Introduction
Esophageal carcinoma (EC) is a common malignant
tumor worldwide, demonstrating the eighth highest incidence rate
and representing the sixth most common cause of cancer associated
mortality (1). The number of EC
cases diagnosed annually in China account for almost half of the
worldwide total (2). Considering
that early symptoms in EC are frequently latent, the majority of
patients are diagnosed at the advanced stage and prognosis is
usually poor, owing to invasion and metastasis (3–5).
Therefore, it is urgent to identify novel and effective molecular
targets for the diagnosis and treatment of EC for improved patient
outcomes.
Epithelial-mesenchymal transition (EMT) is a process
in which epithelial cells lose their polarized organization, which
is accompanied by reduced epithelial (E)-cadherin expression, and
increased cellular mobility, which is accompanied by increased
Vimentin expression (6,7). It has been verified that, via the EMT
program, tumor cells may obtain stronger metastasis and increased
drug-resistance and stemness, which may lead to tumor progression
and poor prognosis (8,9). Therefore, an effective treatment that
targets EMT may inhibit tumor progression.
Cullin7 (CUL7) is a member of the Cullin protein
family; it is a molecular scaffold that organizes an E3 ubiquitin
ligase comprising F-box protein Fbw8, S-phase kinase-associated
protein 1 andring-box 1 finger protein, and regulates cell biology
functions via protein ubiquitination (10,11).
A number of roles for CUL7 have been reported in numerous types of
tumors. For example, in 2007, Kim et al (12) reported that CUL7 may promote human
neuroblastoma cell proliferation by inhibiting p53-dependent or
p53-independent apoptosis. In 2015, Men et al (13) reported high expression levels of
CUL7 in lung cancer and confirmed the proliferation-inducing roles;
however, the role of CUL7 in EC has not yet been reported.
The present study demonstrated for the first time,
to the best of our knowledge, that CUL7 is expressed in elevated
levels in EC tissues, as detected by immunohistochemistry (IHC),
and a close association between this increased expression and
invasion depth, lymph node involvement and advanced clinical stage
were identified. The EMT-promoting roles of CUL7 were also
investigated, and the extracellular signal-regulated kinase
(ERK)-zinc finger protein SNAI2 (SNAI2) pathway was reported to be
involved in this process. In addition, the present study revealed
that CUL7 was positively associated with poor overall survival (OS)
and disease-free survival (DFS) of patients with EC.
Materials and methods
IHC
The present retrospective study was approved by the
review board and Ethics Committee of Yidu Central Hospital of
Weifang (Weifang, China); specimens were collected from 130
patients (53 males and 77 females; 40–73 years old) with primary EC
who had surgical removal between January 2009 and December 2011 at
Yidu Central Hospital of Weifang. Patients did not receive
chemotherapy, radiotherapy or immunomodulatory therapy prior to
surgery. Nontumoral tissues were used as a negative control in the
present study and were obtained from outpatients who
underwentgastroscopy detection.
Paraffin-embedded tissue samples were obtained from
the Yidu Central Hospital of Weifang and then sectioned (4 µm),
placed on slides, deparaffinized in xylene and rehydrated in a
graded ethanol series. Slides were boiled in 10 mmol/l citrate
buffer (pH 6.0) for 3 min at 100°C for antigen retrieval. The
sections were subsequently immersed in 3%
H2O2 for 10 min at room temperature to block
endogenous peroxidase activity and incubated in goat serum blocking
solution (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) at room
temperature for 15 min to block non-specific antigens. Sections
were incubated at 4°C overnight with primary antibodies against
CUL7 (cat. no. ab115304; 1:1,000; Abcam, Cambridge, USA),
E-cadherin (cat. no. ab40772; 1:1,000; Abcam) and SNAI2 (cat. no.
ab85936; 1:500; Abcam), and subsequently washed with PBS and
incubated with horseradish peroxidase (HRP)-conjugated goat
anti-rabbit/mouse immunoglobulin G (IgG) polyclonal antibody (cat.
no. KIT-7710; Fuzhou Maixin Biotechnology Development Co., Ltd.,
Fuzhou, China) at room temperature for 30 min. The slides were
stained with 3,3′-diaminobenzidine for 5–10 sec at room temperature
and counterstained with hematoxylin for 20 sec at room temperature.
The evaluation scoring process was performed by two independent
pathologists who were blinded to the clinical information of
patients. The percentage of positive-stained cells of the total
number of cells was recorded at ×400 magnification in at least 5
random fields using a BX53 microscope (Olympus Corporation, Tokyo,
Japan). A score representing the fraction of positive-staining
tumor cells was as follows: 0, ≤25%; 1, 26–50%; 2, 51–75%; and 3,
>75%). The intensity score represented the average staining
intensity of positive tumor cells (0, negative; 1, weak; 2,
moderate; and 3, strong). The expression levels of CUL7 were
calculated using the product of the proportion score and the
intensity score, and a score <4 was considered low expression,
whereas a score ≥4 was considered as high expression.
Cell lines and culture
The EC cell lines EC1 and EC9706 were purchased from
Guangzhou Jisai Biotech Co., Ltd. (Guangzhou, China) and cultured
in RPMI-1640 Medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS)
(Invitrogen; Thermo Fisher Scientific Inc.).
Reverse
transcription-semi-quantitative polymerase chain reaction (RT-sq
PCR)
Total RNA was extracted from cells
(~2×105/ml) using TRIzol (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. cDNA was
synthesized using a Prime Script RT-PCR kit (Takara Biotechnology
Co., Ltd., Dalian, China), according to the manufacturer's
protocols. The primers were as follows: CUL7 forward,
5′-CCATCTCAGAGTCCCAACAC-3′ and reverse, 5′-TTCAGCACCACGGCATAGFF-3′;
GAPDH forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse,
5′-AGGGGCCATCCACAGTCTTC-3′. The reaction mixture [5 µl mix
including DNA polymerase (cat. no. D7228; Beyotime Institute of
Biotechnology, Haimen, China), 1.4 µl cDNA, 1.6 µl forward primer,
1.6 µl reverse primer and 5.4 µl double distilled H2O]
was amplifiedat 94°C for 5 min, followed by 30 cycles at 94°C for
30 sec, 51°C for 30 sec, 72°C for 30 sec and finally 72°C for 5
min. PCR products were electrophoretically separated using 1.0%
agarose gels and visualized using ethidium bromide. The results
were analyzed using Lab Work software (version 4.0; UVP, Inc.,
Upland, CA, USA). GAPDH was used as an internal control.
Western blot analysis
Protein was extracted from 4×105/ml cells
using radio immune precipitation assay buffer (cat. no. P0013B;
Beyotime Institute of Biotechnology) containing 1% protease
inhibitor. Protein concentration was determined using a
Bicinchoninic Acidkit (Pierce; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. A total of 300 µg protein
was loaded per well separated by 10% SDS-PAGE gel and then
transferred to a nitrocellulose membrane. Following blocking in TBS
with 0.05% Tween-20 containing 5% non-fat dried milk for 1 h at
room temperature, the membrane was incubated with primary
antibodies at 4°C overnight and subsequently with secondary
HRP-conjugated goat anti-rabbit IgG antibodies (cat. no. SA00001-2;
1:5,000; Protein Tech Group, Inc., Chicago, IL, USA) at room
temperature for 1 h. Signals were detected using Enhanced
Chemiluminescence Reagents (Pierce; Thermo Fisher Scientific, Inc.)
and densitometric analysis was performed using Image-Pro software
(version 5.1; Media Cybernetics, Inc., Rockville, MA, USA). The
primary antibodies used were as follows: CUL7 (cat. no. ab115304;
1:1,000; Abcam, Cambridge, MA, USA), E-cadherin (cat. no. ab40772;
1:1,000; Abcam), Vimentin (cat. no. ab45939; 1:1,000; Abcam) SNAI2
(cat. no. ab85936; 1:1,000; Abcam), phosphorylated (p)-ERK (cat.
no. 4370; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA,
USA), ERK (cat. no. 9102; 1:1,000; Cell Signaling Technology, Inc.)
and GAPDH (cat. no. 10494-1-AP; 1:5,000; Protein Tech Group,
Inc.).
Transfection
Two different cell lines exhibiting varying
expression levels of CUL7 were chosen for investigation in the
present study. In the cell line exhibiting high expression of CUL7
(EC1), CUL7 expression was suppressed; in the cell line exhibiting
low CUL7 expression (EC9706), CUL7 expression was over expressed.
Thus, greater changes of CUL7 expression and its effect on EC cells
could be observed. A total of 1 µg pcDNA3.1/CUL7-vector plasmid
(Shanghai Gene Pharma Co., Ltd., Shanghai, China) was transfected
into EC9706 cells (105/ml) to upregulate CUL7
expression, and 1 µg p-GPU6/CUL7-short hairpin (sh)RNA (Shanghai
Gene Pharma Co., Ltd.) was transfected into EC1 cells to silence
CUL7 expression. Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) was used for transfection process and 1 µg
empty negative control (nc) plasmids (Shanghai Gene Pharma Co.,
Ltd.) were used as controls. All procedures were performed
according to the manufacturer's protocol. Following transfection,
cells were incubated at 37°C for 48 h and then collected for
further research. EC1 cells transfected with CUL7-shRNA and
nc-shRNA cells were termed EC1-CUL7-sh and EC1-nc-sh cells,
respectively; EC9706 cells transfected with CUL7-overexpression
vector ornc-vector were designated EC9706-CUL7-vector and
EC9706-nc-vector cells, respectively. The ERK inhibitor U0126 (cat.
no. 9903; Cell Signaling Technology, Inc.) was used to investigate
the molecular mechanism underlying the effect of CUL7 at 37°C for
24 h.
Migration assay
Migration assays were performed using Transwell
chambers (EMD Millipore, Billerica, MA, USA). Cells
(5×104) were seeded into the inserts of Transwell
chambers in serum-free RPMI-1640 medium in the upper chamber, and
RPMI-1640 medium supplemented with 20% FBS was added to the lower
chamber. The cells were incubated for 48 h at 37°C, and the cells
in the upper chamber were removed using a cotton swab; cells that
migrated to the lower surface of the filter were fixed with 4%
formaldehyde at room temperature for 15 min and stained with 0.1%
crystal violet at room temperature for 20 min. Images were captured
and cells were counted in 5 random fields using an XDS-200
microscope to obtain an average number of migrating cells
(magnification, ×200; Olympus Corporation, Tokyo, Japan).
Statistical analysis
Data are expressed as the mean ± standard deviation,
and SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA) was
used for statistical analyses. The difference between two groups
was analyzed using a Student's two-tailed t-test. The association
of CUL7 with clinical parameters was analyzed using the
χ2 test. Survival curves were produced using the
Kaplan-Meier method and compared by means of the log-rank test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
High CUL7 protein expression is
positively associated with invasion depth, lymph node involvement
and advanced clinical stage
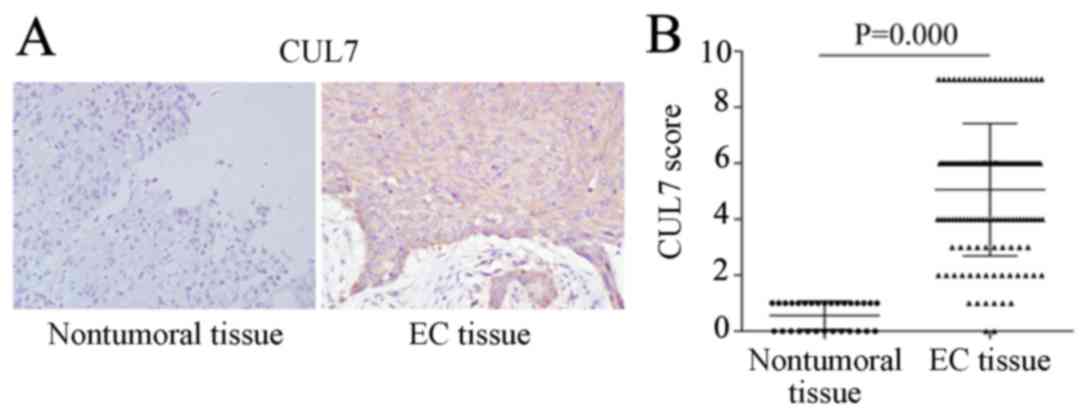
IHC results revealed almost no CUL7-positive
staining in nontumoral tissues, whereas strong CUL7 protein
expression was observed in the cytoplasm of EC tissues (Fig. 1A). Statistical analysis
demonstrated that the overall CUL7 expression score in EC tissues
was significantly higher compared with nontumoral tissues (Fig. 1B). Of the 130 tissue specimens from
patients with primary EC, 98 cases exhibited high expression levels
of CUL7 and 32 cases exhibited low expression. Associations between
CUL7 expression level and clinical parameters were also analyzed
(Table I). In the low CUL7
expression group (32 cases), only 4 cases of T3+T4 were reported,
which was significantly less than the 62 cases of T3 + T4 reported
in the high CUL7 group (98 cases; P=0.000). Furthermore, 5 cases of
N2+N3 were reported in the low CUL7 expression group, which was
significantly less than the 35 cases reported in the high CUL7
group (P=0.033). In addition, 3 cases were reported as stage III in
the low CUL7 expression group, which was significantly less than
the 64 cases reported in the high CUL7 group (P=0.000). However, no
significant differences were identified between the CUL7 expression
level and age, sex, tumor size or differentiation. These results
indicated that CUL7 may serve a role in EC progression.
 | Table I.Association between CUL7 and clinical
parameters of patients with esophageal carcinoma. |
Table I.
Association between CUL7 and clinical
parameters of patients with esophageal carcinoma.
|
|
| CUL7 |
|
|
|---|
|
|
|
|
|
|
|---|
| Parameter | n | Low | High | χ2 | P-value |
|---|
| Age (years) |
|
|
|
|
|
| ≤60 | 70 | 22 | 48 | 3.794 | 0.066 |
|
>60 | 60 | 10 | 50 |
|
|
| Sex |
|
|
|
|
|
| Male | 53 | 16 | 37 | 1.498 | 0.221 |
|
Female | 77 | 16 | 61 |
|
|
| Tumor size (cm) |
|
|
|
|
|
| ≤4 | 64 | 20 | 44 | 2.99 | 0.084 |
|
>4 | 66 | 12 | 54 |
|
|
| Differentiation |
|
|
|
|
|
| Well | 49 | 14 | 35 | 0.663 | 0.415 |
|
Moderate/poor | 81 | 18 | 63 |
|
|
| Invasion depth |
|
|
|
|
|
| T1 +
T2 | 64 | 28 | 36 | 24.873 | 0.000 |
| T3 +
T4 | 66 | 4 | 62 |
|
|
| Lymph node
involvement |
|
|
|
|
|
| N0 +
N1 | 90 | 27 | 63 | 4.57 | 0.033 |
| N2 +
N3 | 40 | 5 | 35 |
|
|
| Clinical stage |
|
|
|
|
|
| I–II | 63 | 29 | 34 | 30.214 | 0.000 |
|
III | 67 | 3 | 64 |
|
|
CUL7 promotes EMT of EC cells
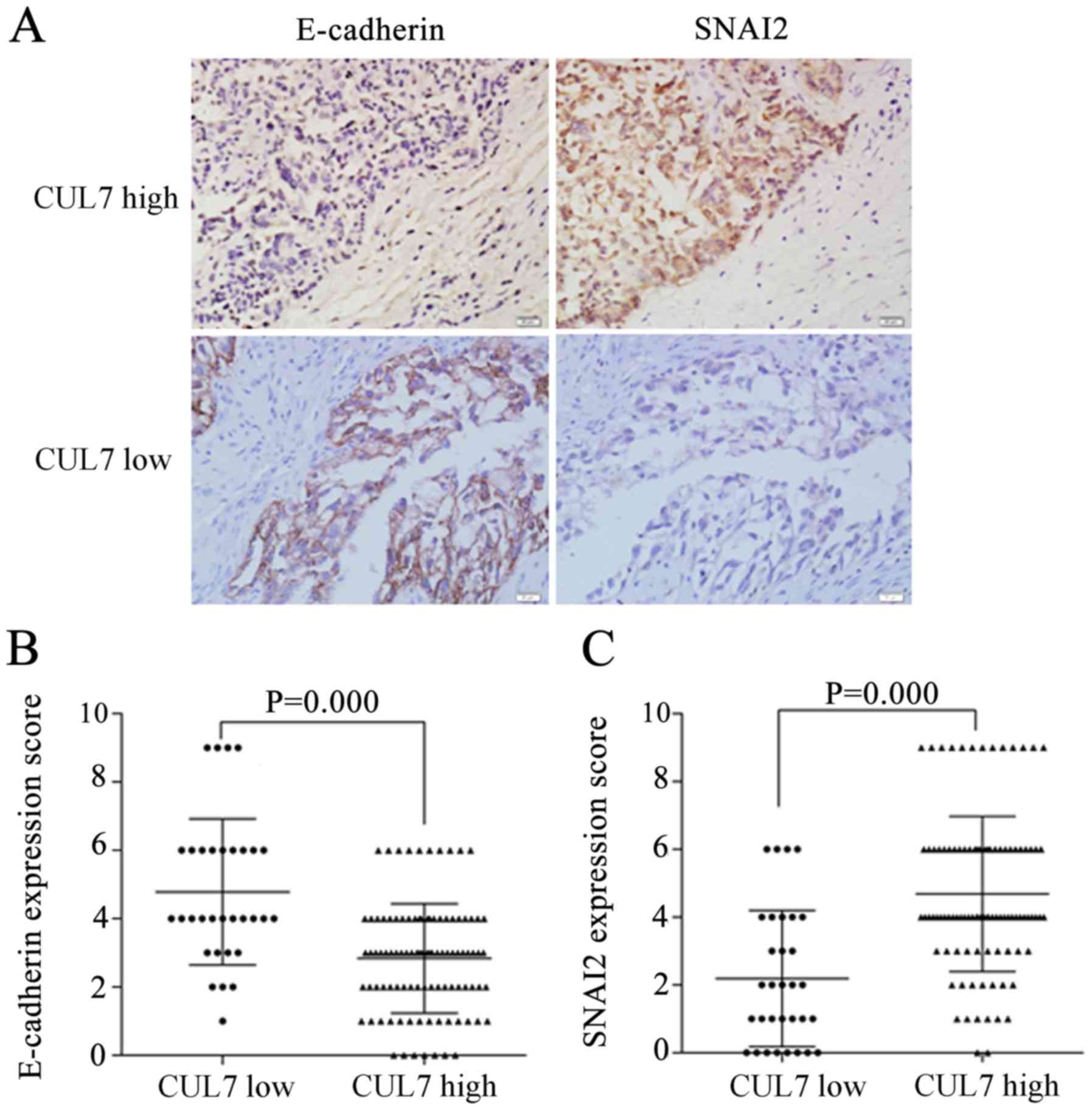
IHC staining of serial tissue sections revealed
that, in the high CUL7 expression group, E-cadherin protein
expression was weak and SNAI2 expression was strong (Fig. 2A); in the low CUL7 group,
E-cadherin expression was strong and SNAI2 expression was weak.
Statistical analysis results indicated that high CUL7 expression
was associated with low E-cadherin expression (P=0.000; Fig. 2B) and with high SNAI2 expression
(P=0.000; Fig. 2C). These data
suggested an association of CUL7 expression with EMT. In cell
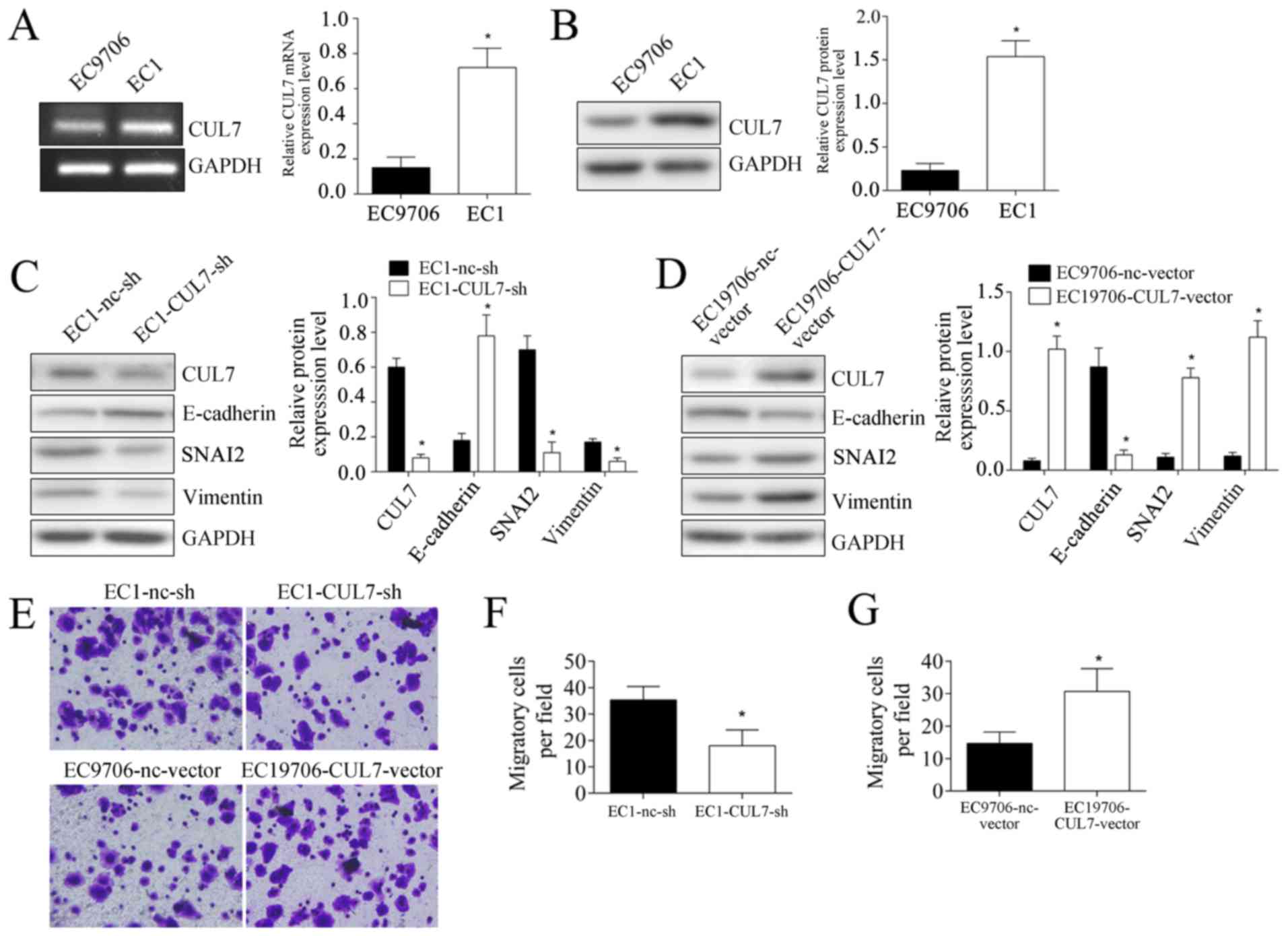
culture experiments, CUL7 mRNA and protein expression levels in EC1
cells were significantly higher compared with EC9706 cells
(P<0.05; Fig. 3A and B). CUL7
expression was altered by transfecting EC1 cells with CUL7-targeted
shRNA, which demonstrated that silencing CUL7 expression
significantly inhibited the protein expression levels of SNAI2 and
Vimentin, whereas E-cadherin expression was upregulated (Fig. 3C). Over expression of CUL7 in
EC9706 cells significantly promoted SNAI2 and Vimentin expression,
but inhibited E-cadherin expression (Fig. 3D). These results provided further
evidence of EMT induction by CUL7. In addition, cell migration
experiments demonstrated that, following CUL7 silencing, the number
of EC1 cells migrating to the lower surface of the filter were
significantly lower compared with nc-sh transfected cells (18±6 vs.
35.33±5.13 cells/field, respectively; p<0.05; Fig. 3E and F). Following CUL7
overexpression, the number of EC9706 cells migrating to the lower
surface of the filter was significantly higher compared with
nc-vector transfected cells (14.67±3.51 vs. 30.67±7.02,
respectively; p<0.05; Fig. 3E and
G).
ERK-SNAI2 pathway participates in
CUL7-induced EMT
EMT is a complicated process and is regulated by
numerous signaling pathways under various biological and
pathological conditions (6,7). ERK
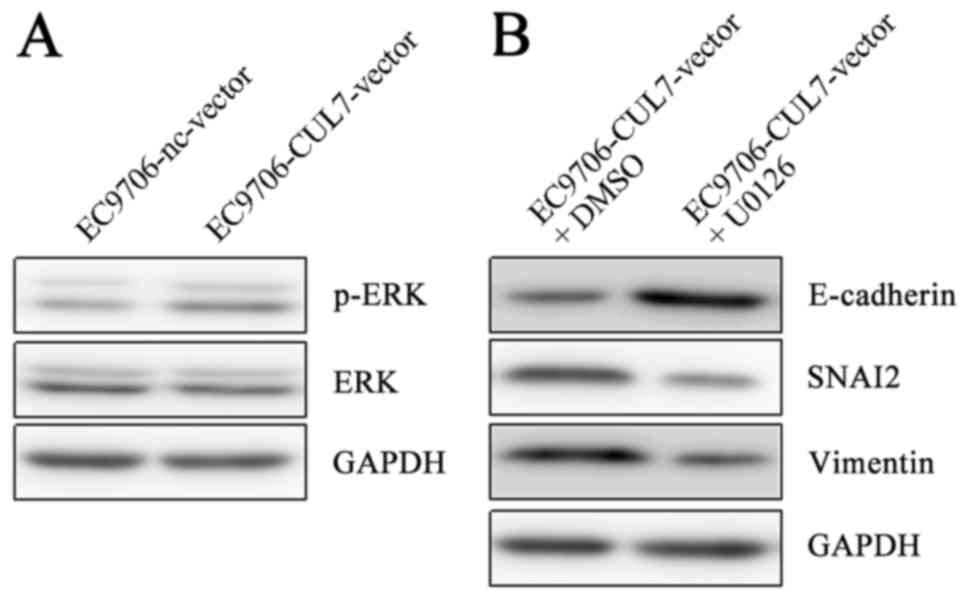
phosphorylation in EC9706 cells was upregulatedin
CUL7-overexpressing cells compared with nc-vector cells (Fig. 4A). Furthermore, in EC9706 cells
co-treated with the CUL7 over expression vector and the ERK
inhibitor U0126 at 37°C for 48 h, Vimentin and SNAI2 protein
expression levels were notably reduced, whereas E-cadherin
expression was notably increased compared with CUL7-vector treated
cells co-treated with dimethylsulfoxide (Fig. 4B). These results suggested that
CUL7 may promote EMT via the ERK-SNAI2 signaling pathway.
CUL7 is associated with poor OS and
DFS
To further investigate the roles of CUL7 in patient
prognosis, survival analysis was performed using the Kaplan-Meier
method. Of the 30 patients in the low CUL7 group, 12 casessuccumbed
to mortality, with an OS rate of 60% at 60 months (Fig. 5A). Of the 40 patients in the high
CUL7 expression group, 30 cases succumbed to mortality with an OS
rate of 25% at 60 months. In addition, 16 of the 30 cases in the
low CUL7 group exhibited recurrence, with a DFS rate of 46.67% at
60 months (Fig. 5B); 32 of the 40
cases in the high CUL7 group exhibited recurrence, with a DFS rate
of 20% at 60 months. The log-rank test revealed that the
differences in OS and DFS rates between the two groups were
significantly different, which further suggested a role for CUL7 in
EC prognosis.
Discussion
The CUL7 gene is located at the 6p21.1 locus
(14); it is a member of Cullin
protein family, and previous studies have examined CUL7 expression
in tumors. For example, it has been reported that CUL7 may promote
the proliferation, invasion and metastasis of liver carcinoma
cells, which suggested that CUL7 is a novel gene that is associated
with liver carcinogenesis and progression (15,16).
Xi et al (17) reported on
the clinical significance of CUL7 in ovarian cancer and suggested
that CUL7 was positively associated with clinical staging and lymph
node metastasis. However, to the best of our knowledge, of the
limited types of tumors that have been used in studies
investigating CUL7, there have been no reports of CUL7 in EC. In
the present study, high expression levels of CUL7 were detected in
EC tissues and this increased level of expression was positively
associated with invasion depth, lymph node involvement and advanced
clinical stage, which was in line with the previously reported
roles of CUL7 in liver cancer and ovarian cancer (15–17).
However, a significant association between CUL7 and tumor size was
not reported in the present study, which was inconsistent with the
pro-proliferation roles of CUL7 reported previously (15,16).
This may be explained by variations in organ origin, experimental
conditions and operation in experiments.
The EMT program is an important factor in tumor
progression (6,7). On the basis of the associations
between CUL7 expression and invasion depth, lymph node involvement
and advanced clinical stage, CUL7 may promote EMT in EC cells.
SNAI2 is a transcription factor that has been reported to be
upregulated in various types of tumors and is associated with
decreased expression of the epithelial marker E-cadherin and
increased expression of the mesenchymal marker Vimentin, and may
thus serve important roles in the regulation of EMT (18). In the present study, results from
IHC and cell culture experiments suggested that CUL7 may enhance
the expression levels of SNAI2 and Vimentin, and inhibit E-cadherin
expression, and increase cell migration. These data provided
evidence for the hypothesis of the present study that CUL7 may
promote EMT in EC cells. This corresponds with previous reports of
the invasion- and metastasis-promoting roles of CUL7 in liver
carcinoma (15,16), which further indicated a close
association between CUL7 and EC progression. The ERK signaling
pathway is also considered an important factor in various types of
cell behaviors, including proliferation, migration and drug
resistance (19,20), and results from the present study
indicated that CUL7 may promote EMT via the ERK-SNAI2 signaling
pathway, which may suggest novel targets for the treatment of
EC.
In clinical work, clinical staging has been
considered to be essential for the prediction of prognosis as well
as representing a basis to decide which therapeuticoption (such as
surgery, chemotherapy or radiotherapy) should be performed
(21,22). As a positive association between
CUL7 expression and clinical stage was identified, the present
study aimed to investigate the relationship between CUL7 and
prognosis. The results revealed that high CUL7 expression was
associated with poorer OS and DFS, which was consistent with the
EMT-promoting roles of CUL7 and suggested that CUL7 may be a marker
of EC progression and poor prognosis.
The present study investigated the roles of CUL7 in
progression, EMT and poor prognosis of EC patients, as well as the
potential molecular mechanism. These data may improve our current
knowledge of the roles of CUL7 in tumors and may provide a novel
target for the treatment of EC.
Acknowledgements
The authors thank all the staff of the Pathology
Department of Yidu Central Hospital of Weifang for their help in
this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Domper Arnal MJ, Ferrandez Arenas A and
Lanas Arbeloa A: Esophageal cancer: Risk factors, screening and
endoscopic treatment in Western and Eastern countries. World J
Gastroenterol. 21:7933–7943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Na MK, Kim CH, Kim JM, Cheong JW, Ryu JI
and Kim HW: Multiple meningocerebral metastasis and extensive skull
metastasis from squamous cell carcinoma of esophagus: A case report
and review of literature. Brain Tumor Res Treat. 4:142–144. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ni PZ, Yang YS, Hu WP, Wang WP, Yuan Y and
Chen LQ: Primary adenosquamous carcinoma of the esophagus: an
analysis of 39 cases. J Thorac Dis. 8:2689–2696. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yazbeck R, Jaenisch SE and Watson DI: From
blood to breath: New horizons for esophageal cancer biomarkers.
World J Gastroenterol. 22:10077–10083. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rhim AD, Mirek ET, Aiello NM, Maitra A,
Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK,
Vonderheide RH, et al: EMT and dissemination precede pancreatic
tumor formation. Cell. 148:349–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao C, Su L, Shan J, Zhu C, Liu L, Liu C,
Xu Y, Yang Z, Bian X, Shao J, et al: IGF/STAT3/NANOG/Slug signaling
axis simultaneously controls epithelial-mesenchymal transition and
stemness maintenance in colorectal cancer. Stem Cells. 34:820–831.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen Z, Che Q, He X, Wang F, Wang H, Zhu
M, Sun J and Wan X: Stem cell protein Piwil1 endowed endometrial
cancer cells with stem-like properties via inducing
epithelial-mesenchymal transition. BMC Cancer. 15:8112015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dias DC, Dolios G, Wang R and Pan ZQ:
CUL7: A DOC domain-containing cullin selectively binds Skp1. Fbx29
to form an SCF-like complex. Proc Natl Acad Sci U S A. 99:pp.
16601–16606. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arai T, Kasper JS, Skaar JR, Ali SH,
Takahashi C and DeCaprio JA: Targeted disruption of p185/Cul7 gene
results in abnormal vascular morphogenesis. Proc Natl Acad Sci U S
A. 100:pp. 9855–9860. 2003; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim SS, Shago M, Kaustov L, Boutros PC,
Clendening JW, Sheng Y, Trentin GA, Barsyte-Lovejoy D, Mao DY, Kay
R, et al: CUL7 is a novel antiapoptotic oncogene. Cancer Res.
67:9616–9622. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Men X, Wang L, Yu W and Ju Y: Cullin7 is
required for lung cancer cell proliferation and is overexpressed in
lung cancer. Oncol Res. 22:123–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Skaar JR, Florens L, Tsutsumi T, Arai T,
Tron A, Swanson SK, Washburn MP and DeCaprio JA: PARC and CUL7 form
atypical cullin RING ligase complexes. Cancer Res. 67:2006–2014.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Paradis V, Albuquerque M, Mebarki M,
Hernandez L, Zalinski S, Quentin S, Belghiti J, Soulier J and
Bedossa P: Cullin7: a new gene involved in liver carcinogenesis
related to metabolic syndrome. Gut. 62:911–919. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang D, Yang G, Li X, Xu C and Ge H:
Inhibition of liver carcinoma cell invasion and metastasis by
knockdown of cullin7 in vitro and in vivo. Oncol Res. 23:171–181.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xi J, Zeng ST, Guo L and Feng J: High
expression of cullin7 correlates with unfavorable prognosis in
epithelial ovarian cancer patients. Cancer Invest. 34:130–136.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li XL, Liu L, Li DD, He YP, Guo LH, Sun
LP, Liu LN, Xu HX and Zhang XP: Integrin β4 promotes cell invasion
and epithelial-mesenchymal transition through the modulation of
Slug expression in hepatocellular carcinoma. Sci Rep. 7:404642017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumari R, Chouhan S, Singh S, Chhipa RR,
Ajay AK and Bhat MK: Constitutively activated ERK sensitizes cancer
cells to doxorubicin: Involvement of p53-EGFR-ERK pathway. J
Biosci. 42:31–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Semaan J, Pinon A, Rioux B, Hassan L,
Limami Y, Pouget C, Fagnere C, Sol V, Diab-Assaf M, Simon A and
Liagre B: Resistance to 3-HTMC-induced apoptosis through activation
of PI3K/Akt, MEK/ERK, and p38/COX-2/PGE2 pathways in human HT-29
and HCT116 colorectal cancer cells. J Cell Biochem. 117:2875–2885.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kokcu A, Kurtoglu E, Celik H, Kefeli M,
Tosun M and Onal M: Is surgical staging necessary for patients with
low-risk endometrial cancer? A retrospective clinical analysis.
Asian Pac J Cancer Prev. 16:5331–5335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ando M, Yamauchi H, Aogi K, Shimizu S,
Iwata H, Masuda N, Yamamoto N, Inoue K, Ohono S, Kuroi K, et al:
Randomized phase II study of weekly paclitaxel with and without
carboplatin followed by cyclophosphamide/epirubicin/5-fluorouracil
as neoadjuvant chemotherapy for stage II/IIIA breast cancer without
HER2 overexpression. Breast Cancer Res Treat. 145:401–409. 2014.
View Article : Google Scholar : PubMed/NCBI
|