Introduction
Sepsis is a systemic inflammatory response syndrome
that occurs when a bacterial, viral or fungal infection spreads to
the bloodstream and induces life-threatening organ dysfunction
(1). Without immediate and
aggressive treatment, sepsis can rapidly cause tissue damage, organ
failure, and even death. More than 5 million people die from sepsis
every year worldwide (2). Despite
numerous advances in fundamental and clinical research, the
mortality rate for sepsis remains high (3–4).
Over the last 30 years, over 100 clinical trials have failed to
indicate a survival benefit for patients with severe sepsis, and
the failure is due, at least partially, to host heterogeneity
(5–8). Patients vary in their circumstances,
and a more precise assessment of sepsis is required.
Risk factors associated with deterioration or
mortality may be used to diagnose and evaluate the severity of
sepsis. Thereby, the subset of patients with a high risk of
mortality for sepsis may receive additional, aggressive therapies.
Thus far, a number of biomarkers have been widely used for the
diagnosis, prognosis and treatment of sepsis (5,10–12).
For example, C-reactive protein (CRP) is an acute phase protein,
which increases rapidly in response to most forms of inflammation,
infection, and tissue damage. High levels of CRP are associated
with the risk of sepsis, cardiovascular disease and stroke
(12–14). While CRP is broadly used for
clinical diagnosis of acute sepsis, it lacks the capacity to
differentiate between infective and non-infective inflammation and
have low specificity for severe sepsis (15,16).
Severe sepsis is often attributed to immune dysregulation, and the
imbalance between pro- and anti-inflammatory cytokines may serve a
crucial role in the pathogenesis of sepsis (17). Patients who express high levels of
interleukin (IL)-6 have an increased risk of mortality (18,19).
Although the combination of IL-6 and CRP plasma biomarkers may be
helpful in sepsis diagnosis, recent studies demonstrated that IL-6
and CRP are not ideal as they lack sufficient sensitivity and
specificity (20–22).
Genome expression profiling is a potential approach
to discover novel risk factors based on microarray technology and
bioinformatics (23). Microarray
technology enables researchers to gain insights into signaling
pathways and gene networks that may participate in sepsis
development (24–26). The present study focused on the
identification of risk factors for predicting sepsis deterioration
by genome-wide expression profiling. Differentially expressed genes
(DEGs) between sepsis survivors and nonsurvivors were analyzed, and
type I interferon (IFN-I) signaling was identified as an important
risk factor for sepsis-associated mortality.
Materials and methods
Gene expression profiles
Two gene expression profiles (GSE54514 and GSE3140)
were downloaded from public functional genomics data repository
Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) (27–28).
The array data from GSE54514 were performed on the platform of
Illumina Human HT-12 v3.0 Expression BeadChip (GPL6947; Illumina,
Inc., San Diego, CA, USA). This dataset contained 53 blood samples,
including 26 samples from sepsis survivors, 9 samples from sepsis
nonsurvivors and 18 samples from healthy controls.
The gene expression profile of GSE3140 was performed
on the platform of an Affymetrix GeneChip Human HG-Focus Target
Array (GPL201; Affymetrix; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Prior to the mRNA expression profiling, blood samples
were collected from 6 healthy adult volunteers and 6 healthy,
full-term infants, and RNA was isolated from cord blood and adult
peripheral blood mononuclear cells of blood samples following
incubation with or without lipopolysaccharide (LPS).
Screening of DEGs
DEGs were identified using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/).
GEO2R is an R-based web tool for performing differential gene
expression analysis in the GEO data repository (29). The adjusted P-values (adj. P) were
adjusted using the Benjamini and Hochberg false discovery rate
method (30). Genes with adj.
P<0.05 and |logFC|>1 were considered to be DEGs, and the
heatmap of DEGs was generated using the heatmap visualization tool
Morpheus (software.broadinstitute.org/morpheus/).
Functional enrichment of DEGs
The Gene Ontology (GO) defines classes used to
describe gene function and associations between biology concepts
(31). It classifies functions
according to three aspects: Molecular Function, Cellular Component
and Biological Process. In the present study, GO enrichment
analysis of DEGs was performed using Gene Ontology Consortium
(www.geneontology.org/), with P<0.05
indicating a significantly enriched term (32).
Protein-protein interaction (PPI)
network
STRING is a database and web resource of
experimental and predicted PPIs. STRING provides a score for each
interaction, and these scores are indicators of confidence and rank
from 0–1. The online STRING 10.5 database (string-db.org/) was used to analyze protein
interactions and a confidence score >0.4 was used as the cut-off
criterion (33).
Results
Identification of DEGs between sepsis
survivors and nonsurvivors
To determine the early risk factors for
sepsis-associated mortality, the GSE54514 dataset was downloaded
from the GEO database and GEO2R was used to identify the DEGs
between blood samples from sepsis survivors and nonsurvivors. A
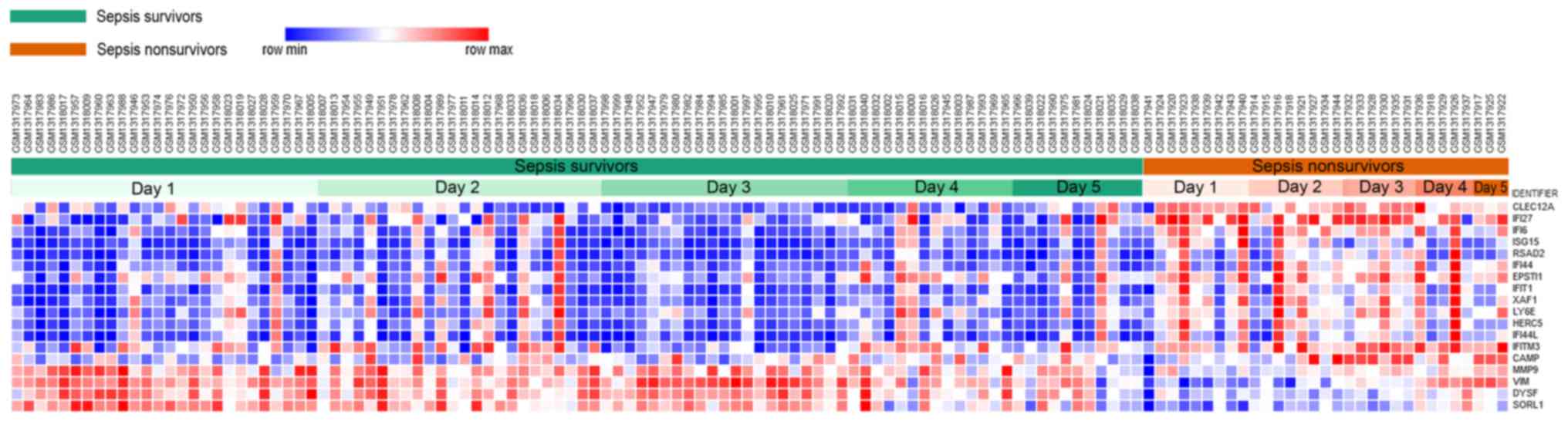
heatmap of DEGs was subsequently generated by Morpheus. The results
demonstrated that a total of 18 DEGs were identified, including 14
upregulated genes and 4 downregulated genes in nonsurvivors
compared with survivors (Fig.
1).
IFN-I signaling is upregulated in
nonsurvivors of sepsis
The identified DEGs were functionally enriched by GO
analysis using the GOC website with P<0.05 as the threshold. As
demonstrated in Table I, DEGs that
were upregulated in the nonsurvivors group compared with the
survivors group were highly enriched in 15 pathways, including
‘type I interferon signaling pathway’, ‘cellular response to type I
interferon’, ‘response to type I interferon’, ‘negative regulation
of viral genome replication’ and ‘regulation of viral genome
replication’. To further investigate the association between the
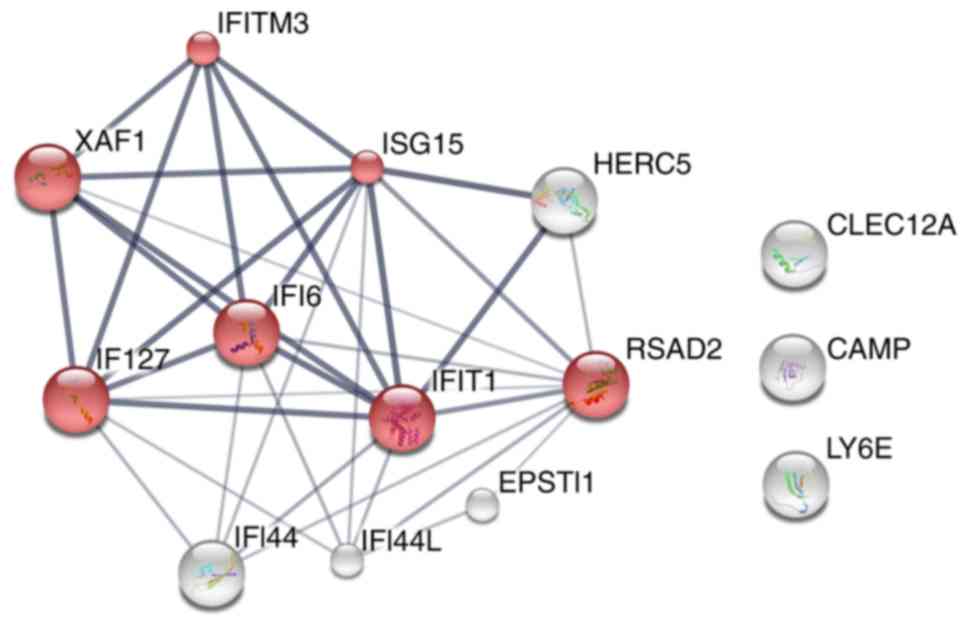
upregulated genes, STRING was used to construct the PPI network. As
demonstrated in Fig. 2, a set of
proteins from IFN-I signaling pathway closely interacted with each
other, which indicates that they may be implicated in sepsis
biology due to their interactions. These results indicated that
upregulated IFN-I signaling may be a risk factor for
sepsis-associated mortality.
 | Table I.Enriched pathways of differentially
expressed genes that were upregulated in the whole blood of sepsis
nonsurvivors compared with survivors. |
Table I.
Enriched pathways of differentially
expressed genes that were upregulated in the whole blood of sepsis
nonsurvivors compared with survivors.
| Pathway ID | Pathway
description | Count | Fold enrichment | P-value |
|---|
| GO:0060337 | Type I interferon
signaling pathway | 7 | >100 |
1.14×10−9 |
| GO:0071357 | Cellular response
to type I interferon | 7 | >100 |
1.14×10−9 |
| GO:0034340 | Response to type I
interferon | 7 | >100 |
1.72×10−9 |
| GO:0045071 | Negative regulation
of viral genome replication | 4 | 84.91 |
1.21×10−3 |
| GO:0045069 | Regulation of viral
genome replication | 4 | 53.84 |
7.33×10−3 |
| GO:1903901 | Negative regulation
of viral life cycle | 4 | 49.61 |
1.01×10−2 |
| GO:0048525 | Negative regulation
of viral process | 4 | 39.42 |
2.51×10−2 |
| GO:0051607 | Defense response to
virus | 6 | 38.96 |
5.96×10−5 |
| GO:0043901 | Negative regulation
of multi-organism process | 5 | 34.07 |
2.48×10−3 |
| GO:0009615 | Response to
virus | 7 | 30.54 |
1.40×10−5 |
| GO:0043903 | Regulation of
symbiosis, encompassing mutualism through parasitism | 6 | 22.53 |
1.49×10−3 |
| GO:0050792 | Regulation of viral
process | 5 | 20.37 |
3.05×10−2 |
| GO:0098542 | Defense response to
other organism | 7 | 17.68 |
5.84×10−4 |
| GO:0043900 | Regulation of
multi-organism process | 6 | 17.03 |
7.61×10−3 |
| GO:0019221 | Cytokine-mediated
signaling pathway | 7 | 16.13 |
1.09×10−3 |
IFN-I signaling is upregulated in
blood samples from LPS-treated healthy neonates
IFN-I signaling is crucial for the host defense;
however, its role in sepsis remains controversial. The results of
the present study indicated that, during early sepsis, upregulated
IFN-I signaling may be a marker for an increased risk of mortality.
Therefore, the present study also investigated whether this
signaling pathway was dysregulated in a subset of patients with a
high risk of sepsis-induced mortality (34). It is widely accepted that neonates
suffer higher sepsis mortality rates compared with adults (35), therefore, DEGs between the cord
blood mononuclear cells of healthy neonates and adult peripheral
blood mononuclear cells were analyzed and numerous DEGs were
identified by GEO2R. However, instead of IFN-1 signaling, DEGs
upregulated in healthy neonates compared with healthy adults were
largely enriched in ‘protoporphyrinogen IX metabolic process’, ‘gas
transport’, ‘oxygen transport’, ‘erythrocyte development’ and
‘porphyrin-containing compound biosynthetic process’, among others
(Table II).
 | Table II.Enriched pathways of differentially
expressed genes that were upregulated in the untreated cord blood
mononuclear cells of healthy neonates compared with the untreated
peripheral blood mononuclear cells of healthy adults. |
Table II.
Enriched pathways of differentially
expressed genes that were upregulated in the untreated cord blood
mononuclear cells of healthy neonates compared with the untreated
peripheral blood mononuclear cells of healthy adults.
| Pathway ID | Pathway
description | Count | Fold
enrichment | P-value |
|---|
| GO:0046501 | Protoporphyrinogen
IX metabolic process | 11 | 33.45 |
3.23×10−4 |
| GO:0015669 | Gas transport | 19 | 25.82 |
1.26×10−5 |
| GO:0015671 | Oxygen
transport | 14 | 21.9 |
3.53×10−2 |
| GO:0048821 | Erythrocyte
development | 24 | 17.89 |
1.59×10−3 |
| GO:0006779 |
Porphyrin-containing compound biosynthetic
process | 26 | 14.15 |
4.58×10−2 |
| GO:0006778 |
Porphyrin-containing compound metabolic
process | 38 | 12.91 |
2.46×10−3 |
| GO:0061515 | Myeloid cell
development | 44 | 12.54 |
5.65×10−4 |
| GO:0046686 | Response to cadmium
ion | 56 |
9.86 |
4.18×10−3 |
| GO:0033013 | Tetrapyrrole
metabolic process | 59 |
9.35 |
6.40×10−3 |
| GO:0034614 | Cellular response
to reactive oxygen species | 119 |
8.24 |
2.01×10−6 |
| GO:0000302 | Response to
reactive oxygen species | 198 |
7.12 |
4.36×10−9 |
| GO:0042542 | Response to
hydrogen peroxide | 107 |
6.88 |
2.46×10−3 |
| GO:0034599 | Cellular response
to oxidative stress | 203 |
6.34 |
3.76×10−7 |
| GO:0009636 | Response to toxic
substance | 210 |
5.84 |
4.50×10−6 |
| GO:0030099 | Myeloid cell
differentiation | 190 |
5.49 |
2.21×10−4 |
Furthermore, upregulated DEGs between the
LPS-treated cord blood mononuclear cells of healthy neonates and
LPS-treated peripheral blood mononuclear cells of healthy adults
were identified and analyzed by GO enrichment. Consistent with the
hypothesis that the IFN-I pathway may be upregulated in a subset of
patients with sepsis that normally exhibit a higher risk of
mortality, ‘type I interferon signaling pathway’ and ‘response to
interferon-α’ were upregulated in the LPS-treated blood mononuclear
cells of healthy neonates compared with adults (Table III). Subsequently, the
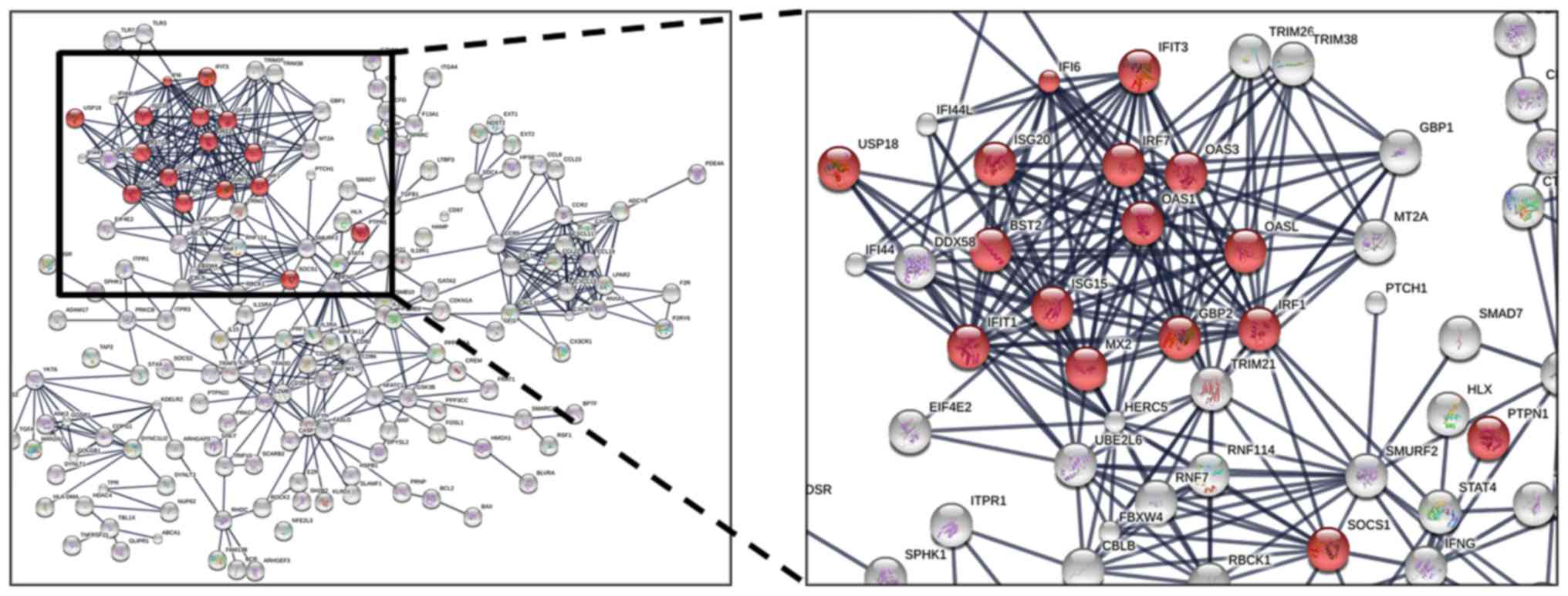
associations among the upregulated genes were analyzed by STRING;
183 proteins were demonstrated to be involved in the interaction
network and were separated into several clusters, and the proteins
involved in IFN-I signaling appeared to exhibit a higher number of
interactions (Fig. 3). These
results indicated that IFN-I signaling may be upregulated in a
subset of patients with sepsis, in this case neonates, that have a
higher risk of mortality.
 | Table III.Enriched pathways of differentially
expressed genes that were upregulated in the LPS-treated cord blood
mononuclear cells of healthy neonates compared with LPS-treated
peripheral blood mononuclear cells of healthy adults. |
Table III.
Enriched pathways of differentially
expressed genes that were upregulated in the LPS-treated cord blood
mononuclear cells of healthy neonates compared with LPS-treated
peripheral blood mononuclear cells of healthy adults.
| Pathway ID | Pathway
description | Count | Fold
enrichment | P-value |
|---|
| GO:0045086 | Positive regulation
of interleukin-2 biosynthetic process | 12 | 15.13 |
3.20×10−2 |
| GO:0045589 | Regulation of
regulatory T cell differentiation | 19 | 12.74 |
2.81×10−3 |
| GO:0035455 | Response to
interferon-α | 20 | 12.11 |
4.12×10−3 |
| GO:0045076 | Regulation of
interleukin-2 biosynthetic process | 18 | 11.77 |
2.57×10−2 |
| GO:0045624 | Positive regulation
of T-helper cell differentiation | 19 | 11.15 |
3.65×10−2 |
| GO:0043372 | Positive regulation
of CD4-positive, α-β T cell differentiation | 25 | 10.89 |
1.91×10−3 |
| GO:0002719 | Negative regulation
of cytokine production involved in immune response | 23 | 10.53 |
1.16×10−2 |
| GO:2000516 | Positive regulation
of CD4-positive, α-β T cell activation | 29 | 10.44 |
6.07×10−4 |
| GO:0002828 | Regulation of type
2 immune response | 27 | 10.09 |
3.60×10−3 |
| GO:0032743 | Positive regulation
of interleukin-2 production | 31 |
9.76 |
1.12×10−3 |
| GO:0042346 | Positive regulation
of NF-κB import into nucleus | 25 |
9.68 |
2.13×10−2 |
| GO:0019835 | Cytolysis | 25 |
9.68 |
2.13×10−2 |
| GO:0032731 | Positive regulation
of interleukin-1β production | 30 |
9.08 |
8.53×10−3 |
| GO:0032663 | Regulation of
interleukin-2 production | 51 | 8.9 |
2.99×10−6 |
| GO:0043370 | Regulation of
CD4-positive, α-β T cell differentiation | 38 |
8.76 |
7.76×10−4 |
| GO:2000514 | Regulation of
CD4-positive, α-β T cell activation | 44 |
8.25 |
3.85×10−4 |
| GO:0045070 | Positive regulation
of viral genome replication | 33 |
8.25 |
1.84×10−2 |
| GO:0045071 | Negative regulation
of viral genome replication | 52 |
8.15 |
3.48×10−5 |
| GO:0046635 | Positive regulation
of α-β T cell activation | 53 |
7.99 |
4.40×10−5 |
| GO:0042108 | Positive regulation
of cytokine biosynthetic process | 61 |
7.94 |
3.97×10−6 |
| GO:0048247 | Lymphocyte
chemotaxis | 42 |
7.93 |
2.07×10−3 |
| GO:0045069 | Regulation of viral
genome replication | 82 |
7.75 |
1.23×10−8 |
| GO:0002704 | Negative regulation
of leukocyte mediated immunity | 47 |
7.73 |
7.78×10−4 |
| GO:0060337 | Type I interferon
signaling pathway | 65 |
7.45 |
9.73×10−6 |
| GO:0071357 | Cellular response
to type I interferon | 65 |
7.45 |
9.73×10−6 |
Discussion
Sepsis is an important cause of mortality from
infection, and although numerous studies have been performed
concerning this life-threatening condition, there remains a lack of
effective treatment and an assessment of risk factors to assist in
the diagnostic process. The present study focused on the
identification of potential risk factors for sepsis-associated
mortality by genome-wide expression profiling. The results
demonstrated that DEGs that were upregulated in sepsis nonsurvivors
compared with survivors were highly enriched in the IFN-I signaling
pathway. The associations between the upregulated genes were
analyzed by STRING and the results demonstrated that the proteins
were also highly associated with IFN-I signaling pathway.
Therefore, it was hypothesized that a dysregulated IFN-I signaling
pathway may be associated with a high risk of sepsis-associated
mortality.
IFN-Is, which include IFN-α and IFN-β, trigger IFN-I
signaling by binding to the IFN-α/β receptor (IFNAR), stimulating
the Janus kinase/signal transducer and activator of transcription
pathway and initiating the transcription of IFN-stimulated genes,
which mediate various anticellular effects by modulating cell
viability and function (36). The
IFN-I signaling pathway is well known for its protective roles in
the majority of viral infections, while its functions in bacterial
infection remain controversial. This controversy arises as certain
studies have reported that IFN-I signaling served a critical role
in host protection against bacterial infection and that the
development of bacteremia during sepsis was enhanced in the mice
that lack IFNAR (37,38), while other studies indicated that
IFNAR-deficient mice were partially protected against lethality in
multiple inflammatory models, including endotoxemia-induced shock,
cecal ligation and puncture-induced sepsis, and colon ascendens
stent peritonitis-induced sepsis (39–41).
In addition, IFN-I signaling may exert toxic effects during sepsis
by negatively regulating neutrophil recruitment and suppressing
adaptive immunity, leading to inefficient control of infections and
eventual mortality (40,42,43).
Although these findings highlight the critical role
of the IFN-I signaling pathway in sepsis, there remains a lack of
reliable evidence from clinical studies. The results of the present
study demonstrated that upregulated IFN-I signaling may be a
high-risk factor for sepsis-associated mortality; however, it
remains to be elucidated whether dysregulated IFN-I signaling may
affect the mortality of patients with sepsis. Although a potential
risk factor for short-term mortality in sepsis is provided, the
survivors still suffer a high risk of long-term mortality for
months or years. Although IFN-I signaling was upregulated in
several survivors compared with the others, no information
concerning their long-term mortality is available due to a lack of
data, therefore a more comprehensive scrutiny of clinical research
concerning the effect of the IFN-I signaling pathway in sepsis is
required.
As the initial results of the present study
indicated that upregulated IFN-I signaling may be a potential
marker for a higher risk of sepsis-associated mortality, it was
further investigated whether this signaling pathway was
dysregulated in a subset of patients that possess a higher risk of
sepsis-associated mortality. It is established that neonates suffer
a higher rate of sepsis-associated mortality compared with adults;
therefore, DEGs between the untreated cord blood mononuclear cells
of healthy neonates and untreated peripheral blood mononuclear
cells of adults were analyzed (44). There were numerous DEGs between
neonates and adults; however, the DEGs were not enriched in the
IFN-1 signaling pathway. Subsequently, upregulated DEGs between
LPS-treated cord blood mononuclear cells of healthy neonates and
LPS-treated peripheral blood mononuclear cells of adults were
identified and, consistent with the hypothesis, ‘type I interferon
signaling pathway’ and ‘response to interferon-α’ were upregulated
in the LPS-treated cells of healthy neonates compared with adults.
Furthermore, the associations among the upregulated genes were
analyzed and the results demonstrated that the proteins associated
with the IFN-I signaling possessed a higher number of interactions
and may function together in pathological processes.
In conclusion, the present findings suggest that
upregulated IFN-I signaling pathway may be a risk factor for
sepsis-associated mortality, but further studies are needed to
confirm the current results.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 31500738 and
81273199), the Health and Family Planning Commission of Jilin
Province (grant no. 2014Z067) and the Union Projects of Jilin
University and Xinjiang Medical University (for Liu Z).
References
|
1
|
Singer CS, Deutschman CS, Seymour CW,
Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche
JD, Coopersmith CM, et al: The third international consensus
definitions for sepsis and septic shock (Sepsis-3). JAMA.
315:801–810. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rudd KE, Delaney A and Finfer S: Counting
sepsis, an imprecise but improving science. JAMA. 318:1228–1229.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rhodes A, Evans LE, Alhazzani W, Levy MM,
Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally
ME, et al: Surviving sepsis campaign: International guidelines for
management of sepsis and septic shock: 2016. Crit Care Med.
45:486–552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shankar-Hari M, Phillips GS, Levy ML,
Seymour CW, Liu VX, Deutschman CS, Angus DC, Rubenfeld GD and
Singer M; Sepsis Definitions Task Force, : Developing a new
definition and assessing new clinical criteria for septic shock:
For the third international consensus definitions for sepsis and
septic shock (Sepsis-3). JAMA. 315:775–787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van der Poll T, van de Veerdonk FL,
Scicluna BP and Netea MG: The immunopathology of sepsis and
potential therapeutic targets. Nat Rev Immunol. 17:407–420. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seeley EJ and Bernard GR: Therapeutic
targets in sepsis: Past, present, and future. Clin Chest Med.
37:181–189. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hotchkiss RS, Monneret G and Payen D:
Immunosuppression in sepsis: A novel understanding of the disorder
and a new therapeutic approach. Lancet Infect Dis. 13:260–268.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dellinger RP: Severe sepsis trials: Why
have they failed? Minerva Anestesiol. 65:340–345. 1999.PubMed/NCBI
|
|
9
|
Gotts JE and Matthay MA: Sepsis:
Pathophysiology and clinical management. BMJ. 353:i15852016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seymour CW and Rosengart MR: Septic shock:
Advances in diagnosis and treatment. JAMA. 314:708–717. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McLean AS, Tang B and Huang SJ:
Investigating sepsis with biomarkers. BMJ. 350:h2542015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vashist SK, Venkatesh AG, Marion Schneider
E, Beaudoin C, Luppa PB and Luong JH: Bioanalytical advances in
assays for C-reactive protein. Biotechnol Adv. 34:272–290. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ridker PM: A test in context:
High-sensitivity c-reactive protein. J Am Coll Cardiol. 67:712–723.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Póvoa P, Coelho L, Almeida E, Fernandes A,
Mealha R, Moreira P and Sabino H: Pilot study evaluating C-reactive
protein levels in the assessment of response to treatment of severe
bloodstream infection. Clin Infect Dis. 40:1855–1857. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fraunberger P, Wang Y, Holler E, Parhofer
KG, Nagel D, Walli AK and Seidel D: Prognostic value of interleukin
6, procalcitonin, and C-reactive protein levels in intensive care
unit patients during first increase of fever. Shock. 26:10–12.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schneider CP, Yilmaz Y, Kleespies A, Jauch
KW and Hartl WH: Accuracy of procalcitonin for outcome prediction
in unselected postoperative critically ill patients. Shock.
31:568–573. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chousterman BG, Swirski FK and Weber GF:
Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol.
39:517–528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mikacenic C, Price BL, Harju-Baker S,
O'Mahony DS, Robinson-Cohen C, Radella F, Hahn WO, Katz R,
Christiani DC, Himmelfarb J, et al: A Two-biomarker model predicts
mortality in the critically III with Sepsis. Am J Respir Crit Care
Med. 196:1004–1011. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qi Y, Ge J, Ma C, Wu N, Cui X and Liu Z:
Activin A regulates activation of mouse neutrophils by Smad3
signalling. Open Biol. 7:1603422017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ettinger M, Calliess T, Kielstein JT,
Sibai J, Brückner T, Lichtinghagen R, Windhagen H and Lukasz A:
Circulating biomarkers for discrimination between aseptic joint
failure, low-grade infection, and high-grade septic failure. Clin
Infect Dis. 61:332–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Buttaro MA, Tanoira I, Comba F and
Piccaluga F: Combining C-reactive protein and interleukin-6 may be
useful to detect periprosthetic hip infection. Clin Orthop Relat
Res. 468:3263–3267. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Elgeidi A, Elganainy AE, Abou Elkhier N
and Rakha S: Interleukin-6 and other inflammatory markers in
diagnosis of periprosthetic joint infection. Int Orthop.
38:2591–2595. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rung J and Brazma A: Reuse of public
genome-wide gene expression data. Nat Rev Genet. 14:89–99. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maslove DM and Wong HR: Gene expression
profiling in sepsis: Timing, tissue, and translational
considerations. Trends Mol Med. 20:204–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wong HR: Clinical review: Sepsis and
septic shock-the potential of gene arrays. Crit Care. 16:2042012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang BM, Huang SJ and McLean AS:
Genome-wide transcription profiling of human sepsis: A systematic
review. Crit Care. 14:R2372010. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parnell GP, Tang BM, Nalos M, Armstrong
NJ, Huang SJ, Booth DR and McLean AS: Identifying key regulatory
genes in the whole blood of septic patients to monitor underlying
immune dysfunctions. Shock. 40:166–174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koch L, Linderkamp O, Ittrich C, Benner A
and Poeschl J: Gene expression profiles of adult peripheral and
cord blood mononuclear cells altered by lipopolysaccharide.
Neonatology. 93:1–100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41(Database Issue):
D991–D995. 2013.PubMed/NCBI
|
|
30
|
Benjamini Y, Drai D, Elmer G, Kafkafi N
and Golani I: Controlling the false discovery rate in behavior
genetics research. Behav Brain Res. 125:279–284. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rhee SY, Wood V, Dolinski K and Draghici
S: Use and misuse of the gene ontology annotations. Nat Rev Genet.
9:509–515. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Munoz-Torres M and Carbon S: Get GO!
Retrieving GO data using AmiGO, QuickGO, API, Files, and tools.
Methods Mol Biol. 1446:149–160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Trinchieri G: Type I interferon: Friend or
foe? J Exp Med. 207:2053–2063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kan B, Razzaghian HR and Lavoie PM: An
immunological perspective on neonatal sepsis. Trends Mol Med.
22:290–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schreiber G and Piehler J: The molecular
basis for functional plasticity in type I interferon signaling.
Trends Immunol. 36:139–149. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
LeMessurier KS, Häcker H, Chi L, Tuomanen
E and Redecke V: Type I interferon protects against pneumococcal
invasive disease by inhibiting bacterial transmigration across the
lung. PLoS Pathog. 9:e10037272013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mancuso G, Midiri A, Biondo C, Beninati C,
Zummo S, Galbo R, Tomasello F, Gambuzza M, Macrì G, Ruggeri A, et
al: Type I IFN signaling is crucial for host resistance against
different species of pathogenic bacteria. J Immunol. 178:3126–3133.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mahieu T, Park JM, Revets H, Pasche B,
Lengeling A, Staelens J, Wullaert A, Vanlaere I, Hochepied T, van
Roy F, et al: The wild-derived inbred mouse strain SPRET/Ei is
resistant to LPS and defective in IFN-beta production. Proc Natl
Acad Sci USA. 103:pp. 2292–2297. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dejager L, Vandevyver S, Ballegeer M, Van
Wonterghem E, An LL, Riggs J, Kolbeck R and Libert C:
Pharmacological inhibition of type I interferon signaling protects
mice against lethal sepsis. J Infect Dis. 209:960–970. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weighardt H, Kaiser-Moore S, Schlautkötter
S, Rossmann-Bloeck T, Schleicher U, Bogdan C and Holzmann B: Type I
IFN modulates host defense and late hyperinflammation in septic
peritonitis. J Immunol. 177:5623–5630. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schwandt T, Schumak B, Gielen GH,
Jüngerkes F, Schmidbauer P, Klocke K, Staratschek-Jox A, van
Rooijen N, Kraal G, Ludwig-Portugall I, et al: Expression of type I
interferon by splenic macrophages suppresses adaptive immunity
during sepsis. EMBO J. 31:201–213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McNab F, Mayer-Barber K, Sher A, Wack A
and O'Garra A: Type I interferons in infectious disease. Nat Rev
Immunol. 15:87–103. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ginsburg AS, Meulen AS and Klugman KP:
Prevention of neonatal pneumonia and sepsis via maternal
immunisation. Lancet Glob Health. 2:e679–e680. 2014. View Article : Google Scholar : PubMed/NCBI
|