Introduction
Systemic lupus erythematosus (SLE) is a fatal
multisystem autoimmune disease with diverse manifestations, which
may lead to severe morbidity and mortality. It is characterized by
a wide profile of autoantibodies, widespread precipitation of
immune complexes and aberrant innate and adaptive immune responses.
SLE preferentially invades skin, joints, kidneys and nervous system
leading to various typical symptoms (1–3).
Recent studies have revealed that genetic factors have an important
role in the susceptibility to SLE (4–6).
However, the precise pathogenic mechanism of SLE remains to be
elucidated. Therefore, it is important to identify the molecular
mechanisms of occurrence and development of SLE.
High-throughput sequencing and microarray technology
have been used to screen for differential gene expression in
disease. Previous gene expression profiling analyses have been
conducted to identify differentially expressed genes (DEGs) and
cellular pathways in SLE (7,8).
However, at present, no specific gene has been identified to act as
a potential diagnosis marker in SLE. The large quantity of data
obtained from high-throughput sequencing and microarray technology
have not been fully used. Ducreux et al (9) collected whole blood samples from SLE
patients and healthy volunteers in order to identify DEGs. However,
interactions among DEGs and key genes involved in signal conduction
pathway of SLE remain to be elucidated. In addition, previous
studies of genetic factors primarily focused on a single gene;
however, interactions among multiple genes may result in the
characteristics of multisystem invasion observed in SLE (10,11).
It is of note that disease-associated gene expression networks have
potential roles in immune response, which highlights their
mechanism and therapeutic values for SLE (12,13).
Therefore, the combination of microarray technology and
bioinformatics analysis may aid in the identification of the gene
regulatory networks, key genes and their associated pathways in
SLE.
The present study analyzed the raw data from the
GSE65391 dataset downloaded from Gene Expression Omnibus (GEO;
www.ncbi.nlm.nih.gov/geo/), which
provides free access to high-throughput gene expression datasets
(14). The present study selected
GSE65391 due to the large sample quantity of the dataset and the
transcriptional data were complemented by demographic, laboratory
and clinical data. Additionally, 24 technical replicates of healthy
samples were included and used for batch correction purposes. DEGs
between SLE and healthy controls were identified in order to
identify the potential mechanisms of SLE. Subsequently, functional
enrichment, protein-protein interaction (PPI) networks and
sub-network analyses were performed to determine the significant
pathogenic genes and their critical pathways involved in the
mechanism of occurrence and development of SLE.
Materials and methods
Microarray data
The gene expression profile of GSE65391 was
downloaded from the GEO database (15). It was established on the platform
of GPL10558 Illumina HumanHT-12 V4.0 expression bead chip (Illumina
Inc., San Diego, CA, USA). The dataset contained 996 whole blood
samples, including 924 SLE samples and 72 healthy controls.
Data preprocessing and identification
of DEGs
The original expression datasets following
background correction, normalization and probe summarization were
converted into expression measures using the R affy package
(www.bioconductor.org/packages/release/bioc/html/affy.html).
The linear models for microarray data package in Bioconductor
(www.bioconductor.org/packages/release/bioc/html/limma.html)
was used to identify DEGs according to the cut-off criteria:
Adjusted P<0.01 and |log2 fold-change
(FC)|>0.8.
Gene ontology (GO) and pathway
enrichment analysis of DEGs
GO is a widely used method for unification of
biology which collected structured, defined and controlled
vocabulary for a large scale of genes annotation (16). Kyoto Encyclopedia of Genes and
Genomes (KEGG) database is a collection of online databases
regarding gene functions, enzymatic pathways and associates genomic
information with higher-order functional information (17). The Database for Annotation,
Visualization and Integrated Discovery (DAVID; david.ncifcrf.gov) provides a comprehensive set of
functional annotation tools to identify GO terms and KEGG pathways
(18,19). To understand the biological
functions and cellular pathways of the DEGs, the present study used
DAVID to identify GO categories and KEGG pathways.
Analysis of protein-protein
interaction (PPI) network and sub-networks
Search Tool for the Retrieval of Interacting Genes
(STRING; string-db.org) (20) is a precomputed global resource
designed to evaluate PPI information. In the present study, the
STRING online tool was used to analyze the PPI of DEGs and
experimentally validated interactions with a combined score >0.4
were selected as significant. Therefore, the degree of connectivity
was statistically analyzed in networks using CytoHubba (version
3.4.0) (www.cytoscape.org) to obtain the
significant nodes or hub genes in the PPI network (21). CytoHubba, a Java plugin for
Cytoscape (version 3.4.0), is capable of predicting important nodes
and subnetworks in a given network by several topological
algorithms. The Molecular Complex Detection (MCODE) algorithm is an
established automated method to find highly interconnected modules
as molecular complexes or clusters in large PPI networks (22). Additionally, the Multi Experiment
Matrix (MEM; biit.cs.ut.ee/mem/index.cgi) (23) is a web-based multi experiment gene
expression query and visualization tool. It gathers hundreds of
publicly available gene expression datasets from the ArrayExpress
database (24). In order to
determine whether these hub genes from different probe sets were
co-expressing or not, the present study compared ISG15
ubiquitin-like modifier (ISG15) and the other hub genes using
MEM.
Results
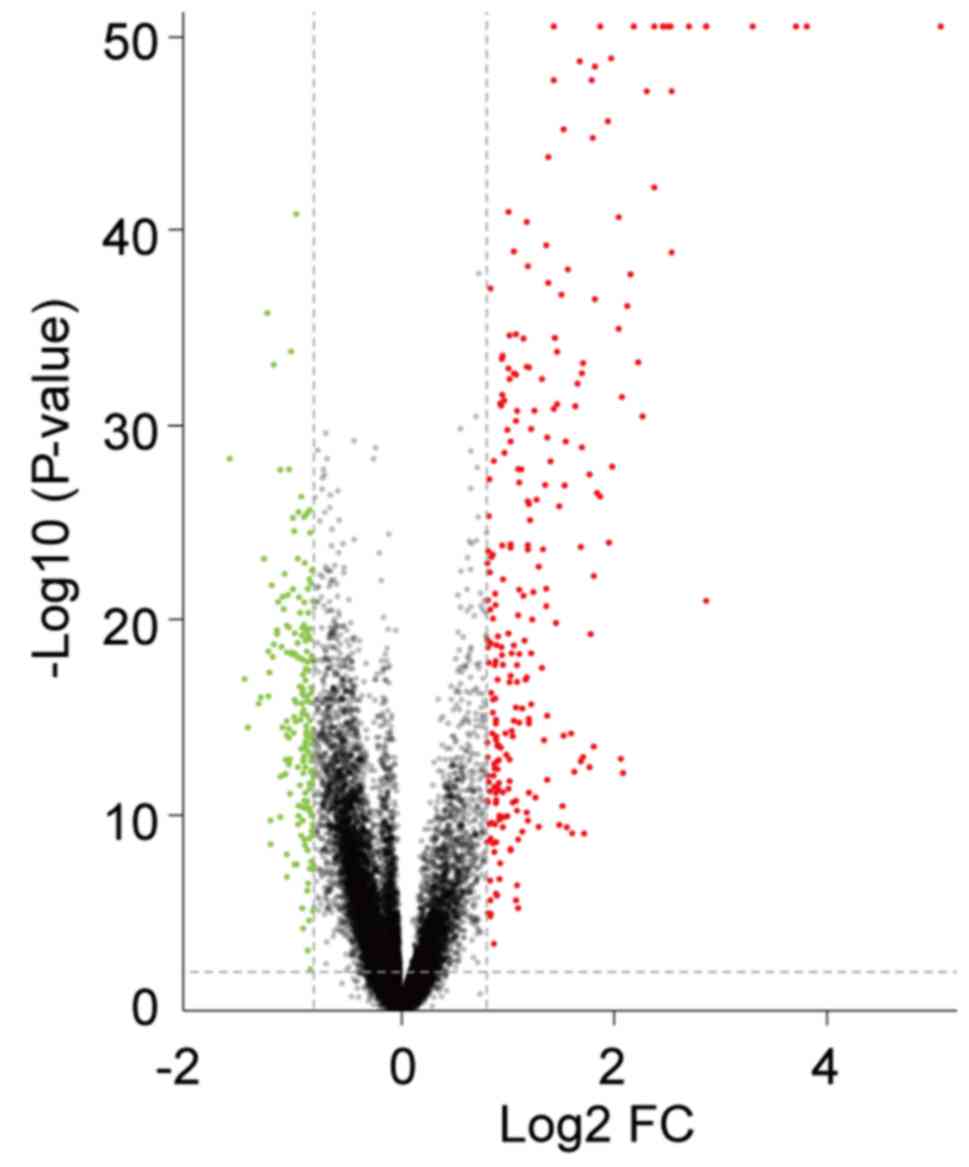
Identification of DEGs in SLE
The present study analyzed the GSE65391 microarray
data, containing 924 SLE samples and 72 healthy controls. Following
data normalization, preprocessing and filtering with the criteria
of adjusted P<0.01 and |log2FC|>0.8 a total of 310
genes were identified as differentially expressed in patients with
SLE compared with healthy controls. Among them, 193 were
upregulated and 117 were downregulated. The volcano plot of all
DEGs is presented in Fig. 1.
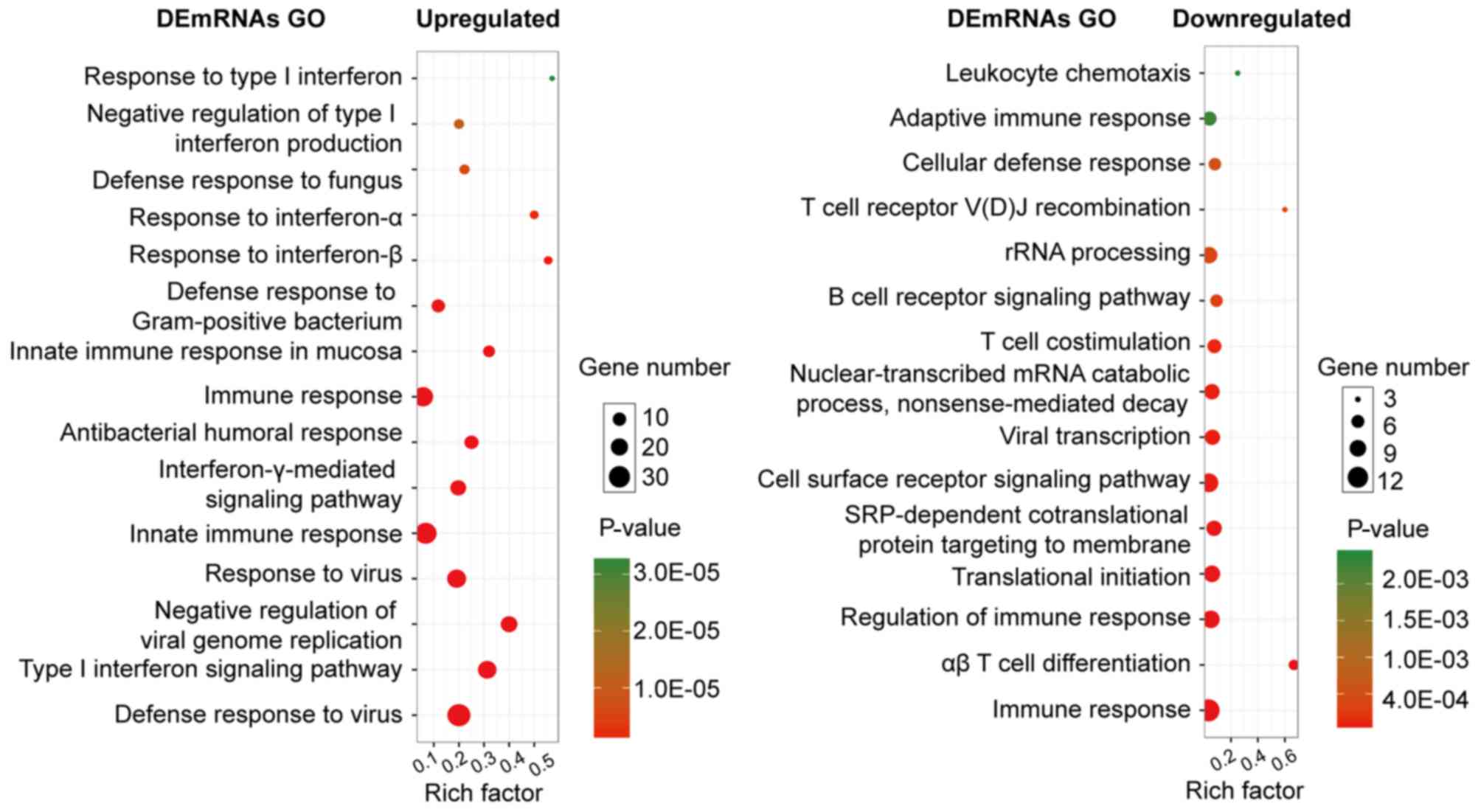
Expression and function of DEGs in
SLE
Several GO categories of upregulated and
downregulated DEGs were enriched in GO terms associated with immune
response and immune cell metabolism. The most significant
biological process (BP) of upregulated and downregulated DEGs was
the immune system process. In addition to the enrichment in immune
response, the upregulated DEGs were significantly enriched in the
BPs of the defense response to other organisms (e.g., virus),
whereas the downregulated DEGs were significantly enriched in
immune cell activities, such as leukocytes and lymphocytes
(Fig. 2).
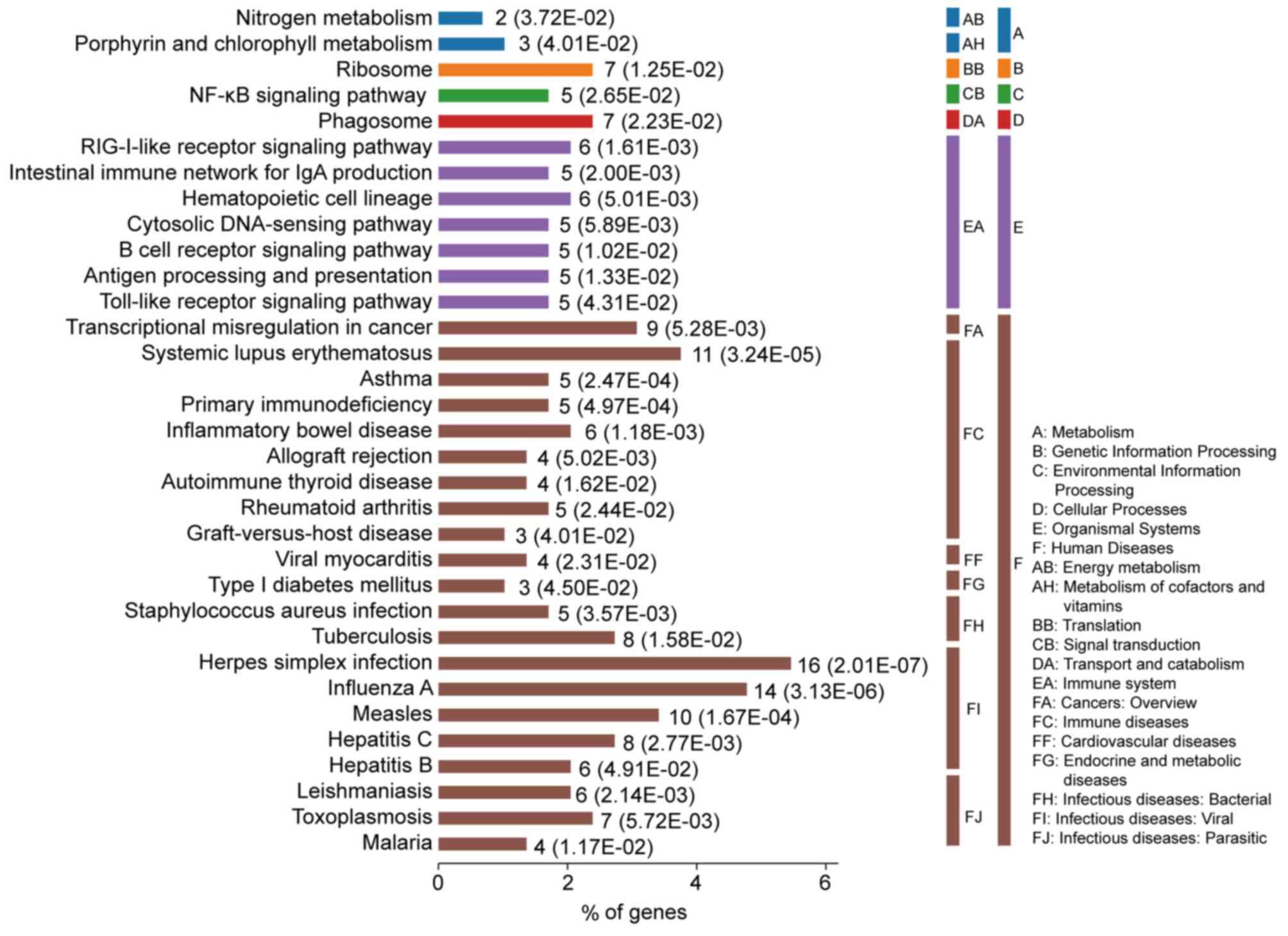
DEGs-associated pathways in SLE are
enriched in multi-system harm diseases
The most significantly enriched pathways of the DEGs
are presented in Fig. 3. The DEGs
were enriched in immune diseases and infectious diseases, including
SLE, inflammatory bowel disease, rheumatoid arthritis, herpes
simplex virus (HSV) infection, influenza A, measles, asthma and
primary immunodeficiency. Furthermore, the DEGs were also enriched
in signaling pathways associated with the immune system, including
the RIG-I-like receptor signaling pathway, intestinal immune
network for IgA, antigen processing and presentation and toll-like
receptor signaling pathway.
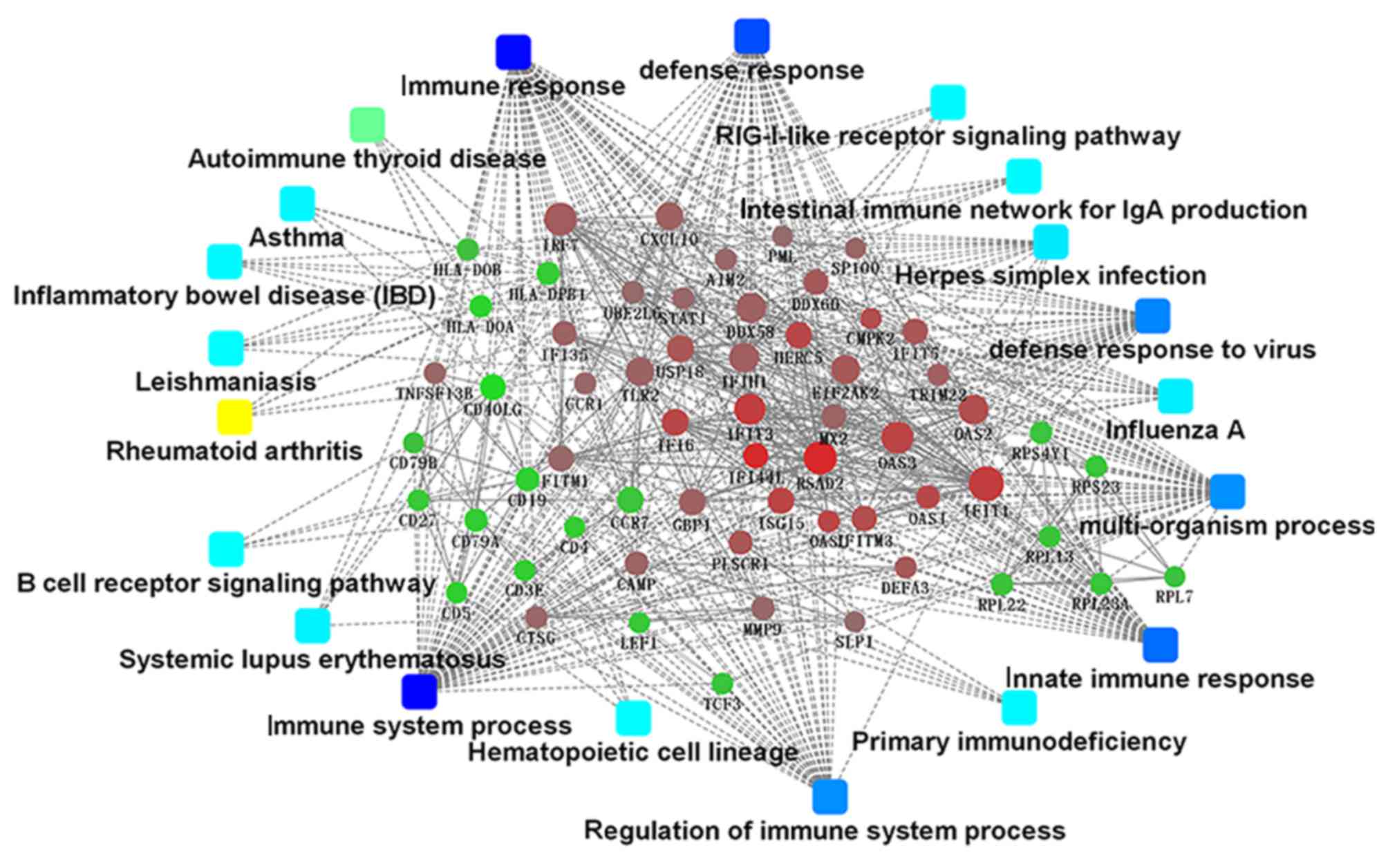
Major PPI networks in SLE
The present study identified 1,514 interactions
among the DEGs. There were 338 interactions matching the condition
of experimentally validated interaction with a combined score
>0.4. The top 15 significant interactions with highest combined
score are presented in Table I.
Based on the information in the STRING database, the PPI network
was constructed (Fig. 4). The
present study screened the top 10 genes with higher degrees as hub
genes in the PPI network using CytoHubba, including
2′-5′-oligoadenylate synthetase 1 (OAS1), MX dynamin like GTPase 2
(MX2), interferon induced protein with tetratricopeptide repeats 1
(IFIT1), interferon regulatory factor 7 (IRF7), interferon induced
with helicase C domain 1 (IFIH1), signal transducer and activator
of transcription 1 (STAT1), ISG15, DExD/H-box helicase 58 (DDX58),
interferon induced protein with tetratricopeptide repeats 3
(IFIT3), and 2′-5′-oligoadenylate synthetase 2 (OAS2).
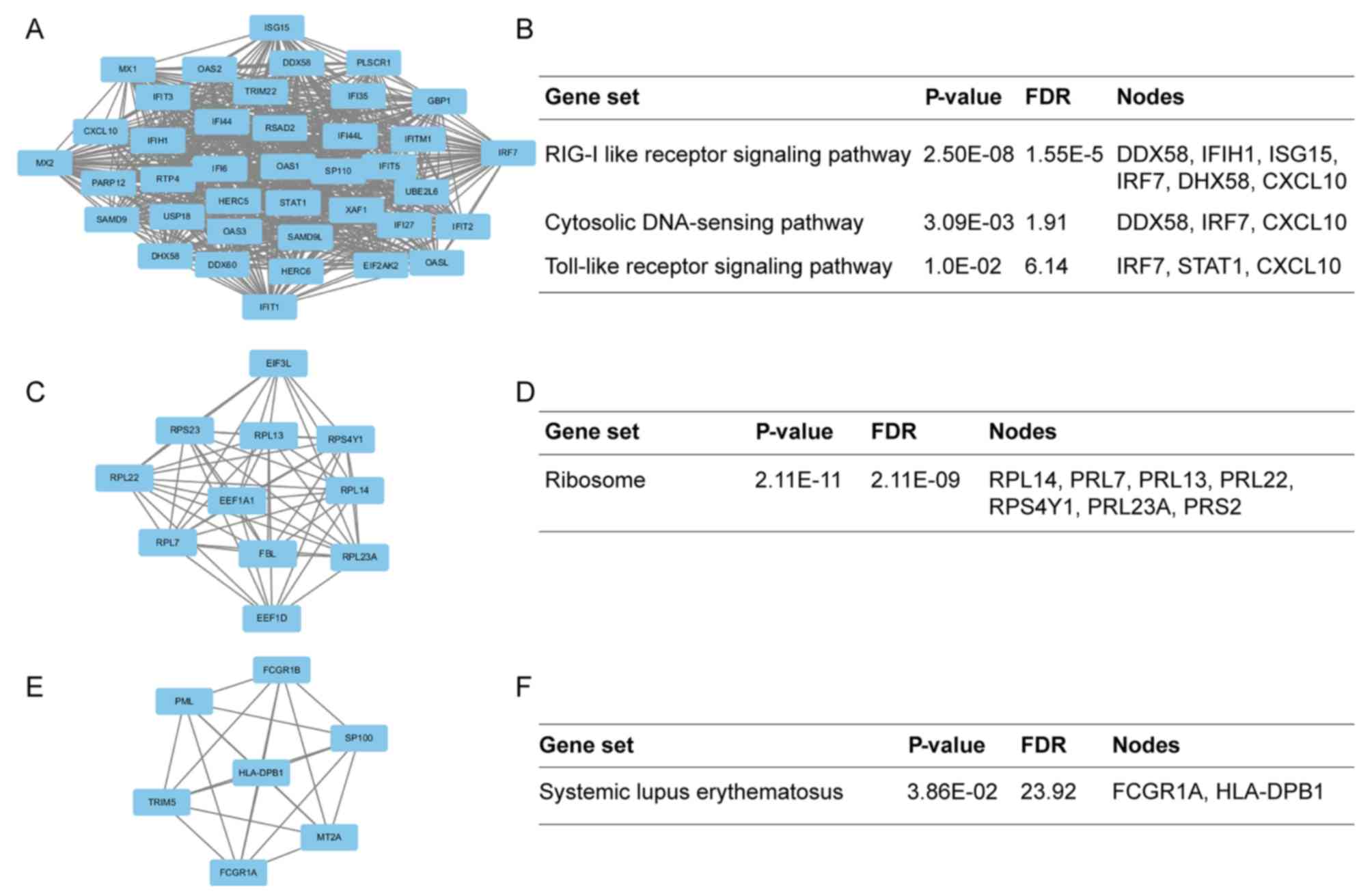
Additionally, the modules of DEGs were obtained using the MCODE
plugin. The presents study selected the top 3 significant modules
and analyzed the cellular pathways of the genes involved in the
modules (Fig. 5). The current
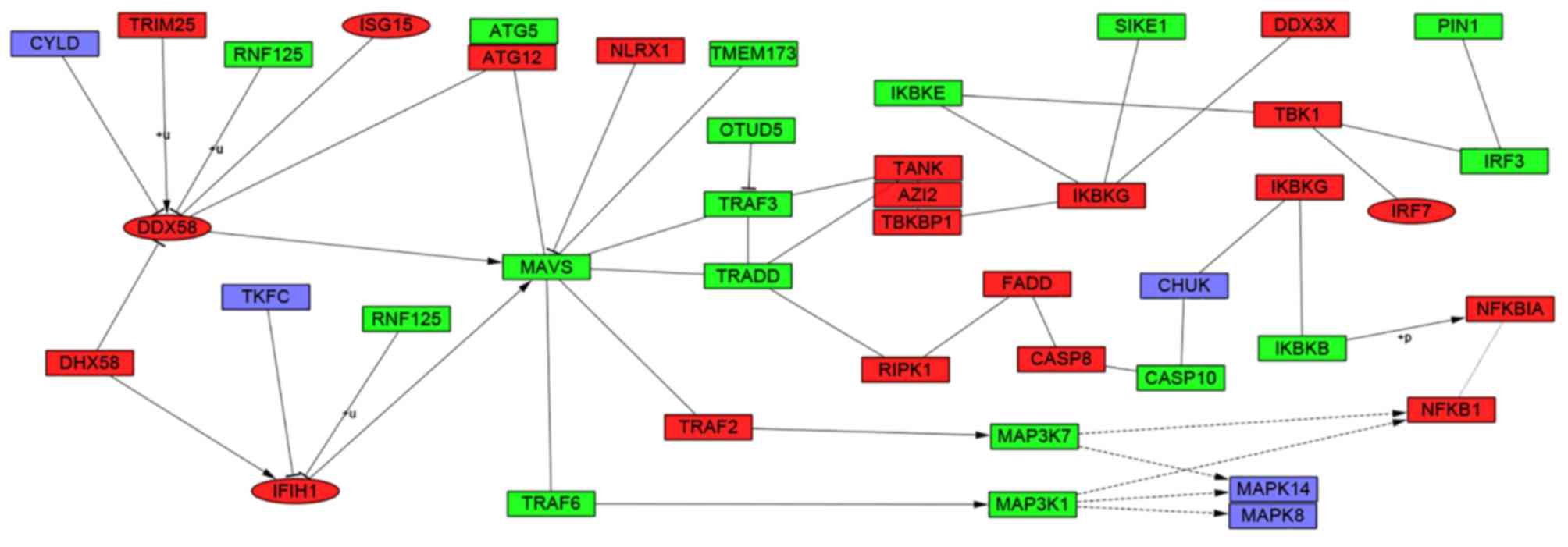
study determined that these hub genes were also involved in the
RIG-I-like receptor signaling (Fig.
6), cytosolic DNA-sensing, toll-like receptor signaling and
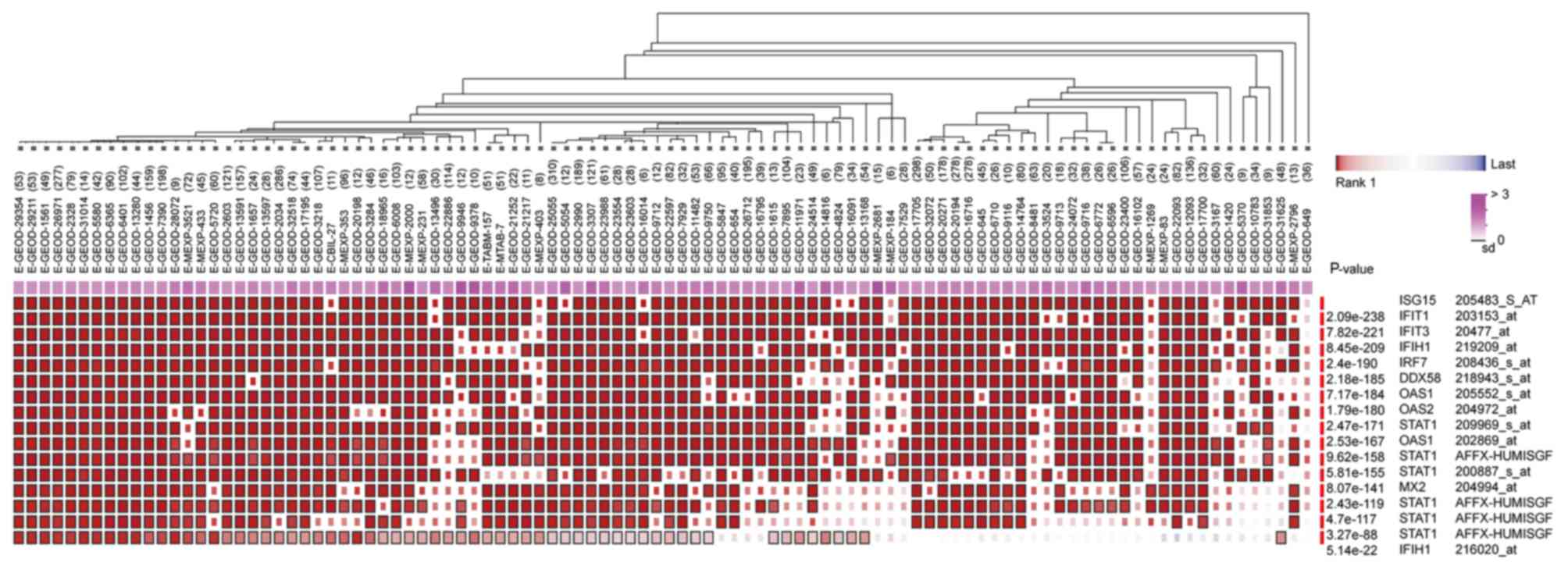
ribosome biogenesis pathways. In addition, it was determined that
the hub genes from different probe sets exhibited significant
co-expression tendency in multi-experiment microarray datasets
(P<0.01; Fig. 7).
 | Table I.Top 15 significant interactions
experimentally validated with highest combined score. |
Table I.
Top 15 significant interactions
experimentally validated with highest combined score.
| Node1 | Node2 | Combined score |
|---|
| RPL23A | RPL7 | 0.999 |
| RPL23A | RPS23 | 0.999 |
| RPL23A | RPL14 | 0.999 |
| RPL14 | RPL7 | 0.999 |
| RPL14 | RPL13 | 0.999 |
| RPL7 | RPL13 | 0.999 |
| RPL23A | RPL13 | 0.999 |
| RPL7 | RPS23 | 0.999 |
| ISG15 | USP18 | 0.999 |
| RPS23 | RPS4Y1 | 0.999 |
| RPL14 | RPS23 | 0.999 |
| RPL13 | RPS23 | 0.999 |
| RPL23A | RPL22 | 0.999 |
| HIST1H2AC | HIST1H2BD | 0.999 |
| RPL22 | RPL14 | 0.998 |
Discussion
The cause of SLE has not been fully elucidated. It
is believed to involve hormonal, environmental and genetic factors.
However, genetic factors are essential to determine the process of
SLE occurrence and development (25). However, no single causal gene has
been identified. Instead, multiple genes may influence a person's
chance of developing SLE when triggered by environmental factors
(26). Microarray and
high-throughput sequencing may be used to quantify the expression
levels of large numbers of genes simultaneously, combined with
bioinformatics analysis, this may allow for the identification of
the associated pathways and key genes with complex diseases. A
total of 310 DEGs that were differently expressed in SLE compared
with healthy controls in the present study using the gene
expression profile contained in GSE65391.
According to BPs identified of the identified
SLE-associated DEGs, a significant portion of upregulated DEGs were
enriched in response to virus, type I interferon signaling pathway
and negative regulation of viral genome replication. Downregulated
DEGs were primarily involved in the immune response, alpha-beta T
cell differentiation and regulation of immune response. These
findings were in accordance with the features of immune
abnormalities belonging to autoimmune diseases. Virus infection is
also one of the primary reasons for onset of SLE in patients.
Additionally, the DEGs were mainly enriched in pathways associated
with susceptibility to immune diseases and infectious diseases
including HSV, influenza A, SLE, measles, inflammatory bowel
disease, tuberculosis and rheumatoid arthritis. The DEGs were also
involved in various signaling pathways, such as RIG-I-like receptor
signaling, cytosolic DNA-sensing and toll-like receptor signaling
pathways. These findings were consistent with the knowledge that
SLE is a type of multiple systemic autoimmune disease which may
affect many organ systems including the skin, joints and internal
organ (27).
Based on the PPI network, the present study
identified the top 10 hub genes: OAS1, MX2, IFIT1, IRF7, IFIH1,
STAT1, ISG15, DDX58, IFIT3 and OAS2. Further analysis revealed that
these hub genes, from different probe sets, exhibited significant
co-expression tendency in multi-experiment microarray datasets
(P<0.01), which indicated that all of these hub genes may work
together in the pathogenesis of SLE. OAS1, an IFN-regulated gene,
had the highest degree of connectivity, is expressed as part of the
innate immune response to viruses and enriched in HSV infection,
hepatitis C, measles, influenza A and the NOD-like receptor
signaling pathway. Rios et al (28) reported that OAS1 may activate RNase
L leading to degradation of cellular and viral RNA, inhibiting thus
viral replication. Previous studies revealed that viral infection
and antiviral immunity contributed to the development of
autoimmunity (29). The second hub
gene MX2 was primarily enriched in interferon-g signaling and
immune system pathways. GO annotations revealed that MX2 has a BP
of GTP binding and GTPase activity. Fernández-Trujillo et al
(30) reported that MX2 protein
has an antiviral activity against RNA and DNA viruses in
vitro. The third hub gene IFIT1, one of IFN-induced antiviral
proteins, was the first gene described as a candidate gene for SLE
(31). It has been previously
reported that IFIT1 may interact with Rho/Rac guanine nucleotide
exchange factor, regulate the activation of Rho/Rac proteins and
contribute to the pathogenesis of SLE (32). IRF7, a member of the interferon
regulatory transcription factor (IRF) family, was involved in
multifarious functions and signaling pathways associated with the
immune system, including the toll-like receptor signaling, NOD-like
receptor signaling, RIG-I-like receptor signaling and cytosolic
DNA-sensing pathways. Furthermore, IRF7 may efficiently activate
IFN-β and the IFN-α and mediate their induction in the
virus-activated, myeloid differentiation primary response 88
(MyD88)-independent pathway and the toll-like receptor-activated,
MyD88-dependent pathway. Previous studies had demonstrated that the
IFN system had an important role in the etiopathogenesis of SLE
(33,34). IFIH1, a member of the RIG-I like
receptor (RLR) family, was involved in immune system and cytosolic
sensors of pathogen-associated DNA pathway. Lincez et al
(35) reported that reduced
expression of IFIH1 may prevent autoimmune diabetes. In addition, a
previous study demonstrated that the gene polymorphism of IFIH1 was
associated with increased sensitivity to IFN-α and serologic
autoimmunity in patients with lupus (36). Another hub gene, STAT1, was
reported to be involved in perturbing regulatory T cell
homeostasis, which may be a possible marker of SLE disease severity
by augmenting STAT1 signaling (37). ISG15, a member of the Ub1 family,
is a 17 kDa secreted protein and has a significant sequence
homology to ubiquitin (38). ISG15
may induce the synthesis and secretion of IFN-γ from
B-cell-depleted lymphocytes. It has been previously reported that
ISG15 acts as a cytokine modulator in immune response (39,40).
It has also been previously reported that IFN-α/IFN-γ may induce
the activation of the JAK-STAT1 pathway, regulate proliferation and
activation of immunocytes and lead to the abnormal activation of
the immune system, which may affect the occurrence and development
of SLE (41–43). Therefore, ISG15 may be involved in
the abnormal immune response of SLE. DDX58 was also enriched in the
RIG-I-like receptor signaling pathway and had a synthetic function
as viral double-stranded RNA recognition and the regulation of
immune response. However, the specific function of DDX58 in SLE
requires further investigation. IFIT3 and OAS2 are
interferon-inducible genes whose encoding proteins are involved in
the innate immune response to viral infection, which may lead to
poor prognosis of SLE.
Through module analysis of the PPI network, the
present study determined that the development of SLE was closely
associated with the RIG-I-like receptor signaling, cytosolic
DNA-sensing, toll-like receptor signaling and ribosome pathways. A
previous study proposed that patients with SLE had an increased
expression of type I IFN and the production of IFN depended on the
RIG-I-like receptor signaling pathway (44). The RIG-I-like receptor signaling
pathway could also trigger innate immune responses against viral
infections that may limit viral replication and stimulate adaptive
immunity (45). Further analysis
revealed that 4 hub genes were involved in the RIG-I-like receptor
signaling pathway and had core positions in the pathway (Fig. 6). The cytosolic DNA-sensing pathway
was reported to signal via stimulator of interferon genes that
mediate immunity to pathogens and promoted autoimmune pathology in
DNase II- and DNase III-deficient mice. Previous studies declared
that polymorphisms of genes involved in toll-like receptor
signaling pathway had a link with the development of SLE (46,47).
Module 2 was presented in Fig. 5
and enriched in ribosome biogenesis pathway. This pathway was
involved in pre-rRNA processing, ribosome assembly and the
synthetic processing of >200 proteins. Ribosomal P protein was a
main component part of ribosome (48). Additionally, molecular biomarkers
for the diagnosis of SLE were anti-ribosomal P antibodies (49), which was sufficient to confirm that
this pathway had a significant effect on the development of
SLE.
In conclusion, these hub genes may have various
roles in the occurrence and development of the SLE, leading to
damage of multiple systems in SLE. Combined with bioinformatics
analysis, the current study identified key genes and cellular
pathways involved in the occurrence and development of SLE. The
present study may provide a basis for an improved understanding of
SLE. However, the current findings are limited by the lack of
experimental verification in vivo and in vitro.
Therefore, future experimental studies should be conducted to
confirm the expression and function of the identified genes at the
protein level, which may be an area of future research.
Acknowledgements
The present study was supported by grants from
Chinese National Natural Science Foundation Grant (grant nos.
81671541, 81273202 and 31400773), Clinical Medicine Science &
Technology Project of Jiangsu Province of China (grant no.
BL2013024), Postgraduate Research & Practice Innovation Program
of Jiangsu Province (grant no. KYCX17_1820) and China Scholarship
Council, Program of Innovative Research Team of Jiangsu Province,
Project of the Priority Academic Program Development of Jiangsu
Higher Education Institutions and Project of the Key Academic
Program Development of Jiangsu University (grant no.
1291270019).
Glossary
Abbreviations
Abbreviations:
|
SLE
|
systemic lupus erythematosus
|
|
DEG
|
differentially expressed genes
|
|
GO
|
gene ontology
|
|
PPI
|
protein-protein interaction
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
BP
|
biological process
|
|
FC
|
fold-change
|
|
IBD
|
inflammatory bowel disease
|
|
HSV
|
herpes simplex virus
|
|
OAS1
|
2′-5′-oligoadenylate synthetase 1
|
|
MX2
|
MX dynamin like GTPase 2
|
|
IFIT1
|
interferon induced protein with
tetratricopeptide repeats 1
|
|
IRF7
|
interferon regulatory factor 7
|
|
IFIH1
|
interferon induced with helicase C
domain 1
|
|
STAT1
|
signal transducer and activator of
transcription 1
|
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
DDX58
|
DExD/H-box helicase 58
|
|
IFIT3
|
interferon induced protein with
tetratricopeptide repeats 3
|
|
OAS2
|
2′-5′-oligoadenylate synthetase 2
|
|
IFN
|
interferon
|
References
|
1
|
Weidenbusch M, Kulkarni OP and Anders HJ:
The innate immune system in human systemic lupus erythematosus.
Clin Sci (Lond). 131:625–634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu CJ, Guo J, Luo HC, Wei CD, Wang CF, Lan
Y and Wei YS: Association of CD40 polymorphisms and haplotype with
risk of systemic lupus erythematosus. Rheumatol Int. 36:45–52.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mok CC, Kwok RC and Yip PS: Effect of
renal disease on the standardized mortality ratio and life
expectancy of patients with systemic lupus erythematosus. Arthritis
Rheum. 65:2154–2160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang W and Lau YL: Solving the genetic
puzzle of systemic lupus erythematosus. Pediatr Nephrol.
30:1735–1748. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dang J, Li J, Xin Q, Shan S, Bian X, Yuan
Q, Liu N, Ma X, Li Y and Liu Q: Gene-gene interaction of ATG5,
ATG7, BLK and BANK1 in systemic lupus erythematosus. Int J Rheum
Dis. 19:1284–1293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Liu Y, Zhao J, Xu J, Li S and Qin
X: P-glycoprotein gene MDR1 polymorphisms and susceptibility to
systemic lupus erythematosus in Guangxi population: A case-control
study. Rheumatol Int. 37:537–545. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Borrebaeck CA, Sturfelt G and Wingren C:
Recombinant antibody microarray for profiling the serum proteome of
SLE. Methods Mol Biol. 1134:67–78. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu H, Luo H, Yan M, Zuo X and Li QZ:
Autoantigen microarray for High-throughput autoantibody profiling
in systemic lupus erythematosus. Genomics Proteomics
Bioinformatics. 13:210–218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ducreux J, Houssiau FA, Vandepapelière P,
Jorgensen C, Lazaro E, Spertini F, Colaone F, Roucairol C, Laborie
M, Croughs T, et al: Interferon α kinoid induces neutralizing
anti-interferon α antibodies that decrease the expression of
interferon-induced and B cell activation associated transcripts:
Analysis of extended follow-up data from the interferon α kinoid
phase I/II study. Rheumatology (Oxford). 55:1901–1905. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith S, Fernando T, Wu PW, Seo J, Ní
Gabhann J, Piskareva O, McCarthy E, Howard D, O'Connell P, Conway
R, et al: MicroRNA-302d targets IRF9 to regulate the IFN-induced
gene expression in SLE. J Autoimmun. 79:105–111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Humrich JY and Riemekasten G: Clinical
trials: The rise of IL-2 therapy-a novel biologic treatment for
SLE. Nat Rev Rheumatol. 12:695–696. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng Y and Tsao BP: Genetic susceptibility
to systemic lupus erythematosus in the genomic era. Nat Rev
Rheumatol. 6:683–692. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bentham J, Morris DL, Graham DSC, Pinder
CL, Tombleson P, Behrens TW, Martín J, Fairfax BP, Knight JC, Chen
L, et al: Genetic association analyses implicate aberrant
regulation of innate and adaptive immunity genes in the
pathogenesis of systemic lupus erythematosus. Nat Genet.
47:1457–1464. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: CytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 Suppl 4:S112014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adler P, Kolde R, Kull M, Tkachenko A,
Peterson H, Reimand J and Vilo J: Mining for coexpression across
hundreds of datasets using novel rank aggregation and visualization
methods. Genome Biol. 10:R1392009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rustici G, Kolesnikov N, Brandizi M,
Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M,
et al: ArrayExpress update-trends in database growth and links to
data analysis tools. Nucleic Acids Res. 41(Database Issue):
D987–D990. 2013.PubMed/NCBI
|
|
25
|
Yusuf JH, Kaliyaperumal D, Jayaraman M,
Ramanathan G and Devaraju P: Genetic selection pressure in TLR9
gene may enforce risk for SLE in Indian Tamils. Lupus. 26:307–310.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Armstrong DL, Zidovetzki R,
Alarcón-Riquelme ME, Tsao BP, Criswell LA, Kimberly RP, Harley JB,
Sivils KL, Vyse TJ, Gaffney PM, et al: GWAS identifies novel SLE
susceptibility genes and explains the association of the HLA
region. Genes Immun. 15:347–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fattal I, Shental N, Mevorach D, Anaya JM,
Livneh A, Langevitz P, Zandman-Goddard G, Pauzner R, Lerner M,
Blank M, et al: An antibody profile of systemic lupus erythematosus
detected by antigen microarray. Immunology. 130:337–343. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rios JJ, Perelygin AA, Long MT, Lear TL,
Zharkikh AA, Brinton MA and Adelson DL: Characterization of the
equine 2′-5′ oligoadenylate synthetase 1 (OAS1) and ribonuclease L
(RNASEL) innate immunity genes. BMC Genomics. 8:3132007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Getts DR, Chastain EM, Terry RL and Miller
SD: Virus infection, antiviral immunity, and autoimmunity. Immunol
Rev. 255:197–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fernández-Trujillo MA, García-Rosado E,
Alonso MC, Castro D, Álvarez MC and Béjar J: Mx1, Mx2 and Mx3
proteins from the gilthead seabream (Sparus aurata) show in vitro
antiviral activity against RNA and DNA viruses. Mol Immunol.
56:630–636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Relógio A, Schwager C, Richter A, Ansorge
W and Valcárcel J: Optimization of oligonucleotide-based DNA
microarrays. Nucleic Acids Res. 30:e512002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ye S, Pang H, Gu YY, Shen N, Chen SL, Chen
XG, Hua J, Qian J and Rao ZH: Using GST-tag to capture protein
interaction of an interferon-inducible systemic lupus associated
gene, IFIT1. Zhonghua Yi Xue Za Zhi. 83:770–773. 2003.(In Chinese).
PubMed/NCBI
|
|
33
|
Rönnblom L and Alm GV: An etiopathogenic
role for the type I IFN system in SLE. Trends Immunol. 22:427–431.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rönnblom L: The importance of the type I
interferon system in autoimmunity. Clin Exp Rheumatol. 34 4 Suppl
98:S21–S24. 2016.
|
|
35
|
Lincez PJ, Shanina I and Horwitz MS:
Reduced expression of the MDA5 gene IFIH1 prevents autoimmune
diabetes. Diabetes. 64:2184–2193. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Robinson T, Kariuki SN, Franek BS, Kumabe
M, Kumar AA, Badaracco M, Mikolaitis RA, Guerrero G, Utset TO,
Drevlow BE, et al: Autoimmune disease risk variant of IFIH1 is
associated with increased sensitivity to IFN-α and serologic
autoimmunity in lupus patients. J Immunol. 187:1298–1303. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Goropevšek A, Gorenjak M, Gradišnik S, Dai
K, Holc I, Hojs R, Krajnc I, Pahor A and Avčin T: Increased levels
of STAT1 protein in blood CD4 T cells from systemic lupus
erythematosus patients are associated with perturbed homeostasis of
activated CD45RA-FOXP3hi regulatory subset and follow-up disease
severity. J Interferon Cytokine Res. 37:254–268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zuo C, Sheng X, Ma M, Xia M and Ouyang L:
ISG15 in the tumorigenesis and treatment of cancer: An emerging
role in malignancies of the digestive system. Oncotarget.
7:74393–74409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Recht M, Borden EC and Knight E Jr: A
human 15-kDa IFN-induced protein induces the secretion of
IFN-gamma. J Immunol. 147:2617–2623. 1991.PubMed/NCBI
|
|
40
|
Care MA, Stephenson SJ, Barnes NA, Fan I,
Zougman A, El-Sherbiny YM, Vital EM, Westhead DR, Tooze RM and
Doody GM: Network analysis identifies proinflammatory plasma cell
polarization for secretion of ISG15 in human autoimmunity. J
Immunol. 197:1447–1459. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong G, You M, Fan H, Ding L, Sun L and
Hou Y: STS-1 promotes IFN-α induced autophagy by activating the
JAK1-STAT1 signaling pathway in B cells. Eur J Immunol.
45:2377–2388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Han C, Fu J, Liu Z, Huang H, Luo L and Yin
Z: Dipyrithione inhibits IFN-gamma-induced JAK/STAT1 signaling
pathway activation and IP-10/CXCL10 expression in RAW264.7 cells.
Inflamm Res. 59:809–816. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Zhao S, Yi M, Hu X, Li J, Xie H, Zhu
W and Chen M: Activation of JAK-STAT1 signal transduction pathway
in lesional skin and monocytes from patients with systemic lupus
erythematosus. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 36:109–115.
2011.PubMed/NCBI
|
|
44
|
Rönnblom L and Pascual V: The innate
immune system in SLE: Type I interferons and dendritic cells.
Lupus. 17:394–399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dixit E and Kagan JC: Intracellular
pathogen detection by RIG-I-like receptors. Adv Immunol.
117:99–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chai HC, Chua KH, Lim SK and Phipps ME:
Insight into gene polymorphisms involved in toll-like
receptor/interferon signalling pathways for systemic lupus
erythematosus in South East Asia. J Immunol Res. 2014:5291672014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Castiblanco J, Varela DC,
Castaño-Rodríguez N, Rojas-Villarraga A, Hincapié ME and Anaya JM:
TIRAP (MAL) S180L polymorphism is a common protective factor
against developing tuberculosis and systemic lupus erythematosus.
Infect Genet Evol. 8:541–544. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kressler D, Hurt E and Bassler J: Driving
ribosome assembly. Biochim Biophys Acta. 1803:673–683. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tan EM, Cohen AS, Fries JF, Masi AT,
McShane DJ, Rothfield NF, Schaller JG, Talal N and Winchester RJ:
The 1982 revised criteria for the classification of systemic lupus
erythematosus. Arthritis Rheum. 25:1271–1277. 1982. View Article : Google Scholar : PubMed/NCBI
|