Introduction
Endometrial carcinoma (EC) is the sixth most common
malignant tumor occurring in females. Approximately 319,600 new EC
cases per year were estimated (1).
Although Asian women have a lower risk of EC than those residing in
the US and other western countries, the morbidity of EC in China
has substantially increased (2).
Several risk factors, including hypertension, obesity, diabetes,
postmenopausal status, infertility, family history of EC and
long-term use of oestrogen, contribute to EC formation and
progression (3,4). EC patients at the early stage
generally have a good prognosis with a 5-year survival rate of up
to 96% (5). By contrast, patients
diagnosed at advanced stage are often associated with a worse
outcome despite the recent advances in surgical treatments and
chemoradiotherapy (6,7). Therefore, determining the underlying
molecular mechanisms of EC formation and development is urgently
needed to create novel therapeutic approaches for EC and to
increase prognosis for patients with this disease.
MicroRNAs (miRNAs) are a family of non-coding RNAs
that are endogenous, short and consist of 18–25 nucleotides
(8). MiRNAs serve as regulators of
gene expression by binding to the 3′-untranslated regions (3′-UTRs)
of their target mRNAs, which leads to either mRNA degradation or
translational repression (9).
miRNAs target mRNA with a semi-complimentary seed sequence (6–9
bp), which guides binding to the response elements; hence, one
miRNA may have many potential targets because each seed sequence
may correspond to many mRNA molecules (10). Growing evidence has shown that
miRNAs play important roles in numerous cellular processes,
including cell proliferation, apoptosis, cell cycle,
differentiation, metabolism and stress response (11–13).
The deregulation and aberrant expression of miRNAs has been
reported in various human diseases, including cancer (14,15).
Downregulated miRNAs may normally act as tumor suppressor genes via
the negative regulation of oncogenes (16), whereas upregulated miRNAs may play
oncogenic roles during tumor development by repressing tumor
suppressor genes (17). These data
emphasise the importance of miRNAs in tumorigenesis and tumor
development, suggesting that the investigation of miRNAs could
provide novel therapeutic targets for anticancer treatment.
miR-381, mapped to the 14q32.31 locus, is aberrantly
expressed in multiple types of human cancer (18–20).
However, the expression pattern, biological roles and underlying
mechanisms of miR-381 in EC are poorly understood. The current
study aimed to detect the expression levels and functions of
miR-381 in EC. The molecular mechanisms involved in the association
of miR-381 with the proliferation and invasion of EC cells were
also investigated.
Materials and methods
Tissue samples
This study protocol was approved by the Ethics
Committee of the First Affiliated Hospital of Nanchang University.
Written informed consent was obtained from each patient. A total of
45 paired EC tissues and corresponding adjacent normal endometrial
tissues were collected from patients who underwent surgical
resection at the Department of Obstetrics and Gynecology, The First
Affiliated Hospital of Nanchang University between June 2014 and
September 2016. All patients had not received chemotherapy or
radiotherapy prior to surgery. Tissues were immediately frozen in
liquid nitrogen and kept in liquid nitrogen until further
analysis.
Cell culture and transfection
Human EC cell lines, HEC-1B, HEC-59, AN3CA and KLE,
were purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Ishikawa cell line was acquired from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). All cells were
grown in Dulbecco's modified Eagle's medium (DMEM) along with 10%
fetal bovine serum (FBS) (both from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and 100
mg/ml streptomycin. Cells were kept in a humidified atmosphere of
5% CO2 at 37°C.
miR-381 mimics and miRNA mimics negative control
(miR-NC) were obtained from GenePharma Co., Ltd. (Shanghai, China).
Insulin-like growth factor receptor 1 (IGF-1R) overexpression
plasmid (pcDNA3.1-IGF-1R) and empty plasmid (pcDNA3.1) were
obtained from Shanghai Genechem Co., Ltd. (Shanghai, China). One
day before transfection, cells were seeded into 6-well plates at a
density of 60–70% confluence. Cell transfection was performed using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to manufacturer's instructions.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. To quantify miR-381
expression, total RNA was reverse transcribed into cDNA using
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The qPCR was performed using
TaqMan MicroRNA PCR kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) on the Bio-Rad CFX96 Real-Time PCR system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). U6 was used as an
internal control for miR-381. To detect IGF-1R mRNA expression
levels, cDNA was synthesized using PrimeScript RT Reagent kit, and
then amplified by using SYBR Premix Ex Taq™ kit (both from Takara
Biotechnology Co., Ltd., Dalian, China) with GAPDH as an internal
control. Primers used in this assay were shown in Table I. Relative expression levels were
analyzed using the 2−∆∆Ct method (21).
 | Table I.Primers for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Sequences
(5′→3′) |
|---|
| microRNA-381 |
|
|
Forward |
GGAGCCTATACAAGGGCAAGC |
|
Reverse |
GCGAGCACAGAATAAATACGACTCACTA |
| U6 |
|
|
Forward |
CTCGCTTCGGCAGCACATATACT |
|
Reverse |
ACGCTTCACGAATTTGCGTGTC |
| IGF-1R |
|
|
Forward |
AGGATATTGGGCTTTACAACCTG |
|
Reverse |
GAGGTAACAGAGGTCAGCATTTT |
| GAPDH |
|
|
Forward |
AGAAGGCTGGGGCTCATTTG |
|
Reverse |
AGGGGCCTCCACAGTCTTC |
3-(4,5-dimethyl-2-Thiazyl)-2,5-diphenyl-2H-tetrazolium bro-mide
(MTT) assay
Cell proliferation was assessed using the MTT assay
(Sigma-Aldrich; Merck KGaA). Transfected cells were collected at 24
h post-transfection. Cells were then resuspend in DMEM medium with
10% FBS, and seeded into 96-well plates at the density of
3×103 cells/well. After incubation for 0, 24, 48 and 72
h respectively, MTT assay was performed following the
manufacturer's protocols. Briefly, 10 µl of MTT solution (5 mg/ml)
was added to each well and the plates were incubated at 37°C for 4
h. The medium was then removed carefully, and 150 µl DMSO
(Sigma-Aldrich; Merck KGaA) was added into each well. Finally, the
optical density (OD) was detected at a wavelength of 490 nm using a
microplate spectrophotometer (Multiskan FC; Thermo Fisher
Scientific, Inc.). All assays were perfomred in quintuplicate
through at least three independent experiments.
In vitro cell invasion assay
Matrigel-coated Transwell cell culture chambers (BD
Biosciences, San Jose, CA, USA) were utilized to perform in
vitro cell invasion assays. Transfected cells were harvested
after 48 h incubation, re-suspended in FBS-free DMEM medium, and
seeded into the upper chamber of Transwell chambers at a density of
5×104 cells/chamber. The lower chambers contained 500 µl
of DMEM medium supplemented with 10% FBS serving as the
chemoattractant. After 24 h incubation at 37°C and 5%
CO2, the non-invasive cells were gently removed with
cotton swabs. The invasive cells were fixed with 4%
paraformaldehyde, stained in 0.5% crystal violet and wash with PBS.
After drying in air, the invasive cells were photographed and
counted under an inverted microscope (IX71; Olympus Corporation,
Tokyo, Japan), with a magnification of ×200, 5 randomly selected
fields for each chamber.
Target prediction and luciferase
reporter assay
To predict the potential targets of miR-381,
bioinformatic analysis was performed using TargetScan (www.targetscan.org) and miRanda (www.microrna.org). IGF-1R was selected as the
candidate target of miR-381. Luciferase reporter vectors,
pmirGLO-IGF-1R-3′-UTR wild-type (Wt) and pmirGLO-IGF-1R-3′-UTR
mutant (Mut), were syntesized and confirmed by GenePharma Co., Ltd.
Cells were seeded into 24-well plates at the density of
2.0×105 cells/well. After incubation overnight, miR-381
mimics or miR-NC was transfected into cells followed by
cotransfection with pmirGLO-IGF-1R-3′-UTR Wt or
pmirGLO-IGF-1R-3′-UTR Mut, using Lipofectamine 2000, following the
manufacturer's instructions. After incubation 48 h, cell lysates
were collected and luciferase activities were examiend using the
Dual-Luciferase Reporter Assay system (Promega, Manheim, Germany)
in accordance with the manufacturer's suggestions. Each experiment
was repeated at least three times.
Western blotting analysis
Total protein Total protein was extracted form
tissues or cells using ice-cold radioimmunoprecipitation assay
buffer containing protease inhibitors, following by quantification
with a BCA protein assay kit (both from Beyotime Institute of
Biotechnology, Jiangsu, Haimen, China). Equal amounts of protein
were separated on 10% SDS-polyacrylamide gel and electrotransferred
to polyvinylidene difluoride membranes (Sigma; Merck KGaA).
Following blocking with 5% non-fat milk in Tris-buffered saline
with 0.1% Tween-20 (TBST), the membranes were incubated overnight
at 4°C with the primary antibody. Subsequently, the membranes were
washed with TBST for three times, and probed with a goat anti-mouse
horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000
dilution; sc-2005; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
at room temperature for 1 h. Immunoreactive protein bands were
visualised using an enhanced chemiluminescence system (GE
Healthcare Life Sciences, Chalfont, UK) according to the
manufacturer's protocol. The intensity of protein bands was
analyzed with Quantity One software version 4.62 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Primary antibodies used in
this research were mouse anti-human monoclonal IGF-1R antibody
(1:1,000 dilution; sc-81464; Santa Cruz Biotechnology, Inc.), mouse
anti-human monoclonal extracellular signal-regulated kinase (ERK;
sc-514302; 1:1,000 dilution), mouse anti-human monoclonal p-ERK
(sc-81492; 1:1,000 dilution), mouse anti-human monoclonal p-AKT
antibody (1:1,000 dilution; sc-271966), mouse anti-human monoclonal
AKT antibody (1:1,000 dilution; sc-81434), and mouse anti-human
monoclonal GAPDH antibody (1:1,000 dilution; sc-69778) (all from
Santa Cruz Biotechnology, Inc.). GAPDH was used as a loading
control.
Statistical analysis
Data were expressed as the mean ± SD. Statistical
analysis was performed with Students t-test or one-way ANOVA using
SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA).
Newman-Keuls method was used to compare between two groups in
multiple groups study. A P-value <0.05 was considered
statistically significant.
Results
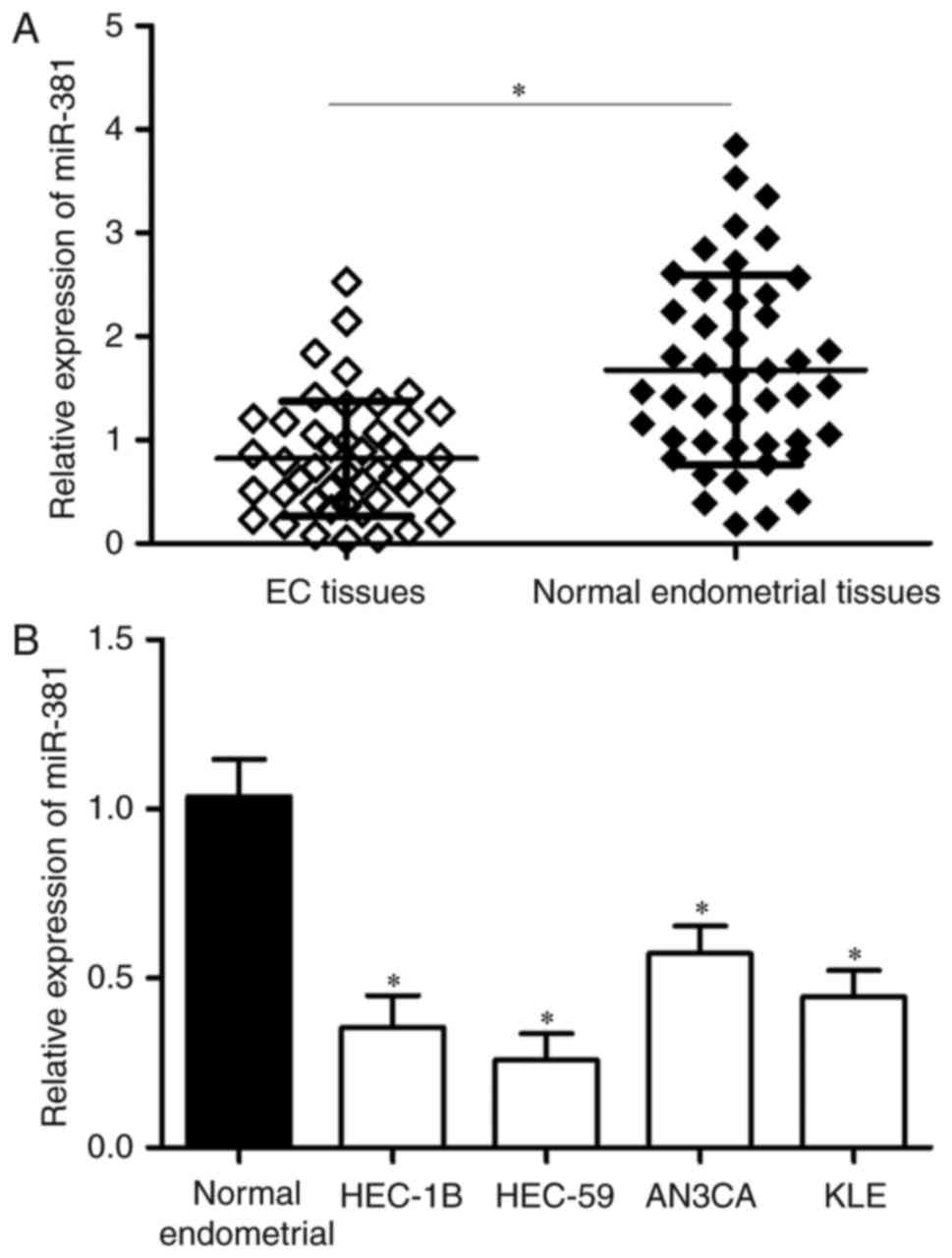
miR-381 is frequently downregulated in
EC tissues and cell lines
The expression levels of miR-381 in 45 paired EC
tissues and corresponding adjacent normal endometrial tissues were
determined by RT-qPCR. Results showed that miR-381 expression was
significantly lower in the EC tissues than in the adjacent normal
endometrial tissues (Fig. 1A,
P<0.05). EC patients were divided into two groups to analyse the
association between miR-381 and the clinicopathological features of
EC. The grouping was based on the median relative miR-381
expression value that was used for the cut-off. As shown in
Table II, the expression level of
miR-381 correlated with the FIGO stage (P=0.001), lymph nodes
metastasis (P=0.004) and myometrial invasion (P=0.021) of EC.
However, miR-381 expression showed no significant correlation with
other clinicopathological factors, including age (P=0.420) and
histological grade (P = 0.286).
 | Table II.Correlation of microRNA-381
expression with different clinicopathological factors of
endometrial carcinoma. |
Table II.
Correlation of microRNA-381
expression with different clinicopathological factors of
endometrial carcinoma.
| Clinicopathologic
factors | No. of cases | Low miR-381
group | High miR-381
group | P-value |
|---|
| Age, years |
|
|
| 0.420 |
|
<50 | 17 | 10 | 7 |
|
|
≥50 | 28 | 13 | 15 |
|
| Histological
grade |
|
|
| 0.286 |
| Well
and moderate | 25 | 11 | 14 |
|
|
Poor | 20 | 12 | 8 |
|
| FIGO stage |
|
|
| 0.001 |
|
I–II | 21 | 5 | 16 |
|
|
III–IV | 24 | 18 | 6 |
|
| Lymph node
metastasis |
|
|
| 0.004 |
| No | 25 | 8 | 17 |
|
|
Yes | 20 | 15 | 5 |
|
| Myometrial
invasion |
|
|
| 0.021 |
| No | 27 | 10 | 17 |
|
|
Yes | 18 | 13 | 5 |
|
The miR-381 expression in four EC cell lines
(HEC-1B, HEC-59, AN3CA and KLE) was also examined. As shown in
Fig. 1B, miR-381 was downregulated
in the EC cell lines compared with the adjacent normal endometrial
tissues. These results suggest that miR-381 is important in EC
progression.
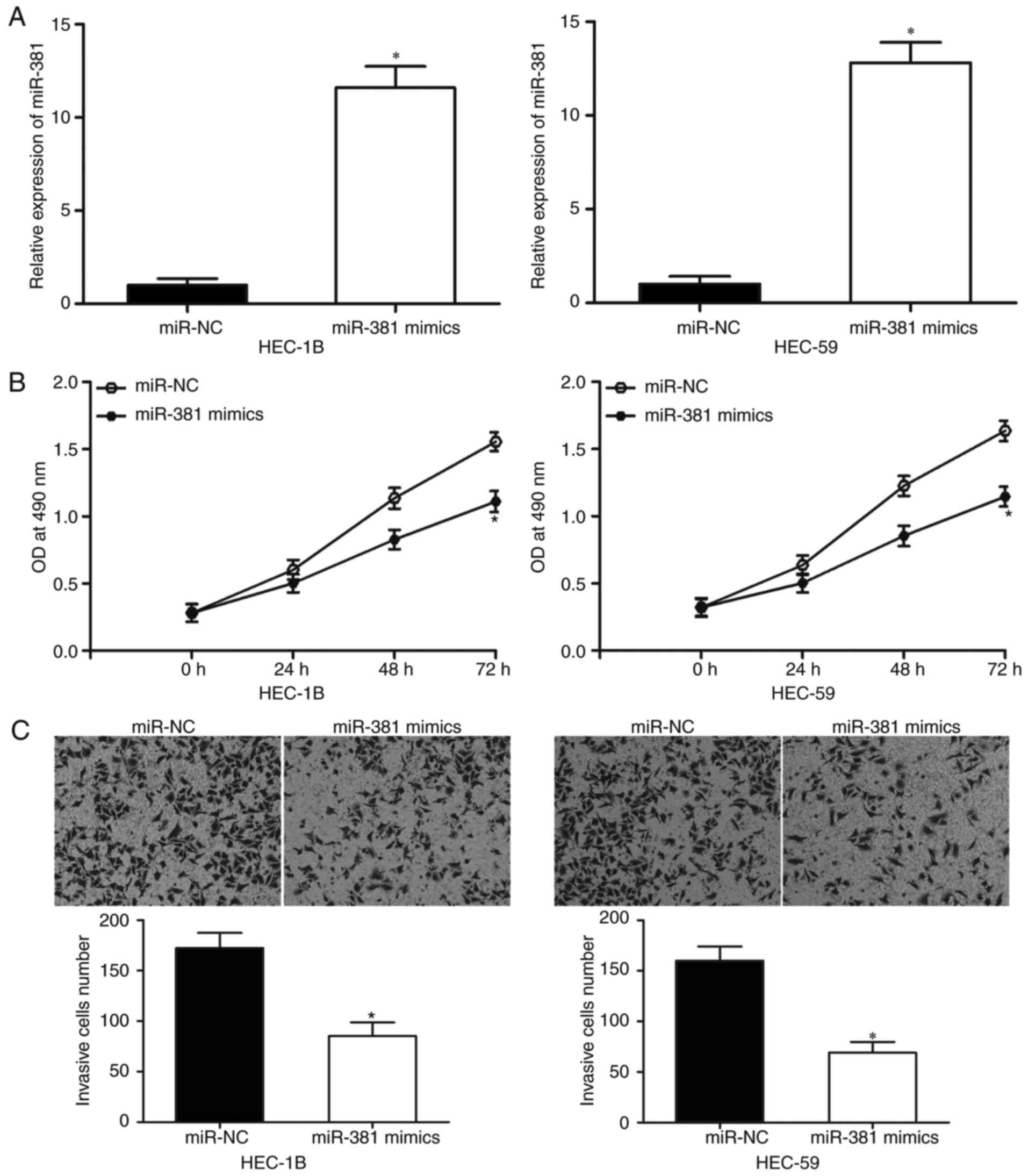
miR-381 overexpression inhibits the
proliferation and invasion of EC cells
HEC-1B and HEC-59 cells were transfected with
miR-381 mimics or miR-NC to elucidate the biological roles of
miR-381 in EC. As shown in Fig.
2A, miR-381 mimics transfection in HEC-1B and HEC-59 cells
markedly increased the expression levels of miR-381 as compared
with miR-NC transfection (P<0.05). MTT assay revealed that the
upregulation of miR-381 significantly inhibited the proliferation
of both HEC-1B and HEC-59 cells as compared with the miR-NC group
(Fig. 2B, P < 0.05).
Furthermore, in vitro cell invasion assay revealed that
miR-381 overexpression reduced the invasion capacities of HEC-1B
and HEC-59 cells (Fig. 2C,
P<0.05). These results suggest that miR-381 acts as a tumor
suppressor in EC progression.
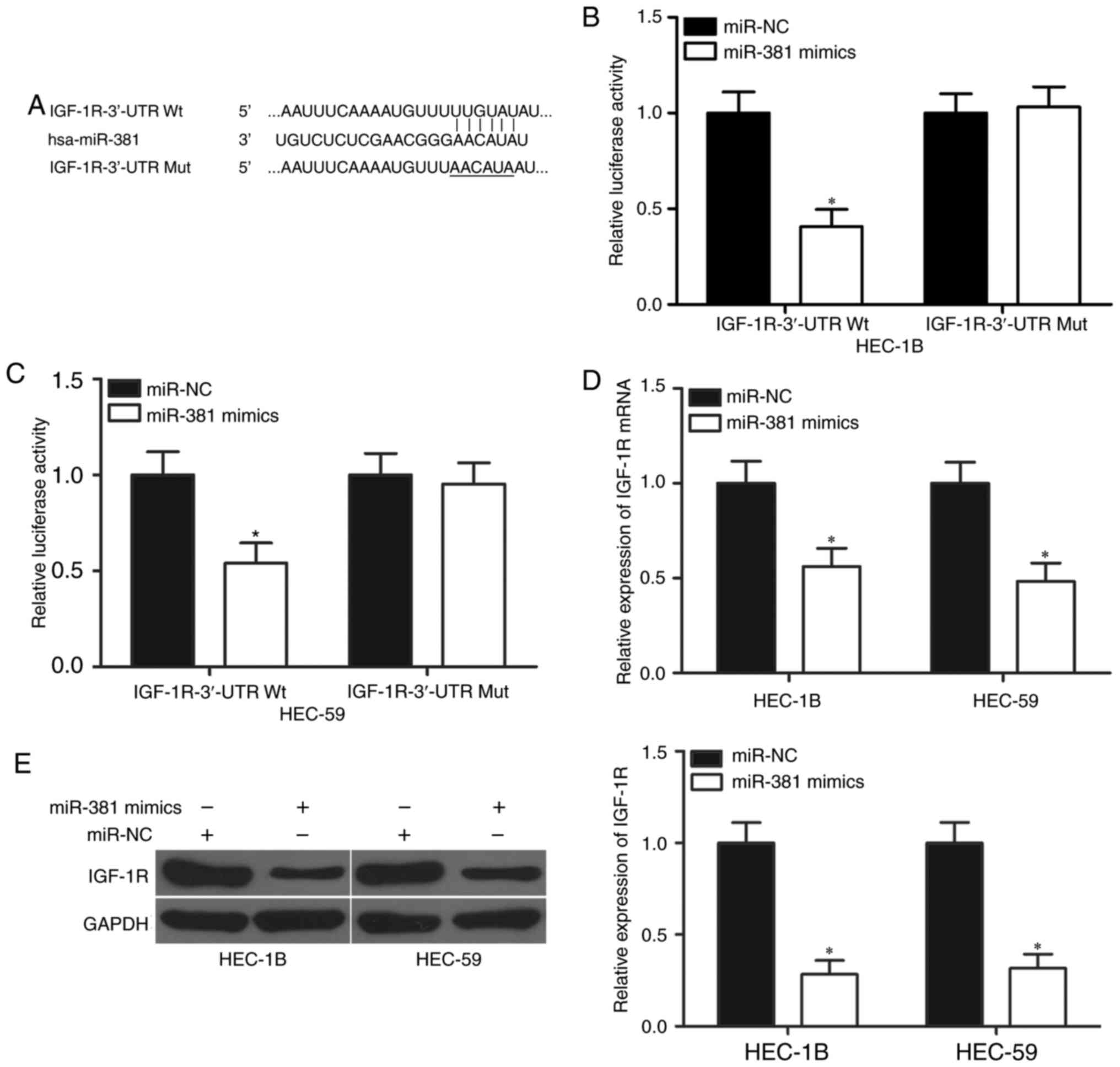
miR-381 directly targets the IGF-1R in
EC
Bioinformatics analysis was performed to analyse the
potential targets of miR-381 and explore the mechanisms underlying
the regulative role miR-381 in EC. IGF-1R harbouring a
miR-381-binding site (Fig. 3A) was
chosen for further validation because of its role in EC formation
and progression (22,23). Luciferase reporter assays were
performed to confirm this hypothesis and examine whether miR-381
interacts directly with the 3′-UTR of IGF-1R. HEC-1B and HEC-59
cells were transfected with miR-381 mimics or miR-NC and
pmirGLO-IGF-1R-3′-UTR Wt or pmirGLO-IGF-1R-3′-UTR Mut. The
co-transfection of miR-381 and wild-type IGF-1R 3′UTR significantly
reduced the luciferase activities (Fig. 3B and C, P<0.05). However, the
co-transfection of the mutant IGF-1R 3′-UTR and miR-381 mimics did
not affect the luciferase activities in both HEC-1B and HEC-59
cells. Furthermore, RT-qPCR and western blot analyses were
conducted to detect the mRNA and protein expression levels of
IGF-1R in HEC-1B and HEC-59 cells after transfection with miR-381
mimics or miR-NC. As shown in Fig. 3D
and E, the restored expression of miR-381 reduced the IGF-1R
expression in HEC-1B and HEC-59 cells at the mRNA (P<0.05) and
protein (P<0.05) levels. In summary, these results suggest that
miR-381 directly targets the 3′-UTR of IGF-1R and thereby represses
the gene expression.
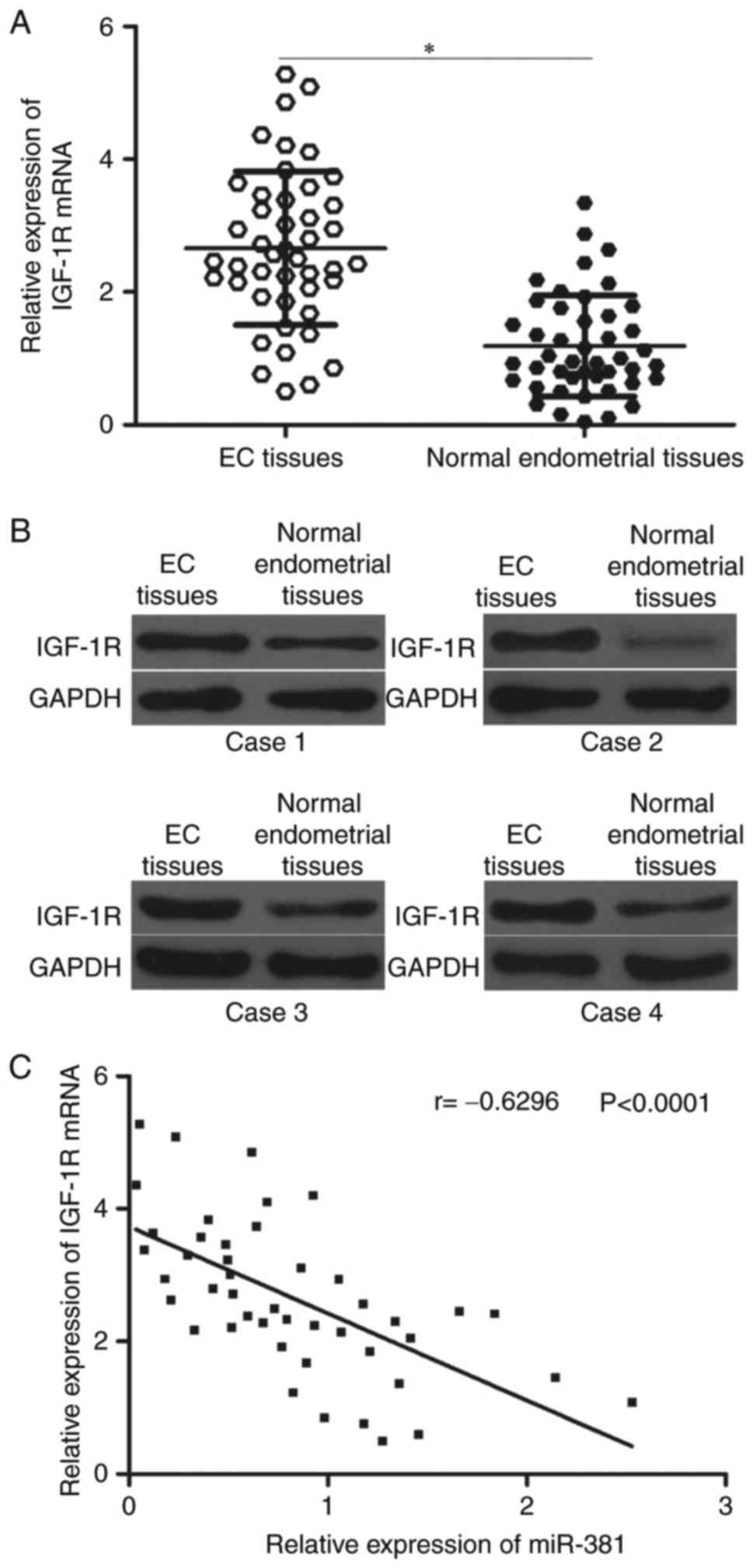
IGF-1R is upregulated in EC tissues
and inversely correlated with miR-381 levels
To further determine the relationship between
miR-381 and IGF-1R, we measured their expression levels in EC
tissues and corresponding adjacent normal endometrial tissues.
Results showed that the mRNA expression of IGF-1R was significantly
higher in EC tissues than in corresponding adjacent normal
endometrial tissues (Fig. 4A,
P<0.05). Western blot analysis also indicated that the protein
expression levels of IGF-1R were upregulated in EC tissues
(Fig. 4B). Furthermore, Spearman's
correlation analysis revealed an inverse association between
miR-381 and IGF-1R mRNA levels in EC tissues (Fig. 4C; r=−0.6296, P<0.0001).
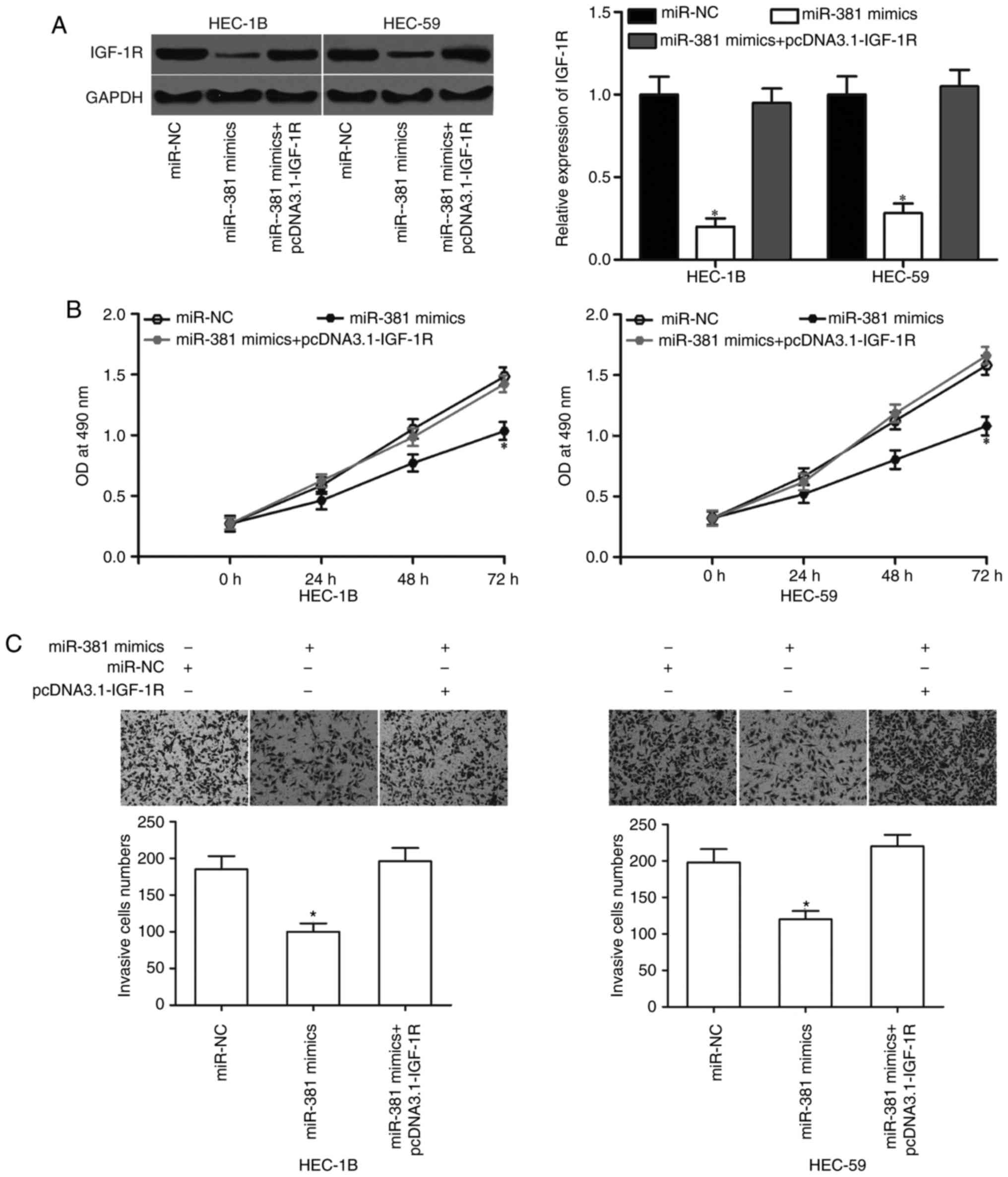
Upregulation of IGF-1R counteracts the
effects of miR-381 in EC cells
A rescue experiment was performed to examine whether
IGF-1R mediates the tumor-suppressing roles of miR-381 in EC cells.
HEC-1B and HEC-59 cells were transfected with miR-381 mimics in the
presence or absence of pcDNA3.1-IGF-1R. Western blot analysis
showed that miR-381 overexpression reduced the protein expression
level of IGF-1R, whereas pcDNA3.1 co-transfection can recover the
IGF-1R expression in HEC-1B and HEC-59 cells (Fig. 5A, P<0.05). MTT and in
vitro invasion assays revealed that the resumed expression of
IGF-1R rescued the suppressive effects of miR-381 on the
proliferation (Fig. 5B, P<0.05)
and invasion (Fig. 5C, P<0.05)
of HEC-1B and HEC-59 cells. These results provide further evidence
that miR-381 partially exerts its tumor-suppressing roles in EC by
negatively regulating IGF-1R.
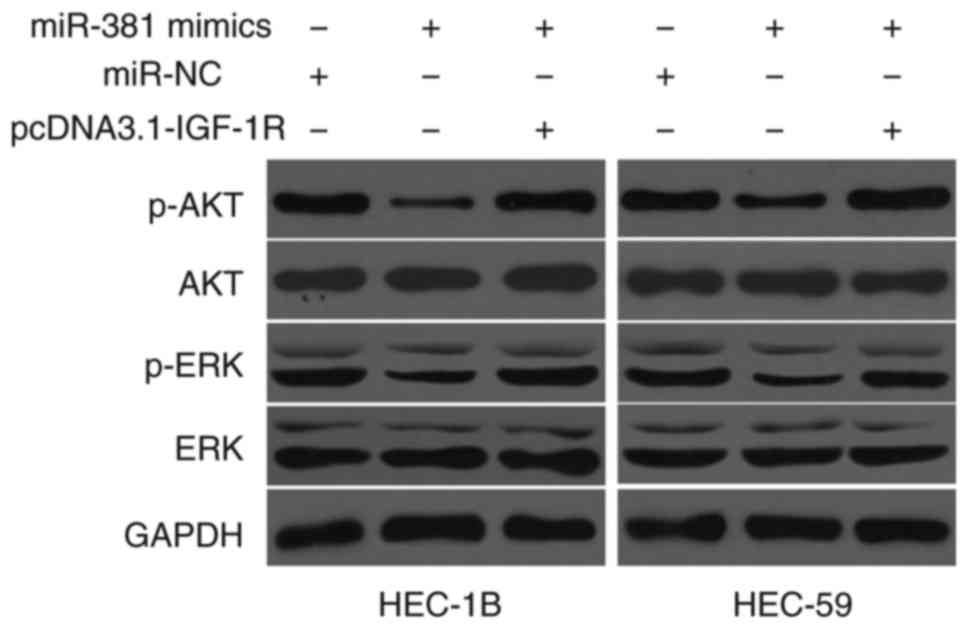
miR-381 targets IGF-1R to inactivate
the AKT and ERK signalling pathway
IGF-1R is involved in the AKT and ERK signalling
pathways (24–26). Therefore, we hypothesised that
miR-381 targets IGF-1R to affect the AKT and ERK signalling pathway
in EC cells. AKT, p-AKT, ERK and p-ERK were detected in HEC-1B and
HEC-59 cells transfected with miR-NC, miR-381 mimics or miR-381
mimics+pcDNA3.1-IGF-1R. As shown in Fig. 6, the ectopic expression of miR-381
reduced the expression levels of p-AKT and p-ERK in HEC-1B and
HEC-59 cells as compared with the cells transfected with miR-NC;
however, the total AKT and ERK levels did not significantly change.
In addition, the expression levels of p-AKT and p-ERK were
recovered in HEC-1B and HEC-59 cells after transfection with
miR-381 mimics and pcDNA3.1-IGF-1R. These results indicate that
miR-381 acts as a tumor suppressor in EC by directly targeting
IGF-1R and indirectly regulating the AKT and ERK signalling
pathways.
Discussion
miRNAs serve as oncogenes or tumor suppressors in
human cancers and play important roles in tumorigenesis and tumor
development by regulating various processes (27,28).
Thus, further investigation of the miRNAs involved in EC occurrence
and progression may contribute to the development of effective
therapeutic strategies for patients with this disease. In this
study, we found that miR-381 was downregulated in EC tissues and
cell lines. The low expression levels of miR-381 correlated with
the FIGO stage, lymph nodes metastasis and myometrial invasion of
EC. The overexpression of miR-381 significantly inhibited the
proliferation and invasion of EC cells in vitro. Moreover,
IGF-1R was validated as a direct target of miR-381. The
upregulation of miR-381 targeted IGF-1R to inactivate the AKT and
ERK signalling pathways. These results suggest that miR-381 can be
further developed as a novel prognostic biomarker for EC and is
potentially a therapeutic target.
miR-381 is aberrantly expressed in several types of
human cancer. For example, miR-381 is significantly downregulated
in lung adenocarcinoma. Low miR-381 expression levels correlate
with poor prognosis for patients with lung adenocarcinoma (29). In colorectal cancer, miR-381 shows
low expression in tumor tissues and is associated with distant
metastasis and TNM stage (30). In
gastric cancer, the expression levels of miR-381 are low in tumor
tissues and cell lines. In addition, miR381 expression is
associated with lymph node metastasis, advanced tumor stage and
poor prognosis (31). miR-381 is
also downregulated in oral squamous cell carcinoma (18), ovarian cancer (19), oesophageal squamous cell carcinoma
(20), hepatocellular carcinoma
(32) and breast cancer (33). However, miR-381 is upregulated in
osteosarcoma tissues. Patients with relatively low miR-381
expression have longer survival time than those with high miR-381
expression (34). These findings
suggest that the expression pattern of miR-381 is tissue-specific,
and the aberrant expression of miR-381 is a potential prognostic
factor in these cancer types.
miR-381 plays tumor-suppressing roles in the
formation and progression of several types of tumors. For instance,
Rothschild et al (29)
reported that miR-381 overexpression suppresses cell migration and
invasion in lung adenocarcinoma. Chen et al (35) found that miR-381 upregulation
attenuates the cell invasion abilities and increases the
chemosensitivity of renal cancer cells to 5-FU. He and Liang et
al (30,36) revealed that the enforced expression
of miR-381 represses cell growth, metastasis and
epithelial-mesenchymal transition in colorectal cancer. Cao et
al found that the restoration of miR-381 expression reduces
cell proliferation and motility in vitro and in vivo
and downregulates the epithelial-mesenchymal transition phenotype
in gastric cancer (31). Yang
et al (18) showed that the
resumption of miR-381 expression inhibits the proliferation and
cell cycle progression while induces the apoptosis of oral squamous
cell carcinoma cells. miR-381 also acts as a tumor suppressor in
ovarian cancer (19),
hepatocellular carcinoma (32) and
breast cancer (33,37). However, miR-381 functions as an
oncogene in osteosarcoma. The re-expression of miR-381 promotes the
proliferation and invasion of osteosarcoma cells and reduces
cisplatin sensitivity (34).
miR-381 overexpression increases the growth and reduces the
chemosensitivity of glioma cells to temozolomide (38,39).
These conflicting findings reveal that miR-381 acts as either an
oncogene in certain types of cancer or as a tumor suppressor in
others, which can be explained by the ‘imperfect complementarity’
of the interactions between miRNAs and their target genes (40). These findings also suggest that
miR-381 is a therapeutic target for patients with these cancer
types.
Some direct targets of miR-381 include WEE1
(35), CBP (41), β-catenin (41), LEF-1 (41) in renal cancer, Twist1 (30), LRH-1 (36) in colorectal cancer, LRRC4 in
osteosarcoma, TMEM16A (31) in
gastric cancer, FGFR2 (18) in
oral squamous cell carcinoma and YY1 (19) in ovarian cancer. In the current
study, IGF-1R was identified as a novel direct target gene of
miR-381 in EC. IGF-1R, a member of the receptor tyrosine kinase
family, contains two extracellular α subunits and two β subunits
(42,43). The highly expressed IGF-1R has been
documented in numerous malignancies, including hepatocellular
carcinoma (44), lung cancer
(45), prostate cancer (46), gastric cancer (47), colorectal cancer (48) and bladder cancer (49). IGF-1R functions as a key oncogene
in the development and maintenance of cancer through the regulation
of cell growth, cycle, apoptosis, migration, invasion and distant
metastasis (50–52). IGF-1R is also upregulated in EC
tissues and cell lines. The high expression level of IGF-1R is
correlated with lymph node metastasis and tumor stage (23,53).
IGF-1R knockdown significantly inhibits the proliferation, induces
the apoptosis in vitro and reduces the tumorigenesis in
vivo of EC cells (22).
Combined with the present findings, the miR-381/IGF-1R pathway
shows a potential to be investigated as a therapeutic strategy for
patients with EC.
In conclusion, our data provide new evidence
supporting the tumor-suppressive roles of miR-381 in EC. We also
revealed that IGF-1R is a novel target of miR-381 in EC. In the
future, miR-381 can be developed as a novel prognostic biomarker
and therapeutic target for the treatment of EC. In our following
experiments, we will focus on the upstream regulation mechanism of
miR-381 in EC.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin F, Devesa SS, Chow WH, Zheng W, Ji BT,
Fraumeni JF Jr and Gao YT: Cancer incidence trends in urban
Shanghai, 1972–1994: An update. Int J Cancer. 83:435–440. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dahlgren E, Friberg LG, Johansson S,
Lindström B, Odén A, Samsioe G and Janson PO: Endometrial
carcinoma; ovarian dysfunction-a risk factor in young women. Eur J
Obstet Gynecol Reprod Biol. 41:143–150. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen YL, Wang KL, Chen MY, Yu MH, Wu CH,
Ke YM, Chen YJ, Chang YY, Hsu KF and Yen MS: Risk factor analysis
of coexisting endometrial carcinoma in patients with endometrial
hyperplasia: A retrospective observational study of Taiwanese
Gynecologic Oncology Group. J Gynecol Oncol. 24:14–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wild PJ, Ikenberg K, Fuchs TJ, Rechsteiner
M, Georgiev S, Fankhauser N, Noske A, Roessle M, Caduff R, Dellas
A, et al: p53 suppresses type II endometrial carcinomas in mice and
governs endometrial tumour aggressiveness in humans. EMBO Mol Med.
4:808–824. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vale CL, Tierney J, Bull SJ and Symonds
PR: Chemotherapy for advanced, recurrent or metastatic endometrial
carcinoma. Cochrane Database Syst Rev. 15:CD0039152012.
|
|
7
|
Boll D, Verhoeven RH, van der Aa MA,
Pauwels P, Karim-Kos HE, Coebergh JW and van Doorn HC: Incidence
and survival trends of uncommon corpus uteri malignancies in the
Netherlands, 1989–2008. Int J Gynecol Cancer. 22:599–606. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moreno-Moya JM, Vilella F and Simoón C:
MicroRNA: Key gene expression regulators. Fertil Steril.
101:1516–1523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dang X, Ma A, Yang L, Hu H, Zhu B, Shang
D, Chen T and Luo Y: MicroRNA-26a regulates tumorigenic properties
of EZH2 in human lung carcinoma cells. Cancer Genet. 205:113–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vandenboom Ii TG, Li Y, Philip PA and
Sarkar FH: MicroRNA and cancer: Tiny molecules with major
implications. Curr Genomics. 9:97–109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A,
Bogunovic D, et al: Aberrant miR-182 expression promotes melanoma
metastasis by repressing FOXO3 and microphthalmia-associated
transcription factor. Proc Natl Acad Sci USA. 106:pp. 1814–1819.
2009; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu D, Zhou Y, Pan H, Zhou J, Fan Y and Qu
P: microRNA-99a inhibiting cell proliferation, migration and
invasion by targeting fibroblast growth factor receptor 3 in
bladder cancer. Oncol Lett. 7:1219–1224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang C, Zhang X, Wang HM, Liu XM, Zhang
XJ, Zheng B, Qian GR and Ma ZL: MicroRNA-18a-5p functions as an
oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis.
8:e27642017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang X, Ruan H, Hu X, Cao A and Song L:
miR-381-3p suppresses the proliferation of oral squamous cell
carcinoma cells by directly targeting FGFR2. Am J Cancer Res.
7:913–922. 2017.PubMed/NCBI
|
|
19
|
Xia B, Li H, Yang S, Liu T and Lou G:
miR-381 inhibits epithelial ovarian cancer malignancy via YY1
suppression. Tumour Biol. 37:9157–9167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou S, Ye W, Ren J, Shao Q, Qi Y, Liang J
and Zhang M: MicroRNA-381 increases radiosensitivity in esophageal
squamous cell carcinoma. Am J Cancer Res. 5:267–277.
2014.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shu S, Li X, Yang Y, Zhang Y, Li T, Liang
C and Wan J: Inhibitory effect of siRNA targeting IGF-1R on
endometrial carcinoma. Int Immunopharmacol. 11:244–249. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pengchong H and Tao H: Expression of
IGF-1R, VEGF-C and D2-40 and their correlation with lymph node
metastasis in endometrial adenocarcinoma. Eur J Gynaecol Oncol.
32:660–664. 2011.PubMed/NCBI
|
|
24
|
Chen G, Fang T, Huang Z, Qi Y, Du S, Di T,
Lei Z, Zhang X and Yan W: MicroRNA-133a inhibits osteosarcoma cells
proliferation and invasion via targeting IGF-1R. Cell Physiol
Biochem. 38:598–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding WZ, Ni QF, Lu YT, Kong LL, Yu JJ, Tan
LW and Kong LB: MicroRNA-497 regulates cell proliferation in
hepatocellular carcinoma. Oncol Lett. 11:1081–1088. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shimizu M, Shirakami Y, Sakai H, Tatebe H,
Nakagawa T, Hara Y, Weinstein IB and Moriwaki H: EGCG inhibits
activation of the insulin-like growth factor (IGF)/IGF-1 receptor
axis in human hepatocellular carcinoma cells. Cancer Lett.
262:10–18. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou XW, Sun X, Yu Y, Zhao HM, Yang ZJ,
Wang X and Cao XC: miR-361-5p suppresses lung cancer cell lines
progression by targeting FOXM1. Neoplasma. 64:526–534. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li P, Wang X, Shan Q, Wu Y and Wang Z:
MicroRNA-130b promotes cell migration and invasion by inhibiting
peroxisome proliferator-activated receptor-γ in human glioma. Oncol
Lett. 13:2615–2622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rothschild SI, Tschan MP, Jaggi R, Fey MF,
Gugger M and Gautschi O: MicroRNA-381 represses ID1 and is
deregulated in lung adenocarcinoma. J Thorac Oncol. 7:1069–1077.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He X, Wei Y, Wang Y, Liu L, Wang W and Li
N: miR-381 functions as a tumor suppressor in colorectal cancer by
targeting Twist1. Onco Targets Ther. 9:1231–1239. 2016.PubMed/NCBI
|
|
31
|
Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W
and Wang L: MicroRNA-381 inhibits the metastasis of gastric cancer
by targeting TMEM16A expression. J Exp Clin Cancer Res. 36:292017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Q, Zhao S, Pang X and Chi B:
MicroRNA-381 suppresses cell growth and invasion by targeting the
liver receptor homolog-1 in hepatocellular carcinoma. Oncol Rep.
35:1831–1840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ming J, Zhou Y, Du J, Fan S, Pan B, Wang
Y, Fan L and Jiang J: miR-381 suppresses C/EBPα-dependent Cx43
expression in breast cancer cells. Biosci Rep. 35:e002662015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Zhao C, Yu Z, Chen J, She X, Li P,
Liu C, Zhang Y, Feng J, Fu H, et al: Low expression of miR-381 is a
favorite prognosis factor and enhances the chemosensitivity of
osteosarcoma. Oncotarget. 7:68585–68596. 2016.PubMed/NCBI
|
|
35
|
Chen B, Duan L, Yin G, Tan J and Jiang X:
miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer
cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J
Chemother. 25:229–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang Y, Zhao Q, Fan L, Zhang Z, Tan B,
Liu Y and Li Y: Down-regulation of microRNA-381 promotes cell
proliferation and invasion in colon cancer through up-regulation of
LRH-1. Biomed Pharmacother. 75:137–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xue Y, Xu W, Zhao W, Wang W, Zhang D and
Wu P: miR-381 inhibited breast cancer cells proliferation,
epithelial-to-mesenchymal transition and metastasis by targeting
CXCR4. Biomed Pharmacother. 86:426–433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Z, Yang J, Xu G, Wang W, Liu C, Yang
H, Yu Z, Lei Q, Xiao L, Xiong J, et al: Targeting miR-381-NEFL axis
sensitizes glioblastoma cells to temozolomide by regulating
stemness factors and multidrug resistance factors. Oncotarget.
6:3147–3164. 2015.PubMed/NCBI
|
|
39
|
Tang H, Wang Z, Liu Q, Liu X, Wu M and Li
G: Disturbing miR-182 and −381 inhibits BRD7 transcription and
glioma growth by directly targeting LRRC4. PLoS One. 9:e841462014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y,
Yu W, Wu X, Ye J, Yang S, et al: Identification of miR-7 as an
oncogene in renal cell carcinoma. J Mol Histol. 44:669–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen B and Liu B: miRNA-381 inhibits the
invasion of renal carcinoma and the underlying mechanisms. Zhong
Nan Da Xue Xue Bao Yi Xue Ban. 40:1053–1059. 2015.PubMed/NCBI
|
|
42
|
LeRoith D and Helman L: The new kid on the
block (ade) of the IGF-1 receptor. Cancer Cell. 5:201–202. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pollak M: Insulin and insulin-like growth
factor signalling in neoplasia. Nat Rev Cancer. 8:915–928. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
E C, Li J, Shao D, Zhang D, Pan Y, Chen L
and Zhang X: The insulin-like growth factor-I receptor inhibitor
picropodophyllin-induced selective apoptosis of hepatocellular
carcinoma cell through a caspase-dependent mitochondrial pathway.
Oncol Res. 21:103–110. 2013.PubMed/NCBI
|
|
45
|
Yeo CD, Park KH, Park CK, Lee SH, Kim SJ,
Yoon HK, Lee YS, Lee EJ, Lee KY and Kim TJ: Expression of
insulin-like growth factor 1 receptor (IGF-1R) predicts poor
responses to epidermal growth factor receptor (EGFR) tyrosine
kinase inhibitors in non-small cell lung cancer patients harboring
activating EGFR mutations. Lung Cancer. 87:311–317. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ma Y, Cheng Q, Ren Z, Xu L, Zhao Y, Sun J,
Hu S and Xiao W: Induction of IGF-1R expression by EGR-1
facilitates the growth of prostate cancer cells. Cancer Lett.
317:150–156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gryko M, Kisluk J, Cepowicz D, Zińczuk J,
Kamocki Z, Guzińska-Ustymowicz K, Pryczynicz A, Czyżewska J, Kemona
A and Kędra B: Expression of insulin-like growth factor receptor
type 1 correlate with lymphatic metastases in human gastric cancer.
Pol J Pathol. 65:135–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shan HB, Zhang R, Li Y, Xu GL, Luo GY, Gao
XY and Yang HL: Expression of IGF-1R in colorectal polyps and its
role in colorectal carcinogenesis. Technol Cancer Res Treat.
10:381–389. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xie QX, Lin XC, Zhang MF, Han CX and Guo
YH: Expression of IGF-I and IGF-IR in bladder cancer. Ai Zheng.
23:707–709. 2004.(In Chinese). PubMed/NCBI
|
|
50
|
Werner H and LeRoith D: The role of the
insulin-like growth factor system in human cancer. Adv Cancer Res.
68:183–223. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pollak M: The insulin and insulin-like
growth factor receptor family in neoplasia: An update. Nat Rev
Cancer. 12:159–169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
King H, Aleksic T, Haluska P and Macaulay
VM: Can we unlock the potential of IGF-1R inhibition in cancer
therapy? Cancer Treat Rev. 40:1096–1105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pavelić J, Radaković B and Pavelić K:
Insulin-like growth factor 2 and its receptors (IGF 1R and IGF
2R/mannose 6-phosphate) in endometrial adenocarcinoma. Gynecol
Oncol. 105:727–735. 2007. View Article : Google Scholar : PubMed/NCBI
|