927–931. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paulson RJ, Sauer MV and Lobo RA: Embryo
implantation after human in vitro fertilization: Importance of
endometrial receptivity. Fertil Steril. 53:870–874. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Margalioth EJ, Ben-Chetrit A, Gal M and
Eldar-Geva T: Investigation and treatment of repeated implantation
failure following IVF-ET. Hum Reprod. 21:3036–3043. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tabibzadeh S and Babaknia A: The signals
and molecular pathways involved in implantation, a symbiotic
interaction between blastocyst and endometrium involving adhesion
and tissue invasion. Hum Repro. 10:1579–1602. 1995. View Article : Google Scholar
|
|
7
|
Somerset DA, Zheng Y, Kilby MD, Sansom DM
and Drayson MT: Normal human pregnancy is associated with an
elevation in the immune suppressive CD25+ CD4+ regulatory T-cell
subset. Immunology. 112:38–43. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fuzzi B, Rizzo R, Criscuoli L, Noci I,
Melchiorri L, Scarselli B, Bencini E, Menicucci A and Baricordi OR:
HLA-G expression in early embryos is a fundamental prerequisite for
the obtainment of pregnancy. Eur J Immunol. 32:311–315. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cavanagh AC and Morton H: The purification
of early-pregnancy factor to homogeneity from human platelets and
identification as chaperonin 10. Eur J Biochem. 222:551–560. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Minhas BS, Ripps BA, Zhu YP, Kim HN,
Burwinkel TH and Gleicher N: Platelet activating factor and
conception. Am J Reprod Immunol. 35:267–271. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roussev RG, Coulam CB and Barnea ER:
Development and validation of an assay for measuring
preimplantation factor (PIF) of embryonal origin. Am J Reprod
Immunol. 35:281–287. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barnea ER, Simon J, Levine SP, Coulam CB,
Taliadouros GS and Leavis PC: Progress in characterization of
pre-implantation factor in embryo cultures and in vivo. Am J Reprod
Immunol. 42:95–99. 1999.PubMed/NCBI
|
|
13
|
Stamatkin CW, Roussev RG, Stout M,
Absalon-Medina V, Ramu S, Goodman C, Coulam CB, Gilbert RO, Godke
RA and Barnea ER: PreImplantation Factor (PIF) correlates with
early mammalian embryo development-bovine and murine models. Reprod
Biol Endocrinol. 9:632011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duzyj CM, Barnea ER, Li M, Huang SJ,
Krikun G and Paidas MJ: Preimplantation factor promotes first
trimester trophoblast invasion. Am J Obstet Gynecol. 203:402.e1–e4.
2010. View Article : Google Scholar
|
|
15
|
Jovanović M and Vićovac L: Interleukin-6
stimulates cell migration, invasion and integrin expression in
HTR-8/SVneo cell line. Placenta. 30:320–328. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barnea ER, Kirk D, Ramu S, Rivnay B,
Roussev R and Paidas MJ: PreImplantation Factor (PIF) orchestrates
systemic antiinflammatory response by immune cells: Effect on
peripheral blood mononuclear cells. Am J Obstet Gynecol.
207:313.e1–11. 2012. View Article : Google Scholar
|
|
17
|
Yang Y, Lim SK, Choong LY, Lee H, Chen Y,
Chong PK, Ashktorab H, Wang TT, Salto-Tellez M, Yeoh KG and Lim YP:
Cathepsin S mediates gastric cancer cell migration and invasion via
a putative network of metastasis-associated proteins. J Proteome
Res. 9:4767–4778. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tong SW, Yang YX, Hu HD, An X, Ye F, Hu P,
Ren H, Li SL and Zhang DZ: Proteomic investigation of
5-fluorouracil resistance in a human hepatocellular carcinoma cell
line. J Cell Biochem. 113:1671–1680. 2012.PubMed/NCBI
|
|
19
|
Yang Y, Toy W, Choong LY, Hou P, Ashktorab
H, Smoot DT, Smoot DT, Yeoh KG and Lim YP: Discovery of SLC3A2 cell
membrane protein as a potential gastric cancer biomarker:
Implications in molecular imaging. J Proteome Res. 11:5736–5747.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
She S, Xiang Y, Yang M, Ding X, Liu X, Ma
L, Liu Q, Liu B, Lu Z, Li S, et al: C-reactive protein is a
biomarker of AFP-negative HBV-related hepatocellular carcinoma. Int
J Oncol. 47:543–554. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Orsi NM and Tribe RM: Cytokine networks
and the regulation of uterine function in pregnancy and
parturition. J Neuroendocrinol. 20:462–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lala PK and Chakraborty C: Factors
regulating trophoblast migration and invasiveness: Possible
derangements contributing to pre-eclampsia and fetal injury.
Placenta. 24:575–587. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
von Rango U: Fetal tolerance in human
pregnancy-a crucial balance between acceptance and limitation of
trophoblast invasion. Immunol Lett. 115:21–32. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Denker HW: Implantation: A cell biological
paradox. J Exp Zool. 266:541–558. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Conti MA and Adelstein RS: Nonmuscle
myosin II moves in new directions. J Cell Sci. 121:11–18. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ouderkirk JL and Krendel M: Non-muscle
myosins in tumor progression, cancer cell invasion, and metastasis.
Cytoskeleton (Hoboken). 71:447–463. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ponti A, Machacek M, Gupton SL,
Waterman-Storer CM and Danuser G: Two distinct actin networks drive
the protrusion of migrating cells. Science. 305:1782–1786. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Giannone G, Dubin-Thaler BJ, Rossier O,
Cai Y, Chaga O, Jiang G, Beaver W, Döbereiner HG, Freund Y, Borisy
G and Sheetz MP: Lamellipodial actin mechanically links myosin
activity with adhesion-site formation. Cell. 128:561–575. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anderson TW, Vaughan AN and Cramer LP:
Retrograde flow and myosin II activity within the leading cell edge
deliver F-actin to the lamella to seed the formation of graded
polarity actomyosin II filament bundles in migrating fibroblasts.
Mol Biol Cell. 19:5006–5018. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nemethova M, Auinger S and Small JV:
Building the actin cytoskeleton: Filopodia contribute to the
construction of contractile bundles in the lamella. J Cell Biol.
180:1233–1244. 2008. View Article : Google Scholar : PubMed/NCBI
|