Introduction
Malignant glioma, a malignant neoplasm of the
central nervous system, is a devastating condition associated with
poor prognosis. Owing to the location and aggressive nature of
glioma, it is extremely difficult to achieve the complete surgical
resection of the tumor, inevitably leading to the recurrence
(1). Statistics revealed that the
five-year survival rate of glioma patients is less than 30%, with
the survival rate of WHO grade IV glioma patients being as low as
six months (2). Considering the
aggressiveness and devastating nature of the disease, it is
critical to develop an effective treatment. In this regards,
identifying the genes that are closely related to the genesis,
development and prognosis of glioma is crucial.
IRX1 gene is a member of Iroquois homeobox gene
family, closely associated with the embryonic development in both
vertebrates and invertebrates. IRX1 protein, encoded by IRX1 gene,
acts as transcription factor, which in combination with other
family proteins, actively participates in nervous system
development (3). A recent study
identified the expression of IRX1 gene during early lung
development in rats, the overexpression of which was correlated to
pulmonary hypoplasia (4). IRX1
also plays a critical role in cancer condition, which may vary from
being tumor suppressor gene, prometastatic gene or tumor oncogene
under different types of cancer (5). However, the role of IRX1 gene in
human glioma is not clear yet, and the same is being addressed in
the present study, by exploring its expression and clinical
significance in glioma.
Materials and methods
This study was approved by the Research Ethics
Committee of Tangdu Hospital, Fourth Military Medical University,
P.R. China. Written informed consent was obtained from the
participating patients. All clinical data/specimens were anonymized
and handled as per the related ethical and legal standards.
Cells and tissue samples
Human glioma cells (U87, U373, LN229 and T98G) were
cultured in Dulbecco's modified eagle's medium (DMEM), supplemented
with 10% fetal calf serum (GIBCO, USA) and 1%
Penicillin/Streptomycin (SIGMA ALDRICH, USA), and incubated at 37°C
with 5% CO2. It should be specially noted that the
origin of the U87 cell line is unknown, but it is a likely
glioblastoma cell line (6). And
U373 cells are known to be a U251 derivative (7), obtained from ATCC (HTB-17).
All the study specific tissue samples were collected
from the glioma patients treated at the department of neurosurgery,
Tangdu hospital, Fourth Military Medical University, P.R. China.
These tissue samples comprised of 4 glioma (1G: WHO IV, 2G: WHO
III, 3G: WHO III–IV, 4G: WHO IV) + 4 adjacent normal brain tissue
specimens, freshly collected from the surgeries during 2015 and 54
glioma only specimens, archived under liquid nitrogen at the
pathology department, from the surgeries during 01/2008 to 12/2010.
None of the patients had received chemotherapy or radiotherapy
before surgery. The fresh specimens were snap-frozen in liquid
nitrogen and stored at −80°C, while archived glioma only specimens
were fixed in 10% buffered formalin and embedded in paraffin, till
further analysis.
Total RNA isolation and cDNA
synthesis
The total RNA from human glioma cell lines (U87,
U373, LN229 and T98G) was purified using TRIzol reagent. Normal
brain tissue RNA was purchased from Clontech. Typically, 1 µg of
the total RNA was used to generate cDNA by employing
SuperScript® II RT (TAKARA, JAPAN) with oligo-dT primer.
The PCR primers for IRX1 and GAPDH were 5′-TCATTGACCTCAACTACATG-3′
(F) and 5′-TCGCTCCTGGAAGATGGTGAT-3′ (R), and
5′-CTCAGCCTCTTCTCGCAGAT-3′ (F) and 5′-TCTTCCTGGTCCTTGCTGC-3′ (R),
respectively (8). Each PCR was
performed for 30 thermal cycles and the PCR products were observed
by electrophoresis on a 1.5% agarose gel, visualized after staining
with ethidium bromide.
The expression of IRX1 protein in
glioma, detected by Western Blot
The IRX1 protein expression in the glioma condition
was assessed using the fresh glioma tissues, along with the
adjacent normal brain tissue. The total protein from fresh tissues,
separated into glioma tissue and adjacent normal tissue, were
extracted via mammalian protein extraction reagent (Pierce,
Appleton, WI, USA) supplemented with protease inhibitors cocktail
(Sigma, USA). The total protein concentrations were determined via
BCA protein assay kit (Boster Systems, Inc., Pleasanton, CA, USA).
The proteins were separated by 10% SDS-PAGE (Genshare biological,
shaanxi, China) and then transferred to PVDF membranes (Millipore,
Boston, USA). The membranes were blocked with TBST buffer (TBS plus
0.1% Tween-20) containing 5% w/v non-fat milk, and hybridized with
IRX1 antibody (Code NO. Ab180860, Abcam, USA) at 1:1500 dilution,
followed by incubation with specific HRP-conjugated secondary
antibody (1:2500, Bioworld, St. Louis, USA). Proteins were
visualized using ECL detection system (Amersham Bio-sciences,
Uppsala, Sweden).
The expression and location of IRX1
protein in glioma tissues via immunohistochemistry (IHC) and tissue
microarray
The 54 glioma only specimens were subject to IHC
analysis, by DAB detection kit (Streptavidin-Biotin; ZSGB-BIO,
Beijing, China), to assess the expression and location of IRX1.
Briefly, following a peroxidase block with 3%
H2O2/methanol for 30 min, specimens were
blocked with 5% normal goat serum. Then the slides were incubated
overnight, at 4°C, with mouse polyclonal antihuman IRX1 primary
antibody (Code NO. BS2291, Bioworld, USA), at 1:50 dilution. Then,
the specimens were briefly washed for 3 times with PBS and
incubated at room temperature with the anti-goat secondary antibody
for 1 h, followed by incubation with HRP/Fab polymer conjugate for
30 min, at 37°C. The signal visualization was performed by treating
with DAB chromogen for 2 to 3 min. After water wash, specimens were
counterstained with Meyer's hematoxylin (ZSGB-BIO).
Tissue microarray assay (NO. BS17017b, alenabio) was
employed to further validate the IHC results. A total of 62 brain
tissue samples were included in the assessment and were classified
as; 24 low-grade gliomas [3 pilocytic astrocytomas (WHO I) and 21
diffuse astrocytomas (WHO II)], 33 high-grade gliomas [9 anaplasia
astrocytomas (WHO III) and 24 primary glioblastomas (WHO IV)] and 5
adjacent normal brain tissue. All the samples were provided and
analyzed, through IHC, by Alenabio biotechnology LTD, Xi'an, China.
The immunostaining was performed using mouse polyclonal anti-human
IRX1 primary antibody (Bioworld, USA), at 1:50 dilution, while the
same was replaced by non-immune IgG antibody for negative control
samples. All specimens were counterstained with Meyer's
hematoxylin.
The qualitative and semi-quantitative evaluation of
the IHC and tissue microarray samples were achieved as follows:
five random zones of the stained glioma samples were observed under
the light microscope (high power field) and assessed the location
of the stain, intensity of staining and the percent of positively
stained cells. Based on the intensity and the percent of positively
stained cells the samples were scored as follows: no staining as
negative (−), <25% positive cells as weakly positive (+),
25%-49% of positive cells as moderately positive (++), and >50%
of positive cells as strong positive (+++/++++). The results were
evaluated according to WHO classifications by two experienced
(>10 years) pathologists, with differences resolved through
careful discussion.
Clinico-pathological features,
prognostic variables and overall patient survival
The clinico-pathological features of the
participating patients were collected from their respective
clinical records and the histopathological classification of their
glioma condition (WHO classification) was re-evaluated using the
study specific H&E staining. The data were correlated to the
IRX1 expression and are listed in Table I. The pathological variables
affecting the prognosis of the glioma condition were also assessed,
through univariate and multivariate analysis.
 | Table I.Characteristic analysis of patients
corresponding to glioma specimen. |
Table I.
Characteristic analysis of patients
corresponding to glioma specimen.
|
|
| IRX1 expression |
|---|
|
|
|
|
|---|
| Clinico-pathological
features | No. of cases | low (n. %) | high (n. %) |
|---|
| WHO grade |
|
|
|
| I | 5 | 4
(80) | 1
(20) |
| II | 16 | 13 (82) | 3
(18) |
| III | 14 | 3
(21) | 11 (79) |
| IV | 19 | 2
(10) | 17 (90) |
| Age |
|
|
|
|
<55 | 30 | 17 (57) | 13 (43) |
| ≥55 | 24 | 5 (21) | 19 (79) |
| Gender |
|
|
|
| Male | 33 | 13 (39) | 20 (61) |
|
Female | 21 | 9 (43) | 12 (57) |
Further, the survival time of all the 54 patients,
from whom glioma only specimens were obtained, was assessed by
retrospectively following up for a median period of 60 months. The
overall survival time, defined as the time from the date of the
initial surgery to death, was calculated based on the Kaplan-Meier
method. Patients who died of reasons not related to glioma were
excluded from the analysis.
Statistical analysis
SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was
applied for all statistical analyses, and a P-value<0.05 was
considered as significant.
Results
The expression level of IRX1 gene in
glioma cells
As per the RT-PCR results, all the glioma cell lines
(U373, LN229 and T98G cells), except U87 cells, expressed IRX1,
while the same was not detected in normal brain tissue (Fig. 1). This suggests of a probable
positive correlation of IRX1 with glioma.
The expression and location of IRX1 in
glioma tissues
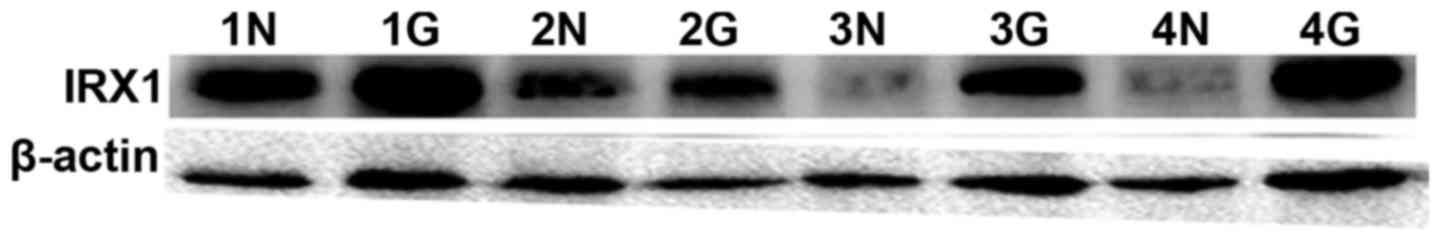
Western blot assay showed an increased expression of
IRX1 protein in glioma tissue, compared to adjacent normal brain
tissue, with the difference reaching statistical significance
(P<0.05) (Fig. 2). The data was
consistent with our finding in cell lines, which further confirmed
a correlation of IRX1 with glioma.
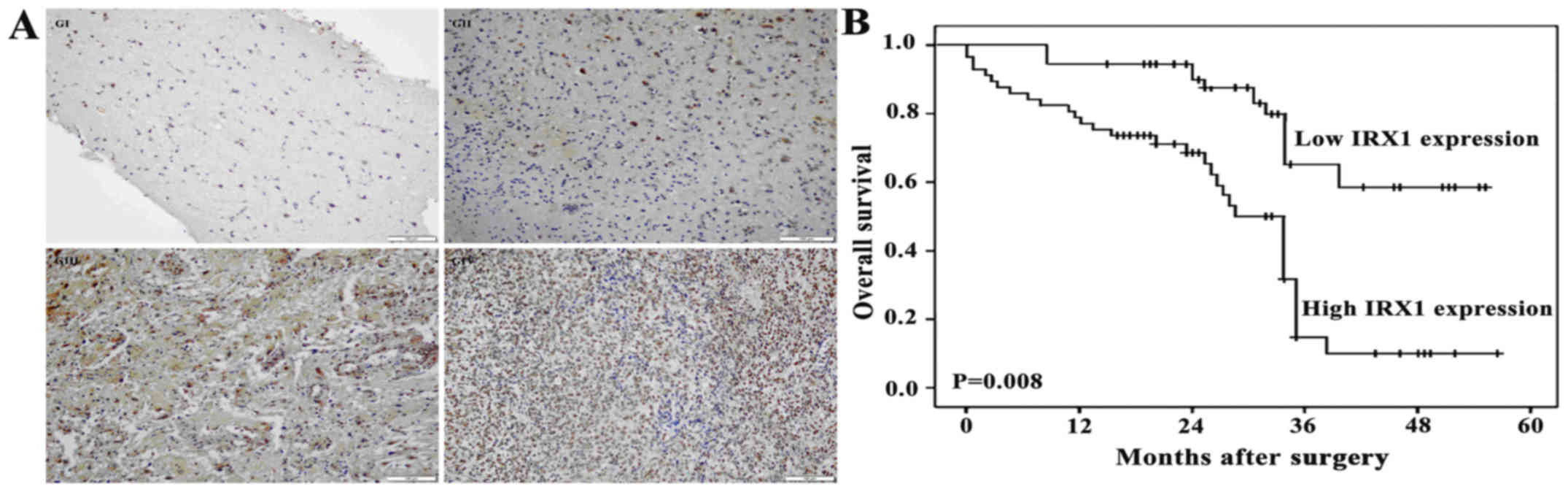
Histologically, among 54 cases of glioma, 25 cases
were classified as low-grade gliomas [6 pilocytic astrocytomas (WHO
I) and 19 diffuse astrocytomas (WHO II)], and 29 cases as
high-grade gliomas [12 anaplasia astrocytomas (WHO III) and 17
primary glioblastomas (WHO IV)]. The IHC analysis of glioma samples
revealed that the IRX1 protein was expressed in all grades of
glioma tissues. Interestingly the localization of IRX1 shifted from
cytoplasm in WHO grade I glioma tissue to nucleus in the higher
grade gliomas (Fig. 3A). Further,
a positive correlation between IRX1 expression and the glioma grade
was observed.
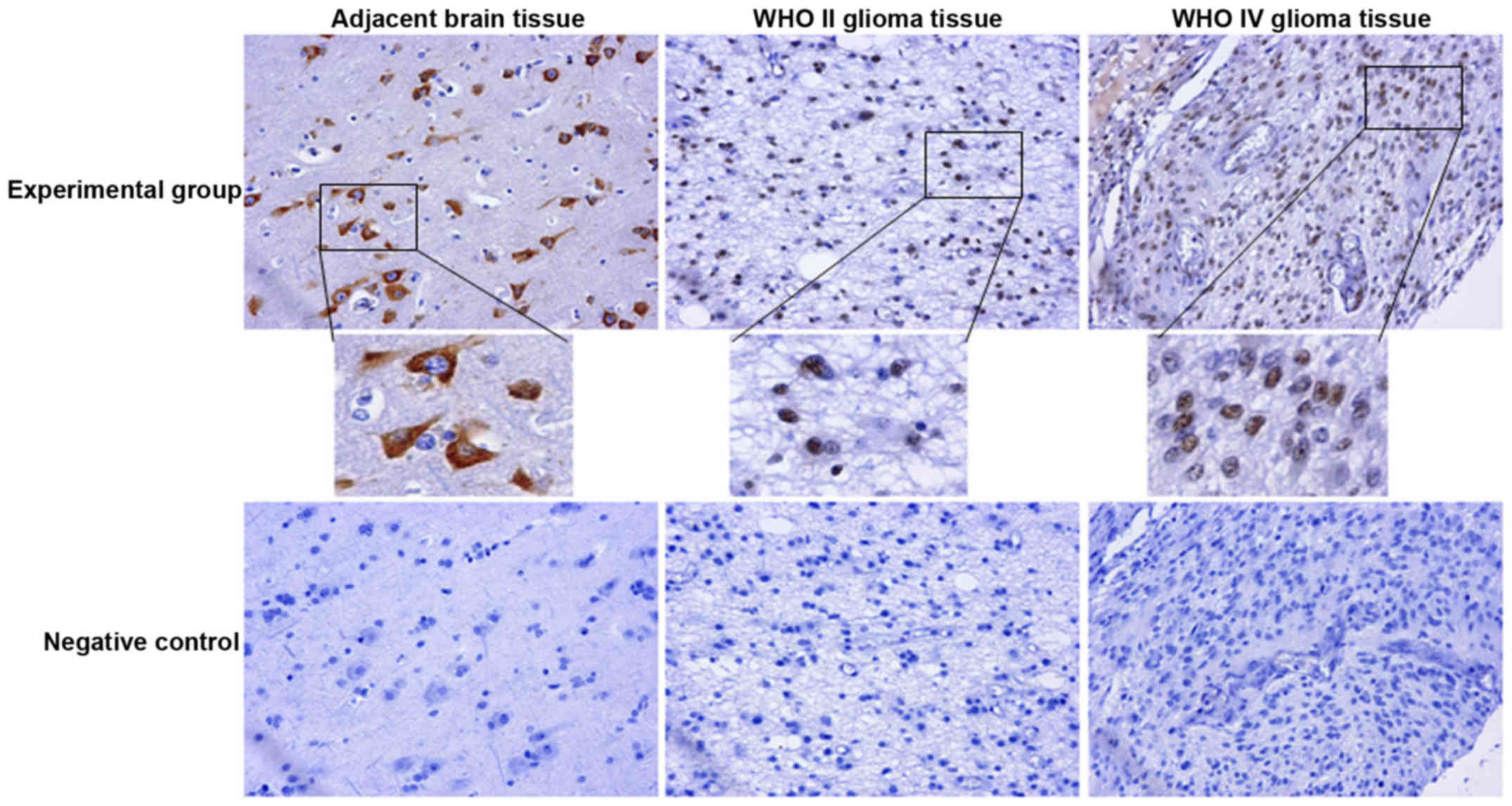
Our IHC results were reconfirmed through tissue
microarray (NO.IHC 160917), which, in addition to confirming the
expression of IRX1 in glioma tissues, also revealed its presence in
the adjacent normal brain tissue. A clear shift in the location of
IRX1 from cytoplasm in adjacent normal brain tissue and WHO grade I
glioma tissue to nucleus in the higher grade gliomas was observed
(Table IIA and Fig. 4), where it took the form of rough
trachychromatic claybank granules. Interestingly, one sample in the
WHO grade II glioma specimen displayed co-localization of IRX1 in
both cytoplasm and nucleus. A few glioma samples with no IRX1
expression was also seen, the number of which sequentially
increased with the increase in glioma grade. Among the assessed
glioma tissue samples, WHO grade I glioma tissues were restricted
to weakly positive IRX1 expression while few WHO grade II and III
samples managed to increase IRX1 expression to moderate positivity.
Compared to the other glioma grades, significant number of WHO
grade IV glioma tissues displayed moderately positive IRX1
expression, with one sample even managing to display strong
positivity (Table IIB). The
localization difference in tissue microarray analysis was more
obvious than that in IHC, and the testing results of normal brain
tissues were added in tissue microarray, which is a supplement and
further verification of IHC. These results reconfirmed a possible
correlation of IRX1 expression with glioma grades.
 | Table II.Cellular location and expression of
IRX1 in brain tissue and glioma tissue. |
Table II.
Cellular location and expression of
IRX1 in brain tissue and glioma tissue.
| A, The cellular
location of IRX1 in glioma and adjacent normal brain tissue |
|---|
|
|---|
|
| Adjacent glioma
tissue brain tissue | Grade I glioma
tissue | Grade II glioma
tissue | Grade III glioma
tissue | Grade IV glioma
tissue |
|---|
| Number of cells
expressing IRX1 in cytoplasm | 5 | 2 | 1(in cytoplasm and
nucleus) | 0 | 0 |
| Number of cells
expressing IRX1 in nucleus | 0 | 0 | 18 | 6 | 20 |
| Number of cells not
expressing IRX1 | 0 | 1 | 2 | 3 | 4 |
| Total number of cells
assessed | 5 | 3 | 21 | 9 | 24 |
|
| B, The expression
of IRX1 in brain tissue and glioma tissue |
|
|
| Negative
(−) |
| Weak positive
(+) | Moderately
positive (++) | Strong positive
(+++) |
|
| Adjacent brain tissue
(cytoplasm) |
| 0 | 1 | 3 | 1 |
| WHO I
(cytoplasm) |
| 1 | 2 | 0 | 0 |
| WHO II (nucleus) |
| 2 | 14 | 5 | 0 |
| WHO III
(nucleus) |
| 3 | 14 | 2 | 0 |
| WHO IV
(nucleus) |
| 4 | 10 | 9 | 1 |
Prognostic variables and correlation
of IRX1 expression with overall survival of glioma patients
Univariate analysis showed that large tumor diameter
(P=0.04), intra-tumor necrosis (P=0.02) and high IRX1 expression
(P=0.008) were significantly associated with poor prognosis.
Further multivariate analysis, based on Cox proportional hazards
model, identified intra-tumor necrosis (P=0.04) and IRX1 expression
(P=0.01) as two independent prognostic variables. Statistical
values of the expression of IRX1 and other clinical parameters
derived from Cox stepwise proportional hazards model are indicated
in Table III.
 | Table III.Univariate and multivariate
analyses. |
Table III.
Univariate and multivariate
analyses.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameter | Risk ratio | 95% confidence
interval | P-value | Risk ratio | 95% confidence
interval | P-value |
|---|
| Age | 0.90 | 0.22–1.97 | N.S. | – | – | – |
| Gender | 1.11 | 0.56–2.69 | N.S. | – | – | – |
| Largest tumor
diameter | 2.08 | 1.03–4.62 | 0.04 | 1.12 | 0.58–2.88 | N.S. |
| Intra-tumor
necrosis | 2.82 | 1.02–6.09 | 0.02 | 2.03 | 1.02–4.92 | 0.04 |
| IRX1
expression | 4.60 | 1.12–10.29 | 0.008 | 3.20 | 1.06–6.98 | 0.01 |
In total, all the 54 patients had a complete
follow-up record. The Kaplan-Meier analysis demonstrated a
significant association of IRX1 expression with the overall
survival of glioma patients (Fig.
3B).
Discussion
In the present study, we assessed the clinical
significance of IRX1 in glioma condition. First, through RT-PCR, we
found that most of the assessed human glioma cell lines (U373,
LN229 and T98G) expressed IRX1, while the same was not detected in
normal brain tissue (Fig. 1).
Western Blot analysis showed an increased expression of IRX1
protein in glioma tissue that was significantly higher than the
adjacent brain tissue (Fig. 2).
Further, the IHC and tissue microarray demonstrated a positive
correlation of the IRX1 expression with the glioma grade. Further,
the localization of IRX1 was found shifted from the cytoplasm in
the WHO I glioma and adjacent normal brain tissue, to nucleus in
the higher grade gliomas (Figs. 3A
and 4). Further to being
established as a prognostic variable, the IRX1 expression was
positively correlated with the overall survival of glioma patients
(Fig. 3B). The above results
indicate that IRX1 might play an important role in the development
of glioma, and be of potential use in assessing the prognosis and
malignancy status of the condition and also as a target for
molecular therapy of glioma. In this study, The normal brain tissue
RNA used for RT-PCR was purchased from Clontech. And there is no
protein in normal brain tissue can be used as controls for western
blot test, the comparative analysis was meaningless. Therefore,
western blot was not used to confirm the PCR results in the cell
lines.
IRX1 is shown to play different roles in different
cancer conditions (5). In
hepatocellular carcinoma condition, Yang et al. identified
IRX1 as a tumor oncogene in hepatocellular cancer, within which the
IRX1 expression was increased and were related to the
differentiation level of the cancer (9). IRX1 behaved as a prometastatic gene
in osteosarcoma, the increased expression of which was associated
with the cancer metastasis, via induction of CXCL14/NF-κB signaling
pathway (10). IRX1 was found down
regulated in gastric cancer, and was labeled as tumor suppressor
gene, the mechanism of which was found associated with the target
genes FGF7, HIST2H2BE, and BDKRB2-dependent downstream gene PAK1
(11,12). The tumor suppressor activity of
IRX1 gene was also acknowledged in head and neck squamous cell
carcinoma and pancreatic cancer (13,14).
In addition, IRX1 is known to induce transcription of HOXB4 gene, a
downstream target gene of WNT, c-KIT and TPO pathways, and inhibit
the function of MLL-AF4 and thus regulate acute lymphoblastic
leukemia (15). MAPK pathway, in
which PAK1 is a key protein, NF-κB pathway and WNT pathway have
demonstrated to influence glioma (16). With IRX1 shown to influence above
pathways, it can be speculated that IRX1 might be involved in the
development and progression of glioma through affecting some key
molecules in these signaling pathways.
As per our study results, demonstrating an increased
expression of IRX1 in glioma condition and its direct correlation
with glioma grade, IRX1 can be labeled as tumor oncogene in glioma.
The underlying molecular mechanism associated with the increased
IRX1 expression in glioma cells and clinical glioma tissues is yet
to be established. It has been reported that the global
hypomethylation is a common mechanism in primary human
glioblastomas (17). In support of
the statement, several studies demonstrate the hypomethylation of
various genes in glioma condition (18–22).
The hypomethylation of IRX1 gene has been observed in metastatic
osteosarcoma and gastric cancer (10,11).
Correspondingly, IRX1 hypomethylation could be a possible
underlying molecular mechanism of the observed increase in its
expression in glioma, which needs further assessments. In further
mechanistic studies, we will focus on investigating how the
increased expression of the protein is associated with the glioma
progression.
The occurrence of glioma is influenced by
multi-factors, multi-stages and multi-genes (23). In-depth study of the tumor gene
expression profile is of great significance in tumor diagnosis,
treatment, and monitoring the recurrence and prognosis of glioma.
Increased expression of IRX1, and its relocation to the nucleus in
higher grade glioma, suggests of a direct influence of IRX1 in the
proliferation, metastasis and development of glioma, though the
underlying mechanism is not clear yet. If hypomethylation triggers
the increase in IRX1 expression and if so, at what stage of glioma,
needs to be determined yet. Further, the molecules/genes among the
MAPK, NF-κB, WNT, and other activated pathways through which the
IRX1 affects glioma progression remains to be studied further.
Acknowledgements
National Natural Scientifc Foundation of China for
Yanyang Tu (No. 81572983), Social Development of Technology
Research Projects in Shaanxi Province for Pengxing Zhang (No.
2015SF027), Social Development of Technology Research Projects in
Shaanxi Province for Hui Liu (No. 2016SF191), and Beijing Key
Laboratory of Brain Major Diseases Open Project for Yanyang Tu
(2015). Foundation of science innovation and development in Tangdu
Hospital, Fourth Military Medical University for Nan Liu (No.
2016JCYJ013).
References
|
1
|
Minniti G, De Sanctis V, Muni R, Filippone
F, Bozzao A, Valeriani M, Osti MF, De Paula U, Lanzetta G,
Tombolini V and Maurizi Enrici R: Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma in elderly patients. J
Neurooncol. 88:97–103. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nakada M, Nakada S, Demuth T, Tran NL,
Hoelzinger DB and Berens ME: Molecular targets of glioma invasion.
Cell Mol Life Sci. 64:458–478. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bürglin TR: Analysis of TALE superclass
homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel
domain conserved between plants and animals. Nucleic Acids Res.
25:4173–4180. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Doi T, Lukošiūtė A, Ruttenstock E,
Dingemann J and Puri P: Expression of Iroquois genes is
up-regulated during early lung development in the nitrofen-induced
pulmonary hypoplasia. J Pediatr Surg. 46:62–66. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang PX, Yang HW, Wang X, Wang L, Cheng
YD, Zhang YS, et al: The genomic organization and function of IRX1
in tumorigenesis and development. Cancer Transl Med. 3:29–33. 2017.
View Article : Google Scholar
|
|
6
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32106. View Article : Google Scholar
|
|
7
|
Torsvik A, Stieber D, Enger PØ,
Golebiewska A, Molven A, Svendsen A, Westermark B, Niclou SP, Olsen
TK, Chekenya Enger M and Bjerkvig R: U-251 revisited: genetic drift
and phenotypic consequences of long-term cultures of glioblastoma
cells. Cancer Med. 3:812–824. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin
W and Zhang Y: MicroRNA-218 inhibits glioma invasion, migration,
proliferation, and cancer stem-like cell self-renewal by targeting
the polycomb group gene Bmi1. Cancer Res. 73:6046–6055. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang HJ, Yu S, Wang J, Chen C, Gu Y and
Bao XY: Expression and clinical significance of Iroquois homebox
gene IRX1 in human hepatocellular carcinoma cells. Chin J
Biologicals. 25:1354–1357. 2010.(In Chinese).
|
|
10
|
Lu J, Song G, Tang Q, Zou C, Han F, Zhao
Z, Yin J, Xu H, Xie X, et al: IRX1 hypomethylation promotes
osteosarcoma metastasis via induction of CXCL14/NF-κB signaling. J
Clin Invest. 25:1839–1856. 2015. View
Article : Google Scholar
|
|
11
|
Guo X, Liu W, Pan Y, Ni P, Ji J, Guo L,
Zhang J, Wu J, Jiang J, Chen X, et al: Homeobox gene IRX1 is a
tumor suppressor gene in gastric carcinoma. Oncogene. 29:3908–3920.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang J, Liu W, Guo X, Zhang R, Zhi Q, Ji
J, et al: IRX1 influences peritoneal spreading and metastasis via
inhibiting BDKRB2-dependent neovascularization on gastric cancer.
Oncogene. 30:4498–4508. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bennett KL, Karpenko M, Lin MT, Claus R,
Arab K, Dyckhoff G, Plinkert P, Herpel E, Smiraglia D and Plass C:
Frequently methylated tumor suppressor genes in head and neck
squamous cell carcinoma. Cancer Res. 68:4494–4499. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei W, Xu L, Wang F, He SS, Yang LJ, Guo
CY and Wang XP: Expression and methylation of Iroquois homeobox
protein 1 in pancreatic cancer. Chin J Pancreat. 11:309–311.
2011.
|
|
15
|
Kühn A, Löscher D and Marschalek R: The
IRX1/HOXA connection: insights into a novel t(4;11)-specific cancer
mechanism. Oncotarget. 7:35341–35352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng YD, Tu YY and Liang P: Promoter
Methylated Tumor Suppressor Genes in Glioma. Cancer Transl Med.
1:123–130. 2015. View Article : Google Scholar
|
|
17
|
Cadieux B, Ching TT, VandenBerg SR and
Costello JF: Genome-wide hypomethylation in human glioblastomas
associated with specific copy number alteration,
methylenetetrahydrofolate reductase allele status, and increased
proliferation. Cancer Res. 66:8469–76. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ting AH, McGarvey KM and Baylin SB: The
cancer epigenome-components and functional correlates. Genes Dev.
20:3215–3231. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Frigola J, Solé X, Paz MF, Moreno V,
Esteller M, Capellà G and Peinado MA: Differential DNA
hypermethylation and hypomethylation signatures in colorectal
cancer. Hum Mol Genet. 14:319–326. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ehrlich M, Jiang G, Fiala E, Dome JS, Yu
MC, Long TI, Youn B, Sohn OS, Widschwendter M, Tomlinson GE, et al:
Hypomethylation and hypermethylation of DNA in Wilms tumors.
Oncogene. 21:6694–6702. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu N and Tu YY: Systematic Review of
MicroRNAs and its Therapeutic Potential in Glioma. Cancer Transl
Med. 1:50–66. 2015. View Article : Google Scholar
|