Introduction
Colorectal carcinoma (CRC) is one of the most common
and lethal cancers worldwide. Although a trend of decline was
observed in CRC due to screening, it still ranked the third in
cancer mortality in the United States regardless of gender
disparity in the year of 2014 and the third in cancer morbidity
worldwide (1,2).
MicroRNAs (miRNAs) are non-coding RNAs participated
in mRNA binding and silencing with approximate length of 22
nucleotides. With revelation of their prominent
post-transcriptional regulation of miRNAs, there is accumulative
evidence demonstrating that miRNAs play significant roles in
numerous biological processes and diseases (3,4), and
CRC is no exception. Various miRNAs were reported to be associated
with CRC, a part of which are oncogenic miRNAs and the others are
tumor suppressors (5,6).
MiR-1-3p, as the first numbered miRNA, undoubtedly
correlated with multiple human diseases. MiR-1-3p is famous for its
regulatory roles both in skeletal and cardiac muscle tissues,
including myogenesis, muscle proliferation and differentiation
(7). The remarkable roles of
miR-1-3p in oncology are also noteworthy. It is widely proved that
miR-1-3p functions as a notable tumor suppressor in various types
of cancers such as prostate cancer (8), hepatocellular carcinoma (9), gallbladder cancer (10), non-small-lung cancer (11) and gastric cancer (12). Inverse expression of oncogenic
factors and miR-1, including NRF2 (13), EGFR (14), MALAT1 (15), etc., indicates targeting
relationships in tumorigenesis and development.
In CRC, the majority of existing researches
unanimously agreed the tumor-suppressive function of miR-1-3p, but
higher expression of miR-1-3p was still observed in brain
metastases CRC than primary CRC (16). While revealing attenuated
expression level of miR-1-3p and its clinical value in CRC,
researchers were also exploring the potential pathways and
coefficient target genes related to miR-1-3p. For instance, Wu
et al (17), stressed that
miR-1-3p could function as an effective diagnostic biomarker of
CRC, with an AUC of 0.806. Furukawa et al (18), discovered that miR-1-3p might
inhibit the migration of CRC cells by directly targeting NOTCH3,
the latter can promote tumorigenesis and migration of CRC. However,
despite of the promising regulatory roles miR-1-3p plays in CRC,
the application of bioinformatics databases to further validate its
function and potential clinical application is yet to be completed.
Although several studies have found a part of target genes and
pathways that associated with miR-1-3p, a more comprehensive map
which extracts data in public databases is essential. Most
importantly, since the inconformity in expression arose, a
confirming meta-analysis is more persuasive to clarify the
characteristics of miR-1-3p.
Hence, we demonstrated the clinical value of
miR-1-3p in CRC by extracting information from Gene Expression
Omnibus (GEO), ArrayExpress and existing literature, combining
microarray data with previous studies. Furthermore, we attained the
potential targets of miR-1-3p by gaining intersection of datasets
that transfected miR-1-3p into CRC cells, online prediction
databases and differentially expressed genes (DEGs) in The Cancer
Genome Atlas (TCGA), and added confirmed targets in literature.
Subsequently we conducted bioinformatics analysis based on
aforementioned selected target genes to unravel the molecular
mechanisms of miR-1-3p in CRC.
Materials and methods
Expression and diagnostic data of
MiR-1-3p in CRC based on literature and microarray from GEO and
ArrayExpress
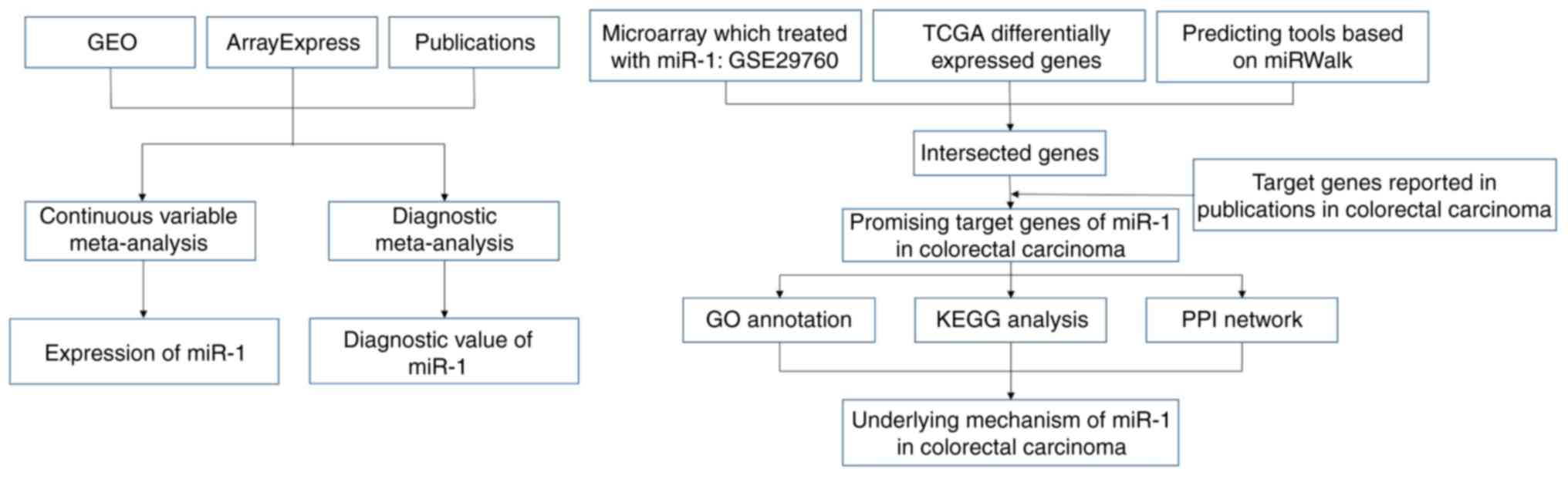
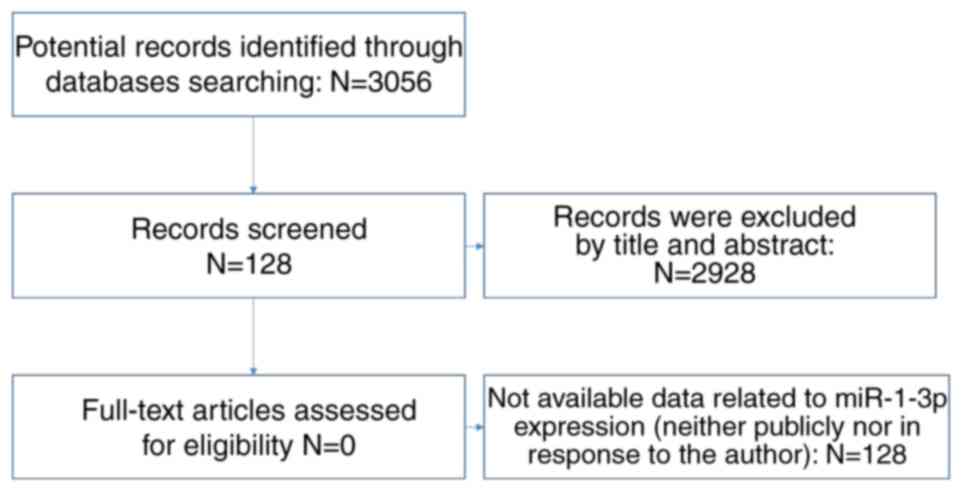
The design of our investigation was shown in
Fig. 1. We searched GEO and
ArrayExpress databases with the following search strategy: (gut OR
intestinal OR colorectal OR colonic OR rectal OR colon OR rectum OR
colon OR colonic OR rectal OR rectum) AND (malignan* OR cancer OR
tumor OR tumour OR neoplas* OR carcinoma). Entry type was filtered
by ‘Series’ and the organism was restricted to Homo sapiens. The
gene chips covering the miR-1-3p level between CRC and non-tumor
controls were included for further analysis. The gene chips whose
precision was less than 14 decimals were excluded. Samples with
other kinds of cancer were eliminated (Fig. 2).
As for literature, we retrieved PubMed, Science
Direct, Google Scholar, Ovid, Wiley Online Library, EMBASE, Web of
Science, Chong Qing VIP, CNKI, Wan Fang and China Biology Medicine
Disc with the same searching strategy. In the aspect of expression
data, studies which we could access the mean, standard deviation
and the case numbers of CRC and non-tumorous group were included
(Fig. 3). With regards of
diagnostic value, we included studies that offered true positive
(TP), false positive (FP), false negative (FN) and true negative
(TN).
Statistical analysis
The data of each individual gene chip was converted
to log2 scale. The number, the mean and the standard deviation of
control group and experimental group were calculated based on each
single gene chip.
We utilized Stata 14 (StataCorp LLC, College
Station, Texas, USA) to display the expression level of miR-1-3p
between neoplastic tissues and non-tumorous tissues in forest
plots. Standardized mean difference (SMD) and 95% confidence
interval (Cl) were calculated to indicate the expression
difference. I2>50% or P<0.05 was considered as
heterogeneous. Fixed effect model was applied at first and then
random effect model was applied when evident heterogeneity still
remained. In order to test the reliability of our analysis, we also
assessed the publication bias of the study by funnel plots, in
which better symmetry suggested less publication bias. Sensitivity
analysis was carried out to explore the source of
heterogeneity.
IBM SPSS v23 (IBM Corp., Armonk, NY, USA) was
applied to perform diagnostic trial based on individual gene chip
to acquire TP, FP, FN and TN. We carried out diagnostic
meta-analysis using Stata 14. Summarized receiver operating
characteristic curve (SROC) were conducted.
Collection of GEO data and literature
about intervening experiments
In order to find the promising target genes of
miR-1-3p, we searched GEO database again about the experiments
which intervened CRC samples or cell lines with miR-1-3p by
knockout or transfection, with the following search strategy:
(malignan* OR cancer OR tumor OR tumour OR neoplas* OR carcinoma)
AND (miR-1 OR miRNA-1 OR microRNA-1 OR miR1 OR miRNA1 OR microRNA1
OR ‘miR 1’ OR ‘miRNA 1’ OR ‘microRNA 1’ OR miR-1-3p OR miRNA-1-3p
OR microRNA-1-3p OR miR-1-1 OR miR-1-2 OR miR1-1 OR miR1-2). The
search range was set as ‘All Fields’. Gene chips after miR-1-3p
intervention which contained mRNAs expression were included.
Meanwhile, in order to collect the target genes of
miR-1-3p, we searched PubMed, Science Direct, Google Scholar, Ovid,
Wiley Online Library, EMBASE, Web of Science, Chong Qing VIP, CNKI,
Wan Fang and China Biology Medicine Disc using the same key words
as in GEO database. The target genes of miR-1-3p which were
verified via experiments were correspondingly recorded.
Obtainment of prospective target genes
of MiR-1-3p
In GSE29760, we considered it significant when
log2FC of down-regulated genes was less than −1.
We downloaded data at the entry of colorectal cancer
from TCGA. There are 459 patients in total contributing 480 tumor
samples and 41 non-neoplasm samples. DESeq data R package was
performed to analyze the DEGs. The up-regulated genes with
log2FC>1 and P<0.05 were considered significant.
As for prediction tools, miRWalk 2.0 was applied to
predict the potential target genes of miR-1-3p based on 12 online
databases, namely miRWalk; Microt4; miRanda; miRBridge; miRDB;
miRMap; miRNAMap; PicTar; PITA; RNA22; RNAhybrid and Targetscan.
For the purpose of acquiring the possible target genes of miR-1-3p
as complete as possible, genes were selected on condition that they
were recorded no less than 2 prediction databases. The genes
selected above were cross-referenced with the DEGs in GEO and TCGA.
We also added the confirmed target genes of miR-1-3p in CRC from
publication databases. The final genes were considered possible
target genes of miR-1-3p.
Analyses in silico via DAVID,
cytoscape and STRING
We performed Gene Ontology (GO) analysis via DAVID
6.8 (https://david-d.ncifcrf.gov/) based upon
aforementioned prospective target genes. The processed GO
categories were biological process (BP), cellular component (CC) as
well as molecular function (MF), by which we could further
understand the enrichment of the prospective target genes. Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis was
applied to explore the significant pathways based on the
prospective target genes. BiNGO and EnrichmentMap plug-in
components in Cytoscape version 3.5.0 were utilized to visualize
the network of GO terms and KEGG pathways. Protein-protein
interaction (PPI) network was built by STRING (http://www.string-db.org) to show the connections
among the possible target genes of miR-1-3p. Disconnected nodes
were hidden in the network.
Results
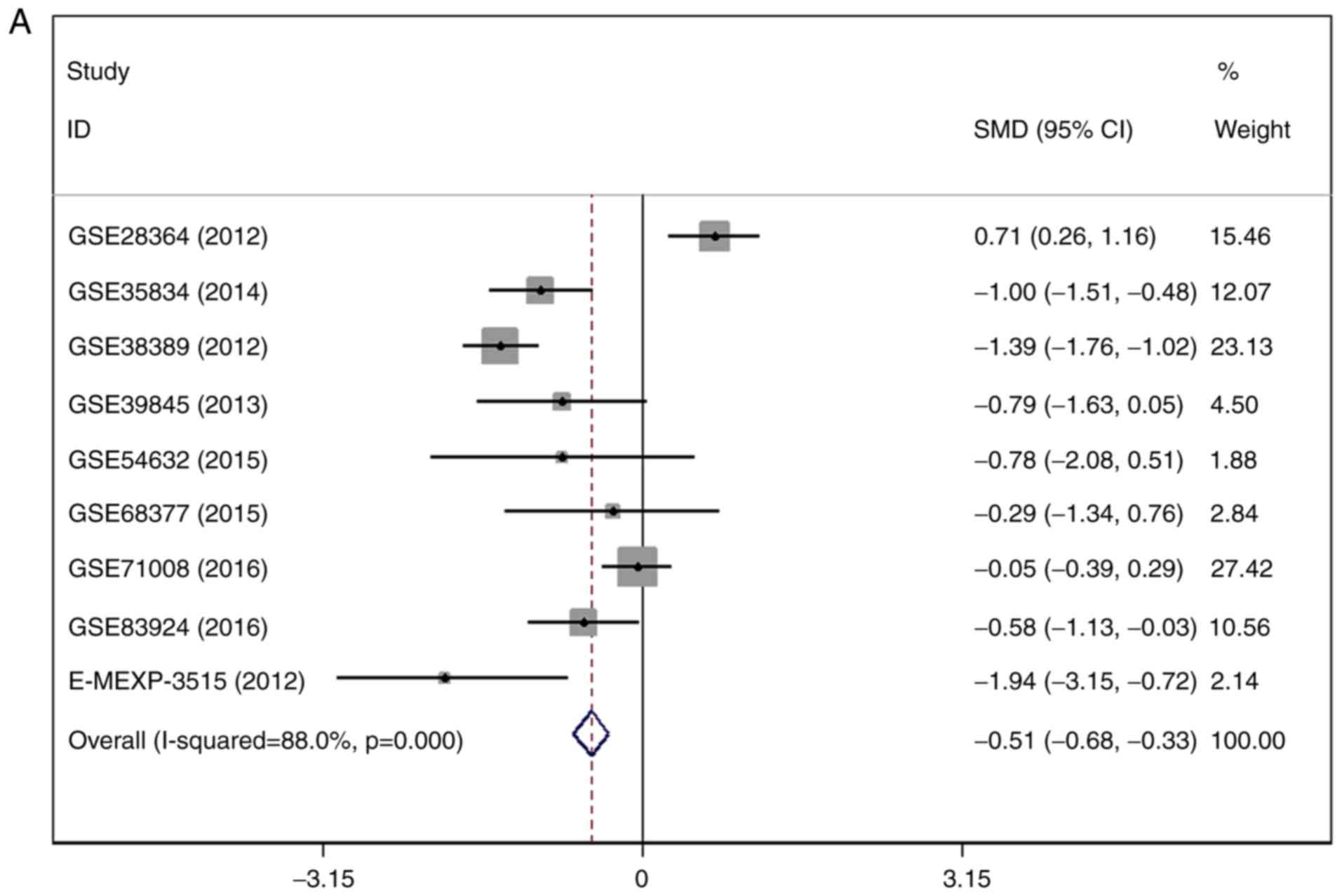
Expression of MiR-1-3p in GEO
There were 7 gene chips from GEO and 1 from
ArrayExpress included, with 589 cases in total. No literature met
our inclusion criteria. The meta-analysis showed the
down-regulation of miR-1-3p in CRC compared to control group. The
combined SMD was −0.51 with 95% confidence interval (CI) of −0.68
to −0.33 and I2=88.0% P=0.000 using fixed effect model
(Fig. 4A). In random effect model,
the SMD changed to −0.63 with 95%CI of −1.19 to −0.07 and
I2=88.0% P=0.000 (Fig.
4B). The SMD in tissue subgroup was −0.67 (95%CI: −0.89, −0.46)
and I2=89.4% (Fig. 4C).
Funnel plot was carried out to test publication bias of our study
(Fig. 4D, P>0.05). Sensitivity
analysis showed that GSE28364 and GSE71008 might be the source of
heterogeneity (Fig. 4E). The SMD
after excluding these two gene chips was −1.06 (95%CI: −1.29,
−0.82) and I2=43.8% (Fig.
4F).
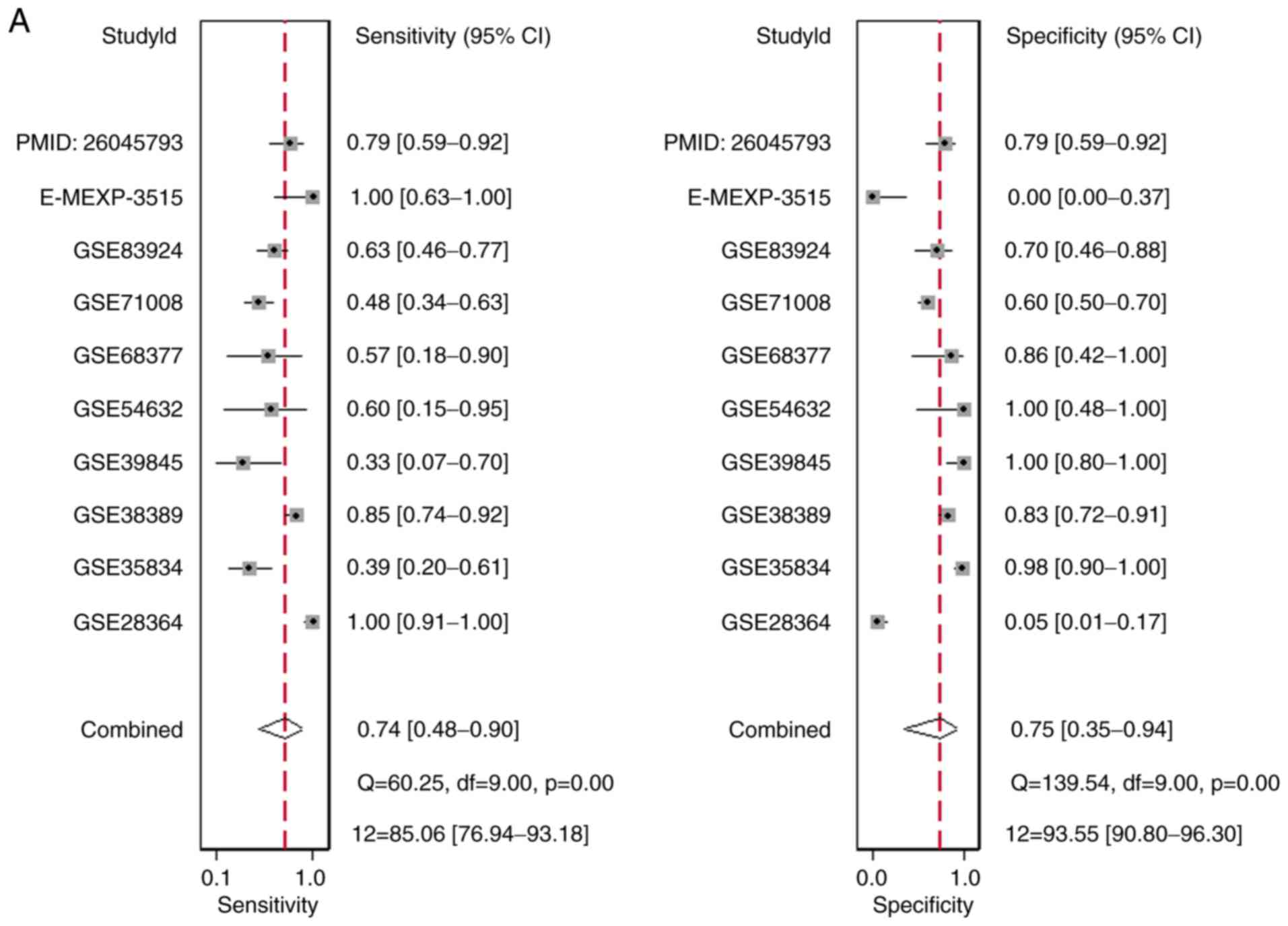
Diagnostic value of MiR-1-3p based on
GEO gene chips and literature
There were 10 studies included, comprising of 9
microarray datasets and 1 publication from Wu et al
(17), containing 645 cases
altogether. The combined sensitivity, specificity, positive
likelihood ratio, negative likelihood ratio, diagnostic score and
odds ratio were 0.74 (95%CI: 0.48, 0.90), 0.75 (95%CI: 0.35, 0.94),
2.94 (95%CI: 1.01, 8.55), 0.34 (95%CI: 0.19, 0.60), 2.15 (95%CI:
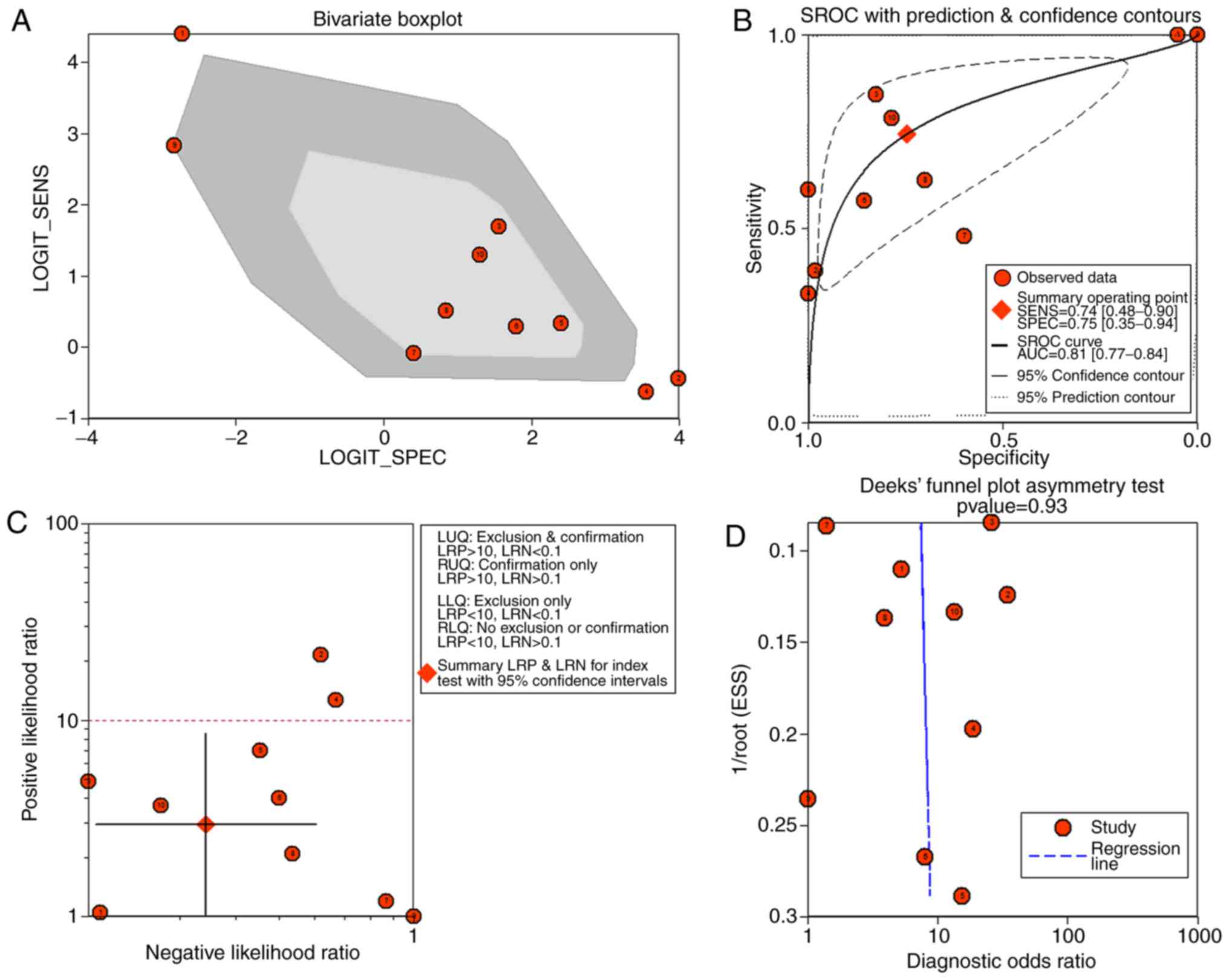
1.06, 3.23) and 8.57 (95%CI: 2.89, 25.36) (Fig. 5). The SROC showed the area under
the curve (AUC) was 0.81 (Fig. 6).
Deek's plot showed P=0.93, which suggested there was no publication
bias (Fig. 6D). We also plotted
Bivariate Boxplot (Fig. 6A),
Likelihood Matrix (Fig. 6C) and
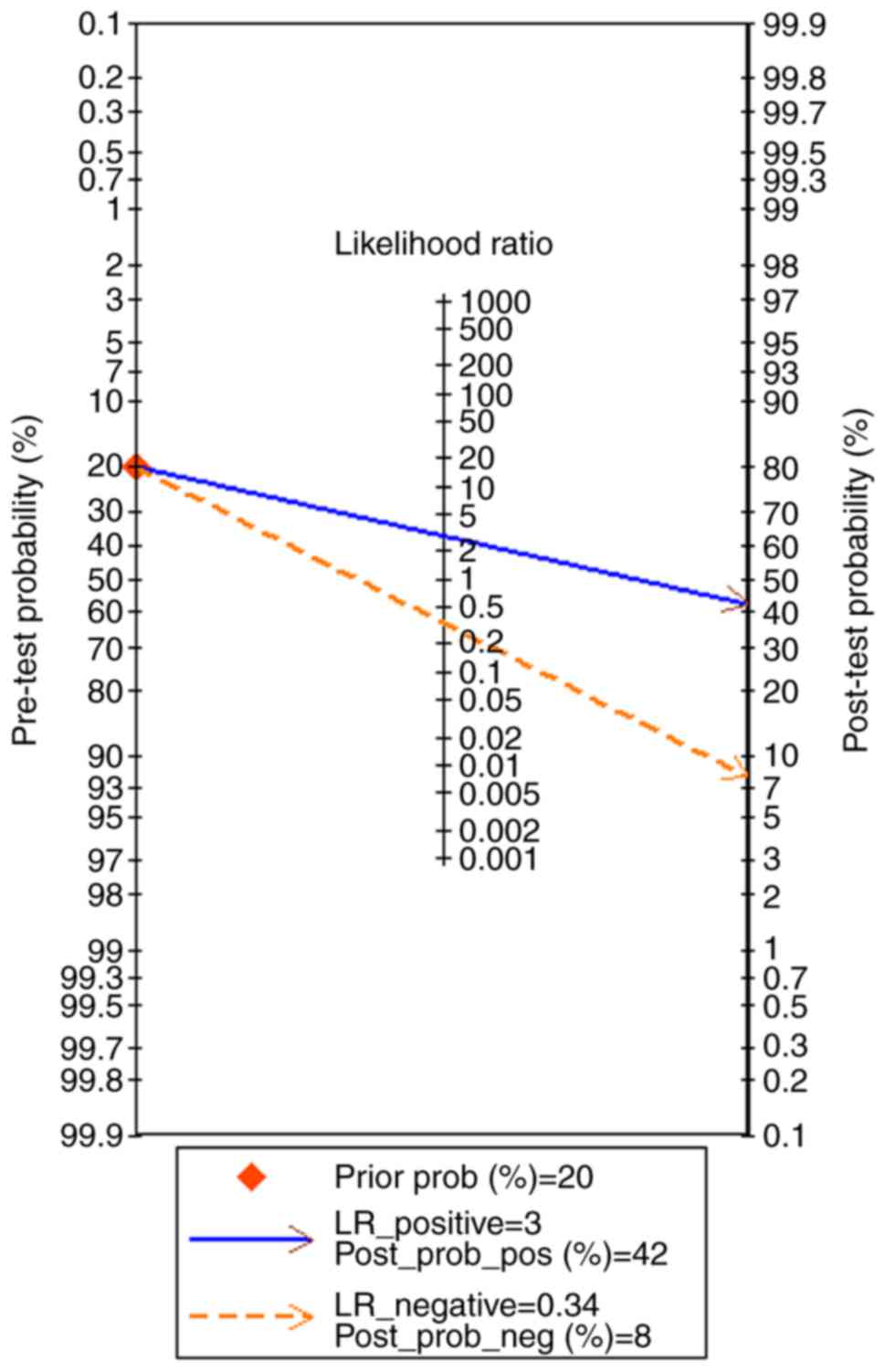
Fagan plot (Fig. 7). To summarize,
the down-regulation of miR-1-3p showed diagnostic potential.
Selection of possible target genes of
MiR-1-3p
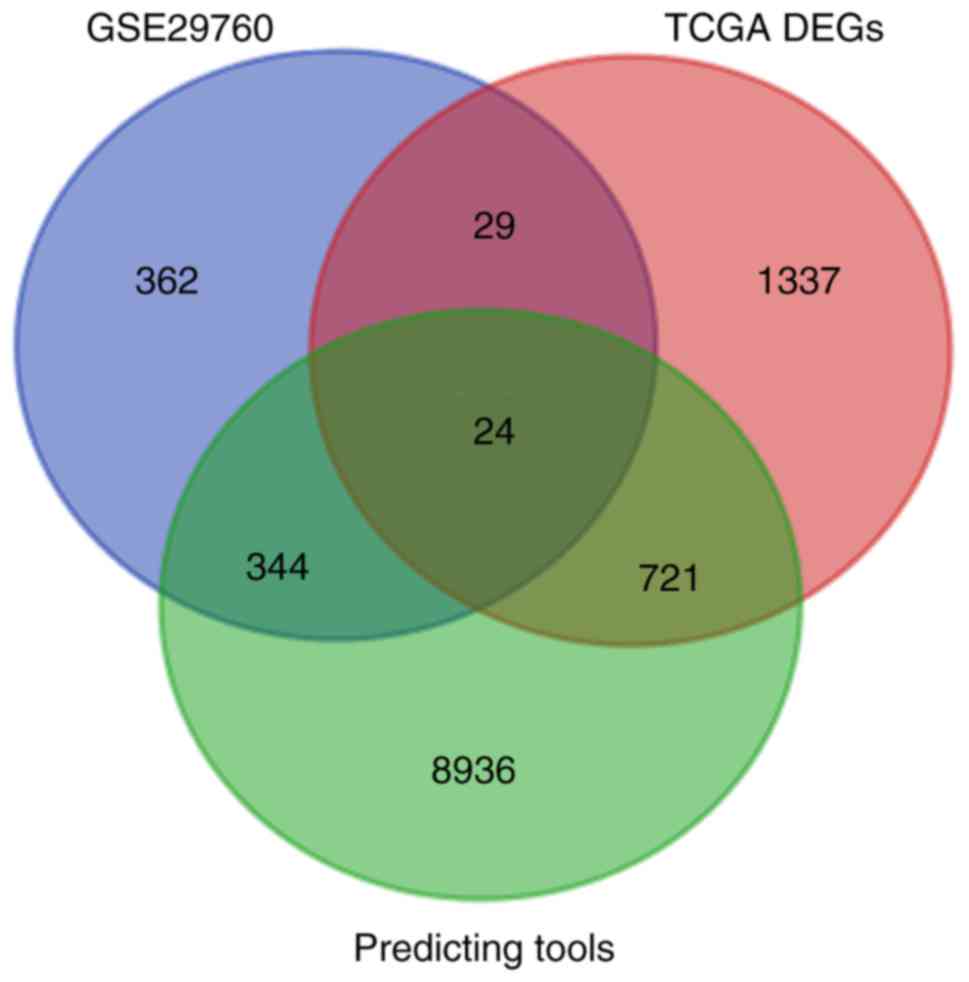
There were 29,834 genes downloaded from miRWalk2.0
with filtration of at least 2 sum scores. In GSE29760, 759 were
selected. In TCGA, there were 2111 DEGs. We obtained 24 overlapping
genes (NEB, SERPIND1, IL1RAP, SPARC, GNG4, FOXA2, SLC25A22, CSF2,
F5, SULT1C2, C2orf61, CREG2, ODAM, PPM1H, TDGF1, E2F5, SH3TC2,
SDR16C5, PF4, IL20RA, GPR143, HOXC11, IL1A and FAM57A) (Fig. 8). Then we added the 6 confirmed
target genes of miR-1-3p in CRC from literature (NOTCH3, LIM,
LASP1, PIK3CA, TWIST1 and GATA4).
Bioinformatics analysis of the
potential target genes of MiR-1-3p
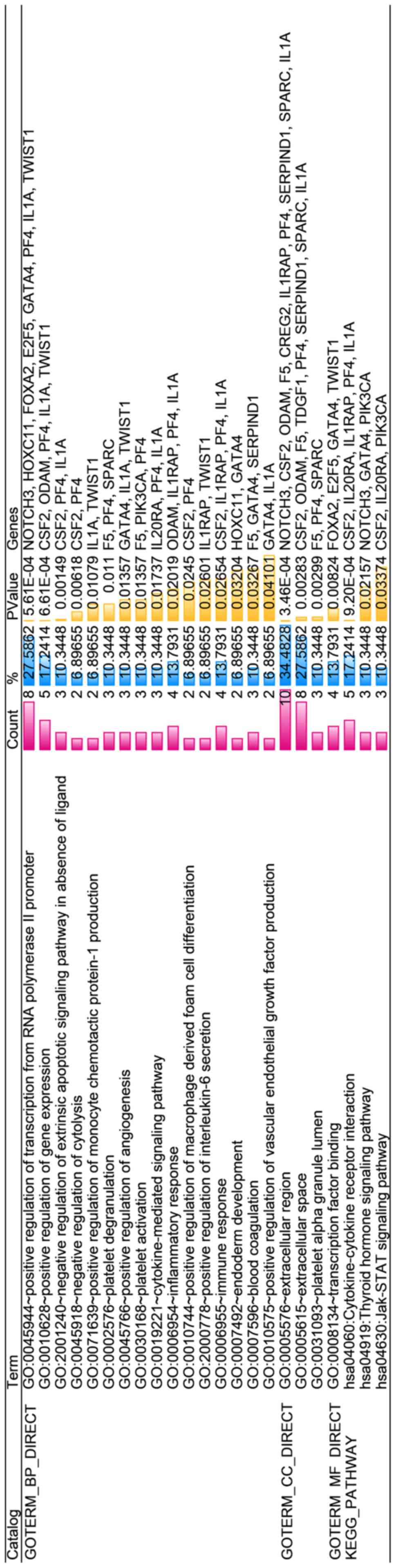
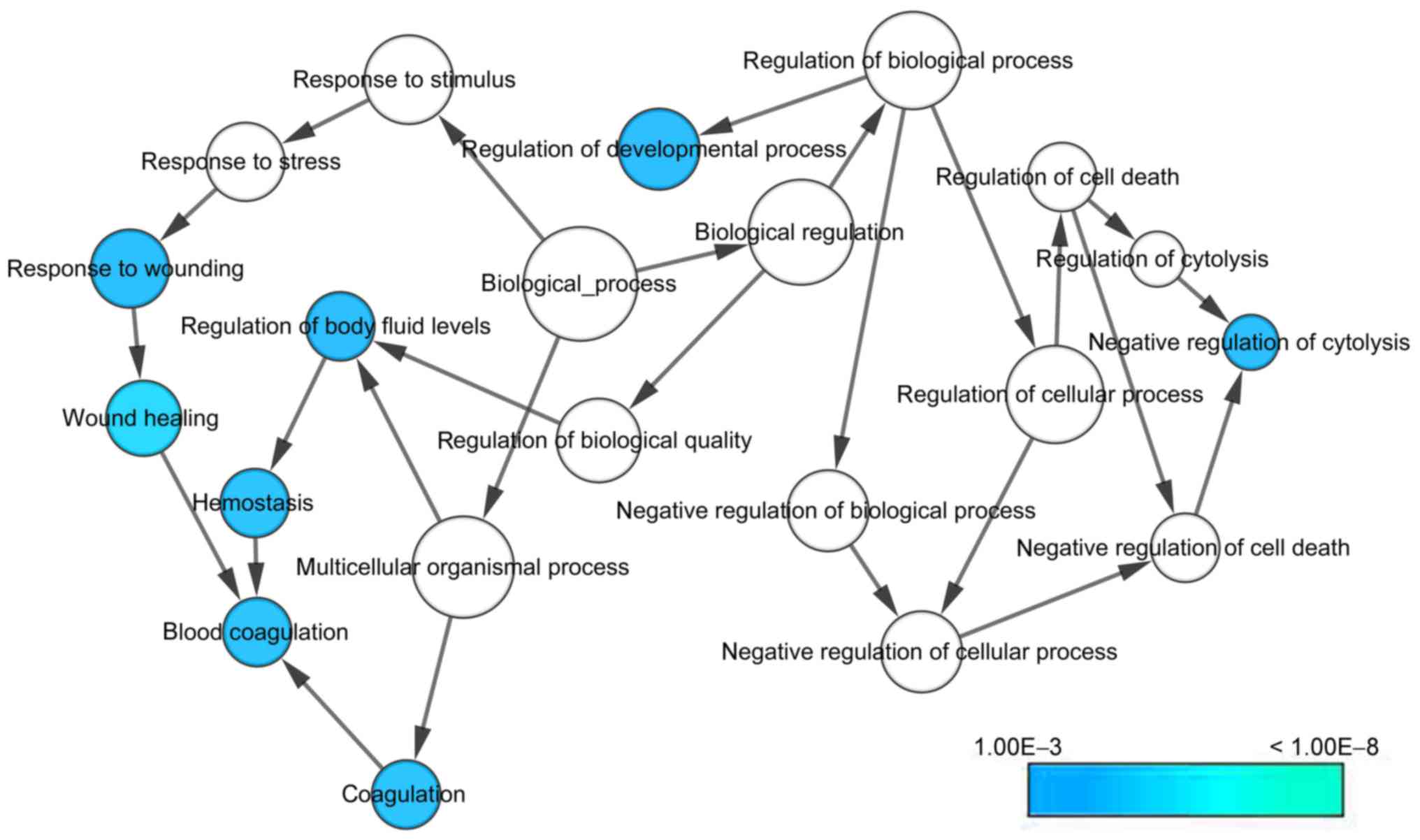
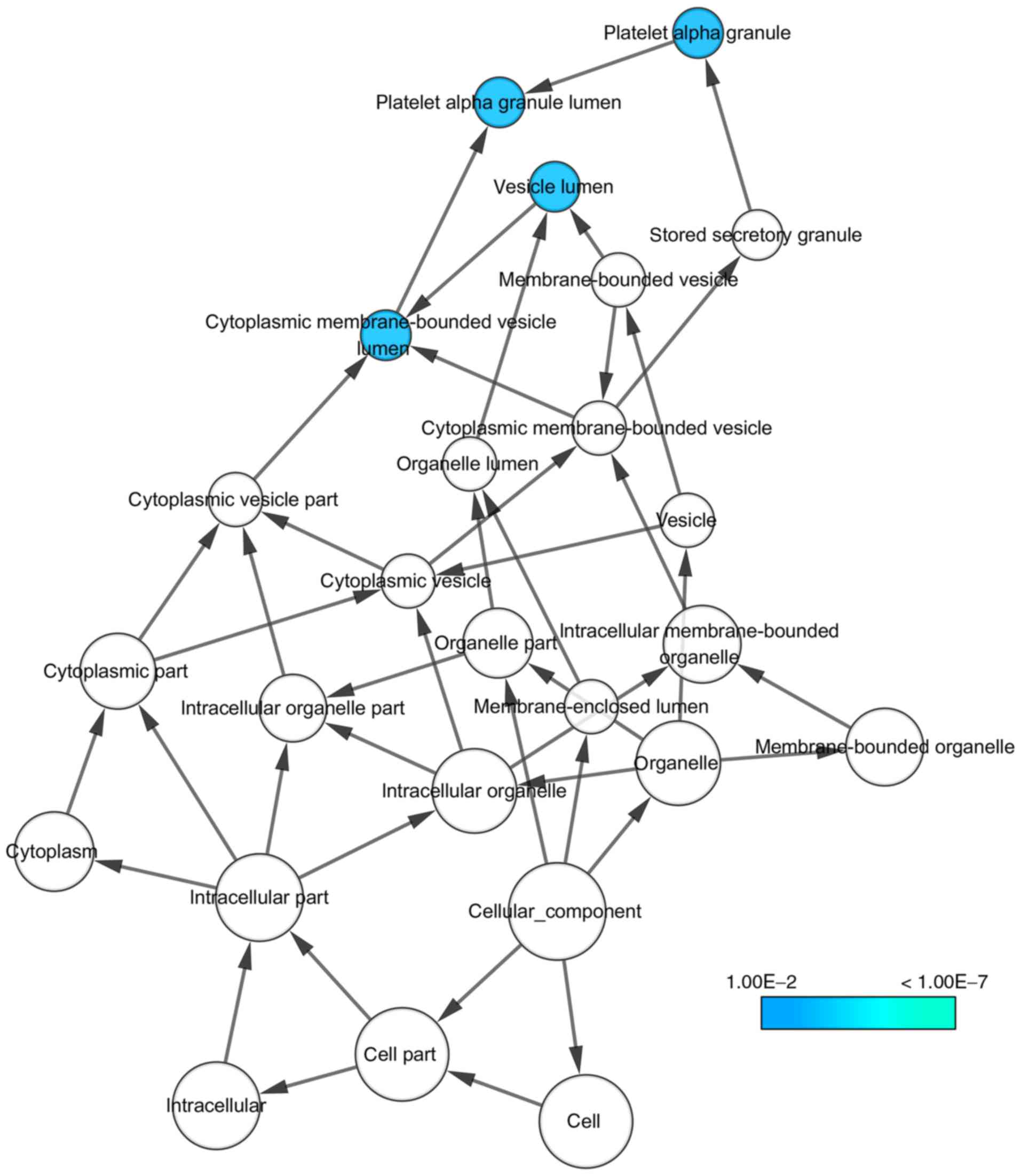
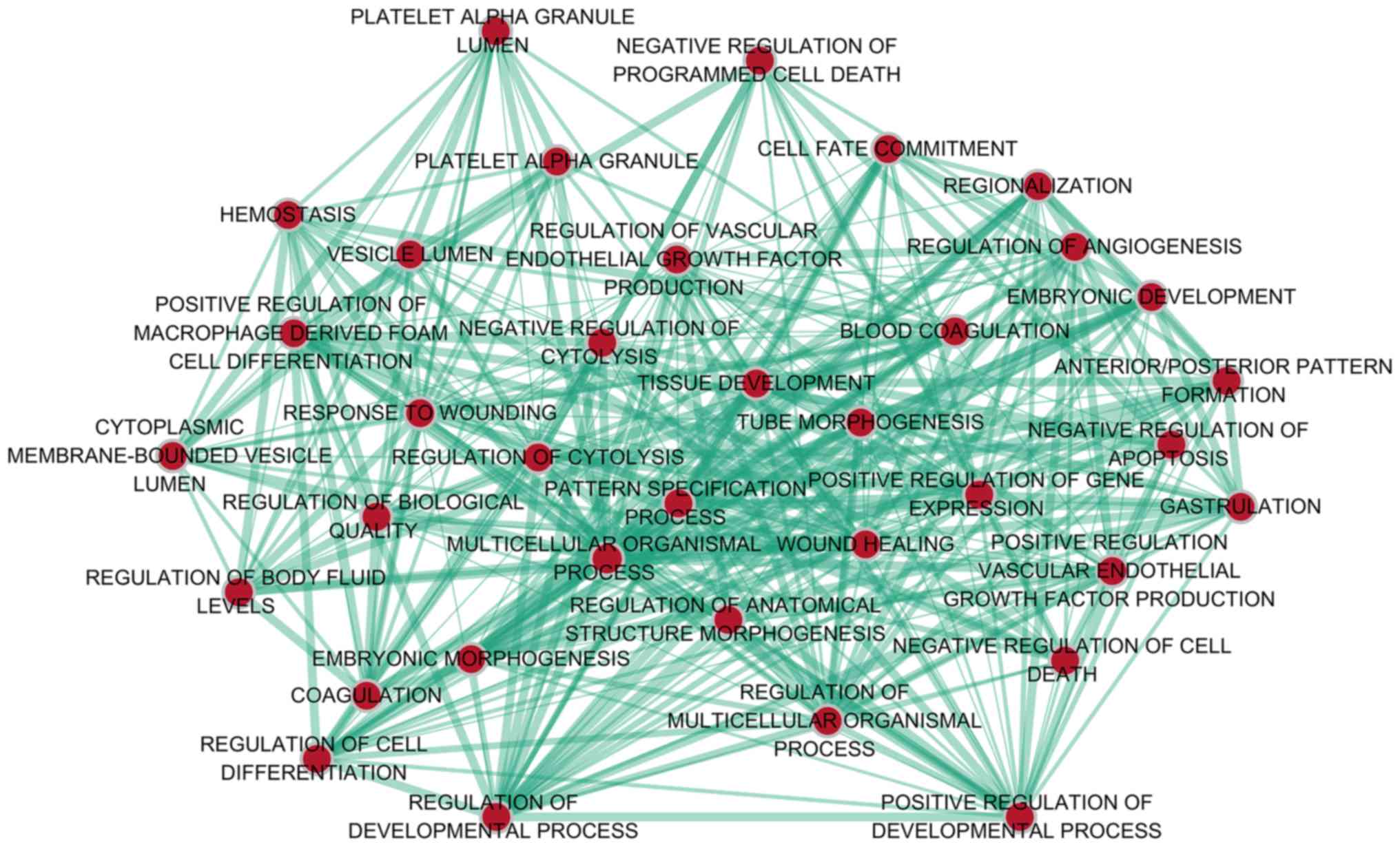
With regard to GO annotation in DAVID, the target
genes were closely related to positive regulation of transcription
from RNA polymerase II promoter in biological process (BP)
(Figs. 9 and 10, P=0.01), extracellular region in
cellular component (CC) (Fig. 9,
P=3.46E-04), transcription factor binding in molecule function (MF)
(Figs. 9 and 11, P=0.01). Moreover, KEGG pathway
analysis (Figs. 9 and 12) revealed that cytokine-cytokine
receptor interaction (P=9.20E-04), thyroid hormone signaling
pathway (P=0.02) and JAK-STAT signaling pathway (P=0.03) were
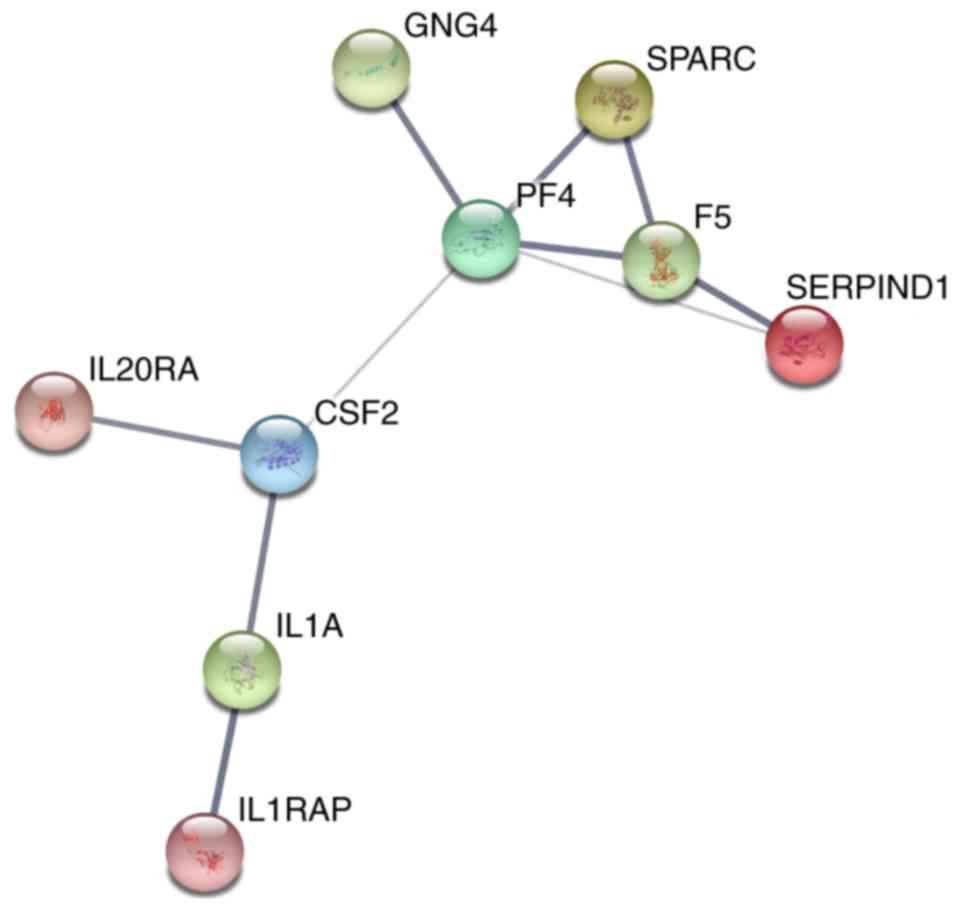
significant. PPI network (Fig.
13) was comprised of GNG4, SPARC, PF4, F5, SERPIND1, IL20RA,
CSF2, IL1A and IL1RAP, with enrichment P-value of 0.000427.
Discussion
In our present study, we indicated lower expression
level of miR-1-3p in CRC than in non-cancerous tissues, which
indicated moderate diagnostic accuracy with AUC of 0.81.
Bioinformatics analyses based on 30 promising target genes
highlighted positive regulation of transcription from RNA
polymerase II promoter, extracellular region and transcription
factor binding in GO analysis; cytokine-cytokine receptor
interaction in KEGG pathway analysis; PF4 in PPI analysis.
At present, the down-regulation of miR-1-3p in CRC
and other types of cancer has been observed extensively.
Nevertheless, exceptional circumstance still exists. As mentioned
in Introduction, we noticed Li et al (16), indicated over-expression of
miR-1-3p in CRC with brain metastasis. Because only 4 patients were
enrolled in this study, more experiments both in vitro and
in vivo are required to verify that whether miR-1-3p
possesses contrary functions in CRC. Concerning how to collect data
and demonstrate the reduced expression, most studies utilized
microarray profiling and quantitative real-time polymerase chain
reaction (qRT-PCR) to extract expression profile from tissue
samples. However, the sample collected by a single research is
limited, and there is no study yet concentrating on collecting
existing cases together and excavating the massive materials in
public gene expression databases. Thus the meta-analysis we
conducted containing literature and microarray data is more
reliable in proving the weakened expression of miR-1-3p in CRC.
Combining results of diagnostic tests, we can deduct that the
down-regulation of miR-1-3p might serve as a more powerful
diagnostic target when grouping together with other possible
biomarkers. The suppressed expression also presented that miR-1-3p
is highly possible to function as a tumor suppressor.
In terms of GO analysis, positive regulation of
transcription from RNA polymerase II (Pol II) promoter was listed
as the most pivotal biological process. Pol II participated in
catalysis of DNA transcription, and studies have found the
degradation of Pol II in cancer cells induced by drugs could lead
to cytotoxic effects (19). In CRC
with microsatellite instability, TAF7, which indirectly impedes Pol
II catalyzing by inhibiting the necessary transcriptional factor
for Pol II to initiate, was found mutated (20). As a result, we infer that by
targeting the genes which can promote the transcription of Pol II
promoter, miR-1-3p could negatively regulate CRC proliferation and
perform antitumor efficacy.
JAK-STAT signaling pathway ranked the third
significant pathway in KEGG pathway analysis. Associated with
malignancies and immune diseases, JAK-STAT signaling pathway
assists cells to respond to cytokines (21,22),
which coincides with ‘cytokine-cytokine receptor interaction’, the
most significant signaling pathway in our analysis. In CRC,
Slattery et al (23),
proved JAK-STAT signaling pathway was intimately connected to CRC
risk and survival in their case-control studies. Wang et al
(24) blocked JAK-STAT signaling
pathway by intervening its up-stream modulator, which contributed
to inhibiting CRC progression and improving CRC survival. In
addition, non-coding RNAs, especially miRNAs, were reported to have
regulatory associations with JAK-STAT signaling pathway (25). For instance, miR-23a and miR-23b
was discovered to target at JAK-STAT signaling pathway in prostate
cancer by decreasing the expression of IL-6R (26). Therefore, it is deducted that
miR-1-3p might be a roadblock in JAK-STAT signaling pathway by
silencing the target genes involved in it, and confront CRC by
inducing apoptosis and reduce proliferation.
PF4 was the hub gene with most interactions with
other genes in PPI network. It mainly participates in blood
coagulating, wound repair and inflammatory (27). With regard of neoplasms, PF4 was
observed having different roles in various cancers. In animal
breast cancer model, PF4 could induce cancer cell apoptosis with
the assistance of rapamycin (28).
However, contrary results appeared in lung cancer. Pucci et
al (29) considered PF4 as an
oncogene for it bettered the microenvironment of tumor and promoted
its proliferation. Jian et al suggested that PF4 was a
protective factor in lung cancer to impede metastasis (30). In CRC, three types of results
emerged. Abbasciano et al (31), didn't notice any different
expression of PF4 in large bowel carcinoma; Peterson et al
(32), claimed PF4 was
up-regulated in CRC and its high expression owned diagnostic
potential; while Maione et al (33), applied an analogue of PF4 to
inhibit CRC development. Apparently, we still have vast space to
explore the functions and mechanisms of PF4, in malignancies
particularly.
Although we have delineate the map of comprehensive
expression and functional annotation with existing data, more
experiments were still needed to further validate and better apply
the clinical value of miR-1-3p in CRC. Clinicopathological
variables that are statistically related to miR-1-3p expression
level and survival statistics also require summarizing and
analyzing to judge whether miR-1-3p is related to prognosis and
other indexes in CRC.
To summarize, miR-1-3p is down-regulated in CRC and
is likely to suppress CRC via multiple biological approaches, which
indicated diagnostic potential and tumor suppressor efficacy.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stewart BW and Wild C; International
Agency for Research on Cancer, : World cancer report 2014. IARC
Nonserial Publication; 2014
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hesse M and Arenz C: MicroRNA maturation
and human disease. Methods Mol Biol. 1095:11–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qu YL, Wang HF, Sun ZQ, Tang Y, Han XN, Yu
XB and Liu K: Up-regulated miR-155-5p promotes cell proliferation,
invasion and metastasis in colorectal carcinoma. Int J Clin Exp
Pathol. 8:6988–6994. 2015.PubMed/NCBI
|
|
6
|
Zhong M, Bian Z and Wu Z: miR-30a
suppresses cell migration and invasion through downregulation of
PIK3CD in colorectal carcinoma. Cell Physiol Biochem. 31:209–218.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang XH: MicroRNA in myogenesis and muscle
atrophy. Curr Opin Clin Nutr Metab Care. 16:258–266. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karatas OF, Guzel E, Suer I, Ekici ID,
Caskurlu T, Creighton CJ, Ittmann M and Ozen M: miR-1 and miR-133b
are differentially expressed in patients with recurrent prostate
cancer. PLoS One. 9:e986752014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li D, Yang P, Li H, Cheng P, Zhang L, Wei
D, Su X, Peng J, Gao H, Tan Y, et al: MicroRNA-1 inhibits
proliferation of hepatocarcinoma cells by targeting endothelin-1.
Life Sci. 91:440–447. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Letelier P, Garcia P, Leal P, Álvarez H,
Ili C, López J, Castillo J, Brebi P and Roa JC: miR-1 and miR-145
act as tumor suppressor microRNAs in gallbladder cancer. Int J Clin
Exp Pathol. 7:1849–1867. 2014.PubMed/NCBI
|
|
11
|
Zhao Q, Zhang B, Shao Y, Chen L, Wang X,
Zhang Z, Shu Y and Guo R: Correlation between the expression levels
of miR-1 and PIK3CA in non-small-cell lung cancer and their
relationship with clinical characteristics and prognosis. Future
Oncol. 10:49–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han C, Zhou Y, An Q, Li F, Li D, Zhang X,
Yu Z, Zheng L, Duan Z and Kan Q: MicroRNA-1 (miR-1) inhibits
gastric cancer cell proliferation and migration by targeting MET.
Tumour Biol. 36:6715–6723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh A, Happel C, Manna SK,
Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW,
Wakabayashi N, Dewi R, et al: Transcription factor NRF2 regulates
miR-1 and miR-206 to drive tumorigenesis. J Clin Invest.
123:2921–2934. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chang YS, Chen WY, Yin JJ,
Sheppard-Tillman H, Huang J and Liu YN: EGF receptor promotes
prostate cancer bone metastasis by downregulating miR-1 and
activating TWIST1. Cancer Res. 75:3077–3086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin C, Yan B, Lu Q, Lin Y and Ma L:
Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1
promotes triple-negative breast cancer development. Tumour Biol.
37:7383–7394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Gu X, Fang Y, Xiang J and Chen Z:
microRNA expression profiles in human colorectal cancers with brain
metastases. Oncol Lett. 3:346–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu X, Li S, Xu X, Wu S, Chen R, Jiang Q,
Li Y and Xu Y: The potential value of miR-1 and miR-374b as
biomarkers for colorectal cancer. Int J Clin Exp Pathol.
8:2840–2851. 2015.PubMed/NCBI
|
|
18
|
Furukawa S, Kawasaki Y, Miyamoto M,
Hiyoshi M, Kitayama J and Akiyama T: The miR-1-NOTCH3-Asef pathway
is important for colorectal tumor cell migration. PLoS One.
8:e806092013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Manzo SG, Zhou ZL, Wang YQ, Marinello J,
He JX, Li YC, Ding J, Capranico G and Miao ZH: Natural product
triptolide mediates cancer cell death by triggering CDK7-dependent
degradation of RNA polymerase II. Cancer Res. 72:5363–5373. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oh HR, An CH, Yoo NJ and Lee SH:
Frameshift mutations of TAF7L gene, a core component for
transcription by RNA polymerase II, in colorectal cancers. Pathol
Oncol Res. 21:849–850. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Harrison DA: The Jak/STAT pathway. Cold
Spring Harb Perspect Biol. 4(pii): a0112052012.PubMed/NCBI
|
|
22
|
O'Shea JJ, Schwartz DM, Villarino AV,
Gadina M, McInnes IB and Laurence A: The JAK-STAT pathway: Impact
on human disease and therapeutic intervention. Annu Rev Med.
66:311–328. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Slattery ML, Lundgreen A, Kadlubar SA,
Bondurant KL and Wolff RK: JAK/STAT/SOCS-signaling pathway and
colon and rectal cancer. Mol Carcinog. 52:155–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang SW, Hu J, Guo QH, Zhao Y, Cheng JJ,
Zhang DS, Fei Q, Li J and Sun YM: AZD1480, a JAK inhibitor,
inhibits cell growth and survival of colorectal cancer via
modulating the JAK2/STAT3 signaling pathway. Oncol Rep.
32:1991–1998. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Witte S and Muljo SA: Integrating
non-coding RNAs in JAK-STAT regulatory networks. JAKSTAT.
3:e280552014.PubMed/NCBI
|
|
26
|
Aghaee-Bakhtiari SH, Arefian E, Naderi M,
Noorbakhsh F, Nodouzi V, Asgari M, Fard-Esfahani P, Mahdian R and
Soleimani M: MAPK and JAK/STAT pathways targeted by miR-23a and
miR-23b in prostate cancer: Computational and in vitro approaches.
Tumour Biol. 36:4203–4212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eisman R, Surrey S, Ramachandran B,
Schwartz E and Poncz M: Structural and functional comparison of the
genes for human platelet factor 4 and PF4alt. Blood. 76:336–344.
1990.PubMed/NCBI
|
|
28
|
Al-Astani Tengku Din TA, Shamsuddin SH,
Idris FM, Ariffin Wan Mansor WN, Abdul Jalal MI and Jaafar H:
Rapamycin and PF4 induce apoptosis by upregulating Bax and
down-regulating survivin in MNU-induced breast cancer. Asian Pac J
Cancer Prev. 15:3939–3944. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pucci F, Rickelt S, Newton AP, Garris C,
Nunes E, Evavold C, Pfirschke C, Engblom C, Mino-Kenudson M, Hynes
RO, et al: PF4 promotes platelet production and lung cancer growth.
Cell Rep. 17:1764–1772. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jian J, Pang Y, Yan HH, Min Y, Achyut BR,
Hollander MC, Lin PC, Liang X and Yang L: Platelet factor 4 is
produced by subsets of myeloid cells in premetastatic lung and
inhibits tumor metastasis. Oncotarget. 8:27725–27739. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abbasciano V, Bianchi MP, Trevisani L,
Sartori S, Gilli G and Zavagli G: Platelet activation and
fibrinolysis in large bowel cancer. Oncology. 52:381–384. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peterson JE, Zurakowski D, Italiano JE Jr,
Michel LV, Connors S, Oenick M, D'Amato RJ, Klement GL and Folkman
J: VEGF, PF4 and PDGF are elevated in platelets of colorectal
cancer patients. Angiogenesis. 15:265–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maione TE, Gray GS, Hunt AJ and Sharpe RJ:
Inhibition of tumor growth in mice by an analogue of platelet
factor 4 that lacks affinity for heparin and retains potent
angiostatic activity. Cancer Res. 51:2077–2083. 1991.PubMed/NCBI
|