Introduction
Cervical cancer is a common form of tumorigenesis
among females globally. The most important risk factor for cervical
cancer is persistent infection with human papillomavirus (HPV)
(1). HPVs with <90% nucleotide
sequence homology in the L1 gene are considered different species
(traditionally referred to as ‘types’), HPVs with 90–98% L1
sequence homology are different subtypes and HPVs with >98% L1
sequence homology are considered mutants of the same subtype
(2,3). Over 150 HPV types have been
identified, of which 60 types are predominantly detected in the
genital tract (2,3). HPV types are divided into high risk
(HPV-16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73
and 83) and low risk (HPV-6, 11, 40, 42, 43, 44, 54, 61, 70, 72 and
81) according to their oncogenic potential. Low risk HPV causes
mild genital warts and no oncogenic risk. The majority of high risk
(HR) types, including 16, 18, 31, 33, 52 and 58, have the potential
to lead to invasive cervical cancer (4–6).
When the HPV DNA integrates it inactivates
tumor-suppressor genes, stimulating oncogene expression (7,8). The
E7 protein of HR HPV binds to and inactivates retinoblastoma
protein (pRB), a tumor suppressor protein (9,10).
E6 protein has been demonstrated to mediate the degradation of p53
through the E6-associated protein (9,11).
High protein expression levels of epidermal growth factor receptor
and erb-b2 receptor tyrosine kinase 4 are promoted by the E5
protein, which has also been demonstrated to promote cell
proliferation and signal transmission (12,13).
The study of HPV variants is on the increase, and
multiple reports have confirmed that HPV variants differ in biology
and etiology (14). At present,
multiple studies have investigated HPV-16, 18, 52 and 58 variants,
however the HPV-31 variant, which is the one of the HR oncogenic
types, has been rarely studied, particularly in China. Due to the
differences in biological characteristics, it is necessary to
identify the HPV-31 variants (15). The results of the present study may
provide an effective reference for further clinical application,
and may aid assessment of the prognosis of patients with HPV.
The distribution of HPV subtypes differs
geographically and across populations, and the primary aim of the
present study was to assess the single nucleotide polymorphisms
and/or amino acid polymorphisms of the E5, E6 E7 and L1 proteins of
HPV-31 in Sichuan (China). To the best of our knowledge, this is
the first analysis of HPV-31 oncogene variants in patients in
Sichuan.
Materials and methods
Ethical statement
The present study was approved by the Ethics
Committee of Sichuan University (Chengdu, China). All participants
provided informed consent prior to the collection of experimental
specimens.
Study subjects and specimen
collection
Between January 1, 2009 and September 14, 2015, a
total of 13,283 patients aged between 18 and 65 years old provided
specimens. They were obtained from maternity hospitals in Sichuan,
China (The Affiliate Reproduction Hospital of Sichuan Genitalia
Hygiene Research Center, Chengdu; Chengdu Medical College Attached
Infertility Hospital, Chengdu; Jinjiang Maternity and Child Health
Hospital, Quanzhou; Angel Women and Children's Hospital, Chengdu;
Chengdu Zongnan Gynecology Hospital, Chengdu; and Chengdu
Songziniao Sterility Hospital, Chengdu) and the patients were
neither in the menstrual period nor had undergone cervical
conization prior to the present study. A brush and a colposcope
were used to collect cervical scrapings, which were placed in Cell
Preservation Liquid (Yaneng Bioscience Co., Ltd., Shenzhen, China),
and stored at −20°C until DNA extraction, HPV detection and typing
were performed. Further experiments primarily took place in the
Institute of Medical Genetics, College of Life Science, Sichuan
University (Chengdu, China).
DNA extraction
DNA was extracted using the Human Papillomavirus
Genotyping kit for 23 Types (PCR-RDB; Yaneng Bio-Technology
(Shenzhen) Co., Ltd., Shenzhen, China) according to the
manufacturer's protocols. The cervical specimens, which were stored
at −20°C, were thawed to room temperature. Cells were suspended in
Cell Preservation Liquid and 1 ml suspension transferred to a 1.5
ml Eppendorf tube (Shanghai Sangon Biological Engineering
Technology and Services Co., Ltd., Shanghai, China) and subjected
to centrifugation for 10 min at 16,155 × g at 4–6°C in a Hema 14D
high speed centrifuge. The supernatant was removed and 100 µl cell
pyrolysis liquid (KCl, Tris-HCl, Triton X-100) was added to the
sediment, which was incubated in a boiling water bath for 10 min.
Centrifugation was conducted again, at 16,155 × g for 10 min at
4–6°C, and the supernatant was removed. The supernatant was stored
at 4°C and used for polymerase chain reaction (PCR) analysis. All
of the extracted DNA samples were stored at −20°C until
examination. None of the samples remained at room temperature for
more than 2 h, or at 4°C for more than 24 h, to avoid DNA
degradation.
HPV genotyping
HPV genotyping was accomplished using the Human
Papillomavirus Genotyping kit for 23 Types (Yaneng Bioscience Co.,
Ltd.), which exploits chip technology and employs reverse membrane
hybridization technology, where the probe is fixed on a membrane
strip, to identify 23 different HPV genotypes in one reaction (18
HR and five low risk subtypes). Viral DNA was extracted, amplified
and genotyped according to the manufacturers' protocol, and
negative and positive specimens were displayed in each reaction.
All HPV-31 positive specimens were subsequently subjected to
variant analysis.
PCR amplification and sequencing
The specific primers used to amplify E5, E6, E7 and
L1 genes in HPV-31 positive DNA are listed in Table I and were designed using Primer 5.0
bioinformatics software (Premier Biosoft, Palo Alto, CA, USA)
according to the published GenBank reference sequence (accession
no. J04353. https://www.ncbi.nlm.nih.gov/nuccore/333048). The
length of the L1 sequence was 1,550 base pairs, so it was divided
into two sections (L1Q and L1H). PCR amplification for each gene
was set up in a 25 µl reaction volume containing 4.0 µl DNA (10–100
ng) 10X PCR buffer, 2.5 mM/l deoxynucleotide triphosphates, 25 mM/l
MgCl2 (TransBionovo Co., Ltd., Beijing, China;
https://www.transbionovo.com), 50 µM/l
of each primer (Shanghai Sangon Biological Engineering Technology
and Services Co., Ltd.) and 2.5 U Taq polymerase
(TransBionovo Co., Ltd.). PCR amplification was conducted at 95°C
for 5 min, 38 cycles of denaturation at 94°C for 45 sec, annealing
at various temperatures for 50 sec (Table I), extension at 72°C for 1 min and
a final extension step at 72°C for 10 min. PCR products were
checked on 2% agarose gels (Shanghai Sangon Biological Engineering
Technology and Services Co., Ltd.), visualized using UV
fluorescence (GeneGreen; Tiangen Biotech Co., Ltd., Beijing, China)
using a WFH-202 fluorometer (Wenzhou Fuhua Instruments, Inc.
wenzhou068795.11467.com) and sequenced
by Sangon Biotech Co., Ltd. (Shanghai, China).
 | Table I.Primer used for the molecular
characterization of HPV-31 E5, E6, E7 and L1. |
Table I.
Primer used for the molecular
characterization of HPV-31 E5, E6, E7 and L1.
| Primer name | Sequence
primers | Sequenced region
(bp) | ORF size (bp) | Annealing
temperature (°C) |
|---|
| HPV-31 E5 F |
5′-gcacaaaccaaacaagggct-3′ | 3,531–4,200
(670) | 255 | 59.5 |
| HPV-31 E5 R |
5′-agtgcgttttgtagcgtt-3′ |
|
|
|
| HPV-31 E6 F |
5′-gaaagtggtgaaccgaaaac-3′ | 41-740 (700) | 450 | 58 |
| HPV-31 E6 R |
5′actgacaacaaaaggtaa-3′ |
|
|
|
| HPV-31 E7 F |
5′-gaccgttgtgtccagaagaa-3′ | 430-963 (534) | 297 | 59 |
| HPV-31 E7 R |
5′-ctctgaaatgttgtcccctg-3′ |
|
|
|
| HPV-31 L1-Q F |
5′-cccctacaacgccacaagt-3′ | 5,441–6,373
(933) | 750 | 58 |
| HPV-31 L1-Q R |
5′-agtagggaccgattcacc-3′ |
|
|
|
| HPV-31 L1-H F |
5′-aatgctattacccctgg-3′ | 6,062–7,098
(1,037) | 800 | 54.5 |
| HPV-31 L1-H R |
5′-atacaatacagcacaagcac-3′ |
|
|
|
Data analysis
Following direct sequencing, all sequences were
aligned with an HPV-31 prototype sequence (GenBank accession no.
J04353) available from the National Center for Biotechnology
Information (NCBI). The samples with mutational sites were
amplified and sequenced again to rule out the possibility of error
due to mismatched bases in the PCR process. All E5, E6, E7 and L1
sequences were separately aligned by Clustal X 2.1 (ftp://ftp.ebi.ac.uk/pub/software/clustalw2/)
(16). Maximum-likelihood
phylogenetic trees of respective HPV-31 E5, E6, E7 and L1 variation
patterns were subsequently constructed by Molecular Evolutionary
Genetics Analysis 6 software (17), using Kimura's two-parameter model
(18). To estimate the selection
pressure acting on the HPV-31 E5, E6, E7 and L1 gene sequences,
non-synonymous and synonymous nucleotide divergence for coding
regions was inferred by the Nei and Gojobori method with
Phylogenetic Analyses by Maximum Likelihood (PAML; http://abacus.gene.ucl.ac.uk/software/paml.html)
software version 4.8 (19–22).
Nucleotide sequence accession
numbers
All sequences of the E5, E6, E7 and L1 genes were
submitted to the NCBI GenBank database and assigned accession
numbers. The HPV-31 E5, E6 and E7 sequences, 31EPL01-31EPL16, were
published with the GenBank accession codes KU163553-KU163568. The
HPV-31 L1 sequences, 31LPL01-31LPL16 are published with the GenBank
accession codes KU163569-KU163575, KU163584 and
KU163576-KU163583.
Results
Distribution of HPV-31
A total of 13,283 specimens from Sichuan (China),
were collected for the present study, and 4,130 (31.1%) were HPV
positive. Of these 4,130 samples, 141/4,130 (3.4%) were positive
for HPV-31. There were 70/141 (49.6%) samples with single HPV-31
infection, 35/141 (24.8%) samples with double infection and 36/141
(25.5%) samples with multiple infection. The ages of the patients
infected by HPV-31 ranged between 18–70, with a median age of 32
years.
E5 sequence variations
In total, 141 HPV-31 positive samples were detected,
of which 87 samples were used for further exploration; 54 samples
were excluded due to incomplete data. In the present study,
sequences of HPV-31 E5, E6, E7 genes and the L1 gene were obtained
from 48/87 and 37/87 patients, respectively. The failure of some
genes to be amplified or sequenced was due to either a short copy
of HPV or instability of the amplicon.
Compared with the HPV-31 reference sequence
(J04353), 11 nucleotide variations of the E5 gene were observed in
the 48 HPV-31 E5 sequences studied (Table II). In total, six missense
mutations of A3828G (1/48), A3957G (15/48), C3981G (1/48), G4005A
(1/48), T4053A (42/48) and A4059G (3/48) were revealed, which
resulted in amino acid changes of N5D, I48V, P56A, V64I, F80I and
S82G, respectively (Table II).
The remaining five variations, A3827G (15/48), G3956A (17/48),
T3980A (11/48), T4052C (16/48) and A4064G (1/48), were synonymous
variations. T4053A was the most common non-synonymous variation,
followed by A3957G. Excluding A4064G, which had only one specimen,
the synonymous mutations were of similar quantity.
 | Table II.Nucleotide sequence mutations of
HPV-31 E5. |
Table II.
Nucleotide sequence mutations of
HPV-31 E5.
|
|
|
|
| Variation of E5 at
nucleotide position |
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| Category | 3827 | 3828 | 3956 | 3957 | 3980 | 3981 | 4005 | 4052 | 4053 | 4059 | 4064 | n |
|---|
| Reference nt | A | A | G | A | T | C | G | T | T | A | A |
|
| 31EPL01 | – | – | A | – | – | – | – | – | A | – | – | 13 |
| 31EPL02 | G | – | – | G | A | – | – | C | A | – | – | 1 |
| 31EPL03 | – | – | – | – | – | – | – | – | A | – | – | 1 |
| 31EPL04 | – | – | – | – | – | – | – | – | A | – | – | 12 |
| 31EPL05 | G | – | – | G | A | – | – | C | A | – | – | 2 |
| 31EPL06 | G | – | – | G | A | – | – | C | – | – | – | 4 |
| 31EPL07 | – | G | – | – | – | G | A | C | A | – | G | 1 |
| 31EPL08 | G | – | – | G | A | – | – | C | A | – | – | 5 |
| 31EPL09 | G | – | – | G | A | – | – | C | – | – | – | 1 |
| 31EPL10 | – | – | – | – | – | – | – | – | A | – | – | 1 |
| 31EPL11 | G | – | – | G | A | – | – | C | – | – | – | 1 |
| 31EPL12 | – | – | – | – | – | – | – | – | A | – | – | 1 |
| 31EPL13 | – | – | A | – | – | – | – | – | A | G | – | 2 |
| 31EPL14 | – | – | A | – | – | – | – | – | A | – | – | 1 |
| 31EPL15 | – | – | A | – | – | – | – | – | A | G | – | 1 |
| 31EPL16 | G | – | – | G | A | – | – | C | A | – | – | 1 |
|
|
| N |
| I |
| P | V |
| F | S |
|
|
| AA mutations |
| 5 |
| 48 |
| 56 | 64 |
| 80 | 82 |
|
|
|
| – | D | – | V | – | A | I | – | I | G | – |
|
E6 sequence variations
The present study detected 14 mutation sites in the
E6 gene in 34/48 (70.8%) samples through comparative analysis with
the HPV-31 reference sequence (J04353). No variations were observed
in 14/48 (29.2%) samples, which were referred to as ‘E6
prototype-like’ sequences. There were six non-synonymous
variations: C285T (15/48), A297G (2/48), A301G (1/48), T312C
(2/48), A475G (2/48) and C520T (17/48), which led to amino acids
changes of H60Y, T64A, K65R, T69L, K123R and A138V (Table III). The frequencies of C285T
(15/48) and C520T (17/48) mutations were 15/48 (31.3%) and 17/48
(35.4%), respectively. These resulted in changes at the same point
in the genetic code, however expressed different amino acids. The
remaining eight mutations were synonymous (Table III).
 | Table III.Nucleotide sequence mutations of
HPV-31 E6. |
Table III.
Nucleotide sequence mutations of
HPV-31 E6.
|
|
|
|
|
|
| Variation of E6 at
nucleotide position |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Category | 134 | 176 | 248 | 285 | 297 | 301 | 312 | 321 | 326 | 335 | 404 | 428 | 475 | 520 | n |
|---|
| Reference nt | T | C | T | C | A | A | T | A | A | T | G | A | A | C |
|
| 31EPL01 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 13 |
| 31EPL02 | – | – | – | T | – | G | – | T | – | – | A | – | – | T | 1 |
| 31EPL03 | – | T | – | – | – | – | – | – | – | – | – | – | – | – | 1 |
| 31EPL04 | – | T | – | – | – | – | – | – | – | – | – | – | – | – | 12 |
| 31EPL05 | – | – | – | T | – | – | – | T | G | – | A | G | – | T | 2 |
| 31EPL06 | – | – | – | T | – | – | – | T | G | – | A | G | – | T | 4 |
| 31EPL07 | – | – | C | – | G | – | – | T | – | – | – | – | G | T | 1 |
| 31EPL08 | – | – | – | T | – | – | – | T | – | – | A | G | – | T | 5 |
| 31EPL09 | – | – | – | T | – | – | – | T | G | C | A | G | – | T | 1 |
| 31EPL10 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 |
| 31EPL11 | – | – | – | T | – | – | – | T | G | – | A | – | – | T | 1 |
| 31EPL12 | – | – | C | – | G | – | – | T | – | – | – | – | G | T | 1 |
| 31EPL13 | A | – | – | – | – | – | – | – | – | – | – | – | – | – | 2 |
| 31EPL14 | A | – | – | – | – | – | C | – | – | – | – | – | – | – | 1 |
| 31EPL15 | A | – | – | – | – | – | C | – | – | – | – | – | – | – | 1 |
| 31EPL16 | – | – | – | T | – | – | – | T | – | – | A | G | – | T | 1 |
|
|
|
|
| H | T | K | F |
|
|
|
|
| K | A |
|
| AA mutations |
|
|
| 60 | 64 | 65 | 69 |
|
|
|
|
| 123 | 138 |
|
|
| – | – | – | Y | A | R | L | – | – | – | – | – | R | V |
|
E7 sequence variations
Compared with the HPV-31 reference sequence
(J04353), six mutation sites were presented in the 48 samples
(Table IV). There were four
non-synonymous variations: C626T (33/48), G695A (15/48), A743G
(47/48) and C737G (1/48) which resulted in amino acid changes of
H23Y, E46K, K62E and Q60E (Table
IV). A743G was the most common non-synonymous mutation across
all specimens. The remaining two mutations were G580A (15/48) and
C670T (15/48), and were synonymous.
 | Table IV.Nucleotide sequence mutations of
HPV-31 E7. |
Table IV.
Nucleotide sequence mutations of
HPV-31 E7.
|
|
| Variation of E7 at
nucleotide position |
|---|
|
|
|
|
|---|
| Category | 580 | 626 | 670 | 695 | 743 | 737 | n |
|---|
| Reference nt | G | C | C | G | A | C |
|
| 31EPL01 | – | T | – | – | G | – | 13 |
| 131EPL02 | A | – | T | A | G | – | 1 |
| 31EPL03 | A | T | – | – | G | – | 1 |
| 31EPL04 | – | T | – | – | G | – | 12 |
| 31EPL05 | A | – | T | A | G | – | 2 |
| 31EPL06 | A | – | T | A | G | – | 4 |
| 31EPL07 | – | T | T | A | G | – | 1 |
| 31EPL08 | A | – | T | A | G | – | 5 |
| 31EPL09 | A | – | T | A | G | – | 1 |
| 31EPL10 | – | T | – | – | G | – | 1 |
| 31EPL11 | A | – | T | A | G | – | 1 |
| 31EPL12 | – | T | – | – | G | – | 1 |
| 31EPL13 | – | T | – | – | G | – | 2 |
| 31EPL14 | – | T | – | – | G | – | 1 |
| 31EPL15 | – | T | – | – | G | – | 1 |
| 31EPL16 | A | – | T | A | G | G | 1 |
|
|
| H |
| E | K | Q |
|
| AA mutations |
| 23 |
| 46 | 62 | 60 |
|
|
| – | Y | – | K | E | E |
|
The specimens were divided into 16 species, termed
31EPL01-31EPL16 (Tables II,
III and IV). According to the frequency of E5, E6
and E7 mutations, based on the statistics from the present study,
31 mutation sites were detected, of which 16 sites were
non-synonymous mutations and the remaining 15 were synonymous
mutations.
Phylogenetic trees of respective HPV-31 E5, E6 and
E7 variation patterns were subsequently constructed by Molecular
Evolutionary Genetics Analysis 6 software (17), using Kimura's two-parameter model
(18). Phylogenetic analysis of
HPV-31 variant lineage distribution (n=48) in Sichuan, China
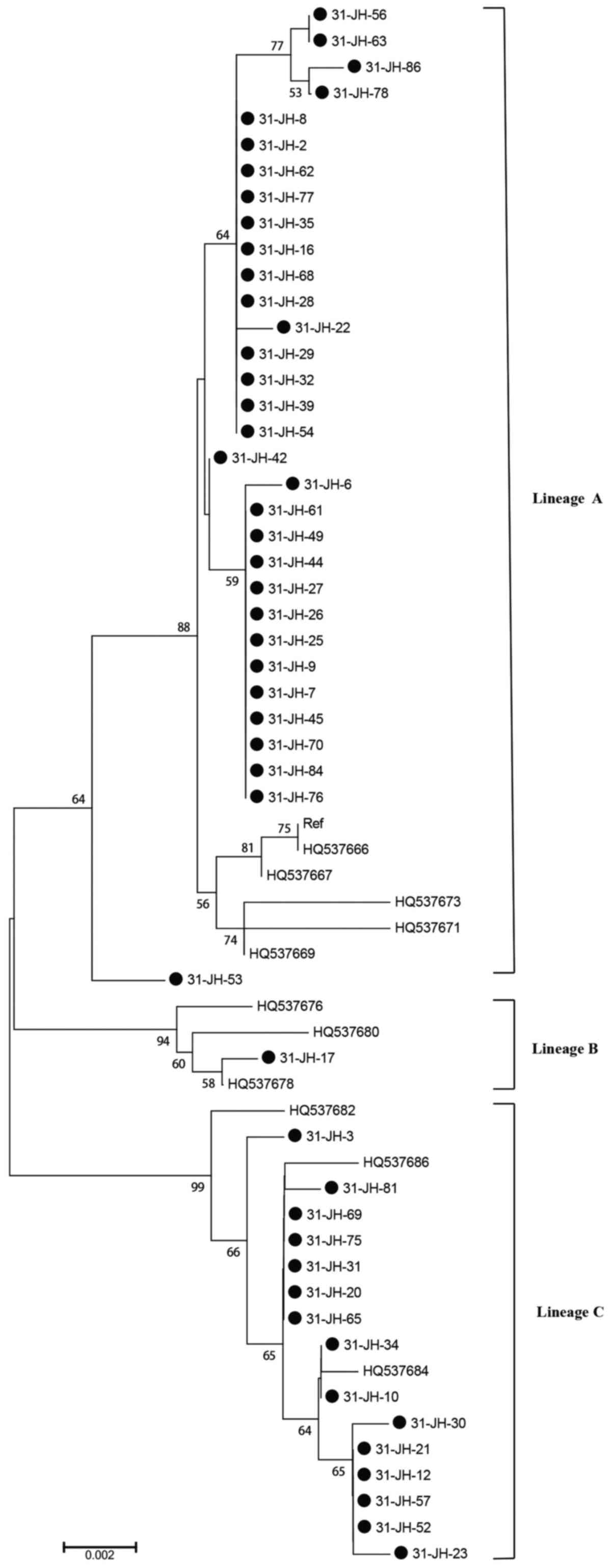
(23), demonstrated that A
variants were most commonly detected (66.7%; Fig. 1), followed by C variants (31.3%;
Fig. 1) and B variants (2.0%;
Fig. 1).
L1 sequence variations
Compared with the HPV-31 reference sequence
(J04353), the specimens were divided into 16 species, named
31LPL01-31LPL16 (Tables V and
VI). In total, 30 nucleotide
variations of the L1 gene were observed in strains from 37
patients, of which four were missense mutations: T6131A (1/37),
A6350G (14/37), C6372A (17/37) and A6840G (1/37), which resulted in
amino acid changes of S206T, T29A, T36N and Q192R, respectively
(Table III). The remaining 26
variations were synonymous, of which C6367T (37/37) and C6817A
(37/37) were detected in samples. The non-synonymous mutations
A6350G (14/37) and C6372A (17/37) were more common than the others.
No nucleotide substitutions resulting in premature termination
codon or frameshift mutations were detected.
 | Table V.Nucleotide sequence mutations of
HPV-31 L1. |
Table V.
Nucleotide sequence mutations of
HPV-31 L1.
|
|
Variation
of L1 at nucleotide position |
|---|
|
|
|
|---|
| Category | 5581 | 5752 | 5797 | 5839 | 5848 | 5866 | 5920 | 5921 | 5998 | 6019 | 6067 | 6085 | 6127 | 6131 | 6199 | n |
|---|
| Reference nt | T | A | C | T | A | T | A | T | A | A | A | C | A | T | C |
|
| 31LPL01 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 12 |
| 31LPL02 | A | G | – | – | – | – | – | C | – | G | G | T | – | – | – | 1 |
| 31LPL03 | A | – | T | – | – | – | – | C | – | G | – | T | – | – | – | 1 |
| 31LPL04 | A | – | – | – | – | – | – | C | – | G | – | T | – | – | – | 2 |
| 31LPL05 | – | – | – | – | G | – | – | C | – | – | – | – | G | – | – | 2 |
| 31LPL06 | – | – | – | – | – | – | – | C | – | – | – | – | G | A | – | 1 |
| 31LPL07 | A | – | – | – | – | – | G | C | – | G | – | T | – | – | – | 2 |
| 31LPL08 | A | – | – | – | – | C | – | C | – | G | – | T | – | – | – | 3 |
| 31LPL09 | A | – | – | – | – | – | – | C | – | G | – | T | – | – | – | 3 |
| 31LPL10 | A | – | – | C | – | – | – | C | – | G | – | T | – | – | – | 1 |
| 31LPL11 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 |
| 31LPL12 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 |
| 31LPL13 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T | 3 |
| 31LPL14 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 |
| 31LPL15 | – | – | – | – | – | – | – | – | G | – | – | – | – | – | – | 2 |
| 31LPL16 | A | – | – | – | – | – | – | C | – | G | – | T | – | – | – | 1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| S |
|
|
| AA mutations |
|
|
|
|
|
|
|
|
|
|
|
|
| 206 |
|
|
 | Table VI.Additional nucleotide sequence
mutations of HPV-31 L1. |
Table VI.
Additional nucleotide sequence
mutations of HPV-31 L1.
|
|
Variation
of L1 at nucleotide position |
|---|
|
|
|
|---|
| Category | 6238 | 6328 | 6350 | 6367 | 6372 | 6379 | 6568 | 6574 | 6586 | 6647 | 6664 | 6772 | 6796 | 6817 | 6840 | n |
|---|
| Reference nt | T | G | A | C | C | A | T | C | T | A | T | G | G | C | A |
|
| 31LPL01 | – | – | – | T | – | – | – | – | – | – | – | – | – | A | – | T |
| 31LPL02 | – | A | G | T | A | – | – | T | G | – | – | – | A | A | – | – |
| 31LPL03 | – | A | G | T | A | – | – | – | G | – | C | – | A | A | – | – |
| 31LPL04 | – | A | G | T | A | – | – | – | G | – | C | – | A | A | – | – |
| 31LPL05 | A | – | – | T | A | G | C | – | – | – | – | A | A | A | – | – |
| 31LPL06 | A | – | – | T | A | G | C | – | – | – | – | A | A | A | – | A |
| 31LPL07 | – | A | G | T | A | – | – | – | G | – | C | – | A | A | – | A |
| 31LPL08 | – | A | G | T | A | – | – | – | G | C | C | A | A | A | – | – |
| 31LPL09 | – | A | G | T | A | – | – | T | G | – | – | – | A | A | – | – |
| 31LPL10 | – | A | G | T | A | – | – | – | G | – | C | – | A | A | – | – |
| 31LPL11 | – | – | – | T | – | – | – | – | – | – | – | – | – | A | G | – |
| 31LPL12 | – | – | – | T | – | – | – | – | – | C | – | – | – | A | – | – |
| 31LPL13 | – | – | – | T | – | – | – | – | – | – | – | – | – | A | – | – |
| 31LPL14 | – | – | – | T | – | – | – | – | G | – | – | – | – | A | – | – |
| 31LPL15 | – | – | – | T | – | – | – | – | – | – | – | – | – | A | – | – |
| 31LPL16 | – | A | G | T | A | – | – | – | G | – | – | – | A | A | – | – |
|
|
|
| T |
| T |
|
|
|
|
|
|
|
|
| Q |
|
| AA mutations |
|
| 29 |
| 36 |
|
|
|
|
|
|
|
|
| 192 |
|
|
|
|
| A |
| N |
|
|
|
|
|
|
|
|
| R |
|
Phylogenetic trees of respective HPV-31 L1 variation
patterns were subsequently constructed by Molecular Evolutionary
Genetics Analysis 6 software (17), using Kimura's two-parameter model
(18). Phylogenetic analysis of
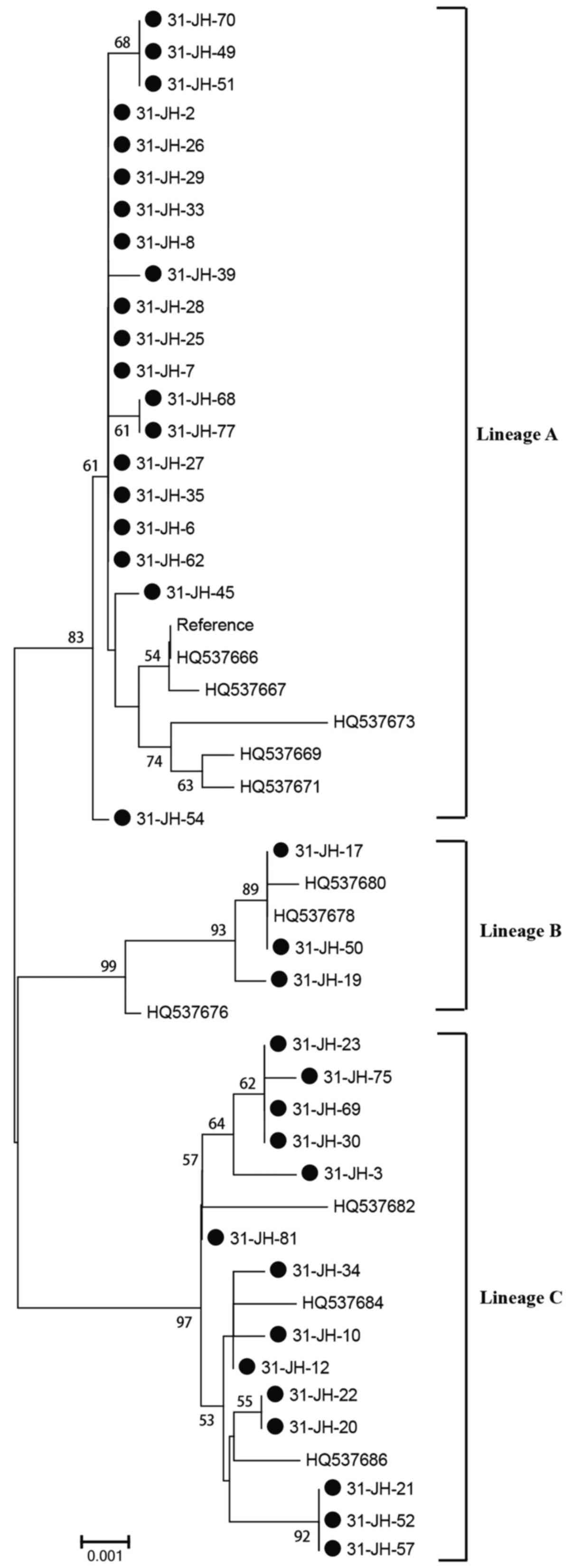
HPV-31 variant lineage distribution (n=37) in Sichuan, China,
revealed that A variants were most commonly detected (54.0%;
Fig. 2), followed by C variants
(37.8%; Fig. 2), and B variant
(8.2%; Fig. 2).
Selective pressure analysis of all
sequences
PAML 4.8 software was used to test for variable
dN/dS rate ratios among the lineages. There was no evidence of
negative selection in the sequence alignment of HPV-31 E5, E6 and
E7 genes or L1 genes (P>0.05 and P>0.05, respectively;
Tables VII and VIII, respectively).
 | Table VII.Site-specific tests for positive
selection on HPV-31 E5-E6-E7. |
Table VII.
Site-specific tests for positive
selection on HPV-31 E5-E6-E7.
| Model | InL | Estimates of
parameters | 2Δ1 | Positively selected
sites |
|---|
| M1 | −1731.91 | P=0.774, 0.226;
w=0.000, 1.000 |
| NA |
| M2 | −1724.28 | P=0.919, 0.000,
0.0815; | 15.26 | 138 Aa,
195 Ea |
|
|
| w=0.000, 1.000,
5.227 | P<0.05 |
|
 | Table VIII.Site-specific tests for positive
selection on HPV-31 L1. |
Table VIII.
Site-specific tests for positive
selection on HPV-31 L1.
| Model | InL | Estimates of
parameters | 2Δl | Positively selected
sites |
|---|
| M7 | −2556.01 | P=0.009;
q=0.149 |
| NA |
| M8 | −2544.39 | P0=0.995; P=1.600;
q=99.000 | 23.24 | 267 Ta,
274 Ta |
|
|
| P1=0.005;
w=11.648 | P<0.05 |
|
Discussion
The present study demonstrated that 31.1% of the
females with cervical cancer studied were infected with at least
one subtype of HPV. Globally, ~2–20% of healthy females have
detectable levels of HPV DNA in their cervical tissue, as detected
by epidemiological studies (24).
The higher rate recorded in the present study may reflect the
design of the study, which selected only females with active
cervical cancer. Previous studies have demonstrated that smoking
habits, the number of sexual partners, history of sexually
transmitted diseases and abnormal cervical cytology collectively
increase the risk of HPV infection (25).
Knowledge of HPV genetic variants may aid in the
understanding of the pathogenic mechanisms and progression of
cervical cancer. It has previously been suggested that variants of
the same HPV type are biologically distinct and may have different
pathogenic risks (26). HPV-16 is
the most frequent HPV type globally, followed by HPV-18 (8). In Sichuan, China, HPV-16 is also the
most frequent, however the second most frequent is HPV-58 (27). The incidence of HR HPV types in
Sichuan were demonstrated to be as follows: HPV-16 (28.1%), HPV-58
(16.0%), HPV-33 (9.2%), HPV-52 (8.4), HPV-18 (7.3%) and HPV-31
(3.4%), with HPV-18 only being the fifth most common (27). In the present study, the observed
HPV-31 prevalence was 1.1%. This was similar to the prevalence
reported by Xi et al (15),
which enrolled 5,060 females from the ASC-US and LSIL Triage study
in the US and observed a prevalence of 1.1%. However, this differs
from the rate (0.4%) reported by a meta-analysis of females with
normal cytology from Asia (28),
which was in accordance with an international study that reported
that the prevalence of HPV-31 is lower in Asia (0.3%) compared with
the global average (0.8%), particularly in European (2.3%) and
Latin American (1.2%) females with normal cytology (29). However, the rate of HPV-31
prevalence in Sichuan, China, differed from the 0.52% previously
reported in Northern Chinese females (30). Future studies regarding geographic
variation and ethnic differences may be worthwhile.
HPV E5, E6 and E7 proteins are important for
replication and transcription of viral DNA, and are involved in
interacting with the cytoskeleton network, cell immortalization and
transformation (31). E6 and E7
are known for their ability to bind to and inactivate p53 and pRB,
respectively, however they also interact with a wide range of
cellular proteins (11,32,33).
To the best of our knowledge, the present study was
the first to examine gene mutations of HPV-31 E5. All the mutation
sites detected by the present study were novel. Nucleotide changes
in the HPV-31 E6 oncogene at positive 134, 301, 312 and 335 were
discovered, to the best of our knowledge, for the first time in the
present study. When considering the HPV-31 E7 oncogene, a novel
nucleotide change at positive 737 was reported by the present
study. According to the frequency of E5, E6, E7 gene mutations, the
samples were divided into six, nine and five species, respectively.
However, when the E5, E6 and E7 sequences of the samples were
integrated, 16 species were detected. E5, E6 and E7 oncogenes
displayed characteristics of the alternative model, Kimura's two
parameter model, suggesting that these mutations may be cyclic in
frequency in the Sichuan population. In these genes, the
non-synonymous mutations C285T, A297G, A475G, C520T, C626T, G695A,
A743G, C737G, A3975G, C3981G, G4005A, T4053A and A4059G can lead to
changes to polarity, hydropathic potential and the amino acid side
chain, which potentially altered the folding of the oncoprotein
(34).
In the L1 region, one or more amino acid changes may
lead to a conformational change of the capsid protein, and
interfere with the conformation of epitopes relevant to viral
neutralization. A previous study in Central Brazil reported six
mutations (34), fewer than the 30
mutations reported by the present study, of which four were
non-synonymous mutations. This previous study reported the presence
of the mutation C6862T in Brazil, however the present study did not
observe this mutation in Sichuan, China.
Xi et al (15) reported the proportion of A, B and C
variants were 41.7, 21.1 and 37.2% in the USA female population
(28). Chagas et al
(35) examined five HPV-31
positive specimens from Brazil and reported that the proportions of
A, B and C variants were 57.2, 5.7 and 37.1%, respectively
(35). However, previous
investigations have only observed variant lineages A (64.3%) and C
(35.7%) in Southern China (30).
As above, in the present study the variant lineage A of HPV-31 was
the most commonly detected variant, followed by C. However, the
present study also observed the presence of variant lineage B,
which has not been previously reported from Southern China. The
results of one previous study by Ferenczi et al (36) differ substantially from the above;
when 41 HPV-31 positive specimens were collected from Italian
females, the proportions of A, B and C variants were 4.9, 29.3 and
65.8%, respectively (36).
Therefore, geographic variation and ethnic differences cannot be
ignored.
In conclusion, the present study reported sequence
variations in the E5, E6, E7 and L1 genes of HPV-31 isolates from
Sichuan, China. The sequence variations in the genes examined may
contribute to HPV oncogenesis. These data will provide a solid
foundation for further biological and clinical studies, and may
also be employed in epidemiological studies where sequence
variations are used as markers for monitoring HPV infections in
target populations.
Acknowledgements
The present study was supported by the Bio-Research
and Utilization Joint Key Laboratory of Sichuan and Chongqing,
Institute of Medical Genetics, College of Life Sciences, Sichuan
University. The authors would like thank the patients and
volunteers who participated in the study.
References
|
1
|
Bosch FX, Manos MM, Muñoz N, Sherman M,
Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R and Shah KV:
Prevalence of human papillomavirus in cervical cancer: A worldwide
perspective. International biological study on cervical cancer
(IBSCC) Study Group. J Natl Cancer Inst. 87:796–802. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Villiers EM, Fauquet C, Broker TR,
Bernard HU and Zur Hausen H: Classification of papillomaviruses.
Virology. 324:17–27. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bernard HU, Burk RD, Chen Z, van Doorslaer
Kzur Hausen H and de Villiers EM: Classification of
papillomaviruses (PVs) based on 189 PV types and proposal of
taxonomic amendments. Virology. 401:70–79. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Munoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ;
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group, : Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li N, Franceschi S, Howell-Jones R,
Snijders PJ and Clifford GM: Human papillomavirus type distribution
in 30,848 invasive cervical cancers worldwide: Variation by
geographical region, histological type and year of publication. Int
J Cancer. 128:927–935. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smith JS, Lindsay L, Hoots B, Keys J,
Franceschi S, Winer R and Clifford GM: Human papillomavirus type
distribution in invasive cervical cancer and high-grade cervical
lesions: A meta-analysis update. Int J Cancer. 121:621–632. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ojesina AI, Lichtenstein L, Freeman SS,
Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio
L, Cibulskis K, Bertelsen B, et al: Landscape of genomic
alterations in cervical carcinomas. Nature. 506:371–375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rusan M, Li YY and Hammerman PS: Genomic
landscape of human papillomavirus-associated cancers. Clin Cancer
Res. 21:2009–2019. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pande S, Jain N, Prusty BK, Bhambhani S,
Gupta S, Sharma R, Batra S and Das BC: Human papillomavirus type 16
variant analysis of E6, E7, and L1 genes and long control region in
biopsy samples from cervical cancer patients in north India. J Clin
Microbiol. 46:1060–1066. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boulenouar S, Weyn C, Van Noppen M, Moussa
Ali M, Favre M, Delvenne PO, Bex F, Noël A, Englert Y and Fontaine
V: Effects of HPV-16 E5, E6 and E7 proteins on survival, adhesion,
migration and invasion of trophoblastic cells. Carcinogenesis.
31:473–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zehbe I, Wilander E, Delius H and
Tommasino M: Human papillomavirus 16 E6 variants are more prevalent
in invasive cervical carcinoma than the prototype. Cancer Res.
58:829–833. 1998.PubMed/NCBI
|
|
12
|
Kast WM, Brandt R, Sidney J, Drijfhout JW,
Kubo RT, Grey HM, Melief CJ and Sette A: Role of HLA-A motifs in
identification of potential CTL epitopes in human papillomavirus
type 16 E6 and E7 proteins. J Immunol. 152:3904–3912.
1994.PubMed/NCBI
|
|
13
|
Conrad M, Bubb V and Schlegel R: The human
papillomavirus type 6 and 16 E5 proteins are membrane-associated
proteins which associate with the 16-kilodalton pore-forming
protein. J Virol. 67:6170–6178. 1993.PubMed/NCBI
|
|
14
|
Giannoudis A and Simon Herrington CS:
Human papillomavirus variants and squamous neoplasia of the cervix.
J Pathol. 193:295–302. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xi LF, Schiffman M, Koutsky LA, Hulbert A,
Lee SK, Defilippis V, Shen Z and Kiviat NB: Association of human
papillomavirus type 31 variants with risk of cervical
intraepithelial neoplasia grades 2–3. Int J Cancer. 131:2300–2307.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thompson JD, Gibson TJ, Plewniak F,
Jeanmougin F and Higgins DG: The CLUSTAL_X windows interface:
Flexible strategies for multiple sequence alignment aided by
quality analysis tools. Nucleic Acids Res. 25:4876–4882. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tamura K, Stecher G, Peterson D, Filipski
A and Kumar S: MEGA6: Molecular evolutionary genetics analysis
version 6.0. Mol Biol Evol. 30:2725–2729. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kimura M: A simple method for estimating
evolutionary rates of base substitutions through comparative
studies of nucleotide sequences. J Mol Evol. 16:111–120. 1980.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hamza AA, Robene-Soustrade I, Jouen E,
Lefeuvre P, Chiroleu F, Fisher-Le Saux M, Gagnevin L and Pruvost O:
MultiLocus Sequence Analysis- and Amplified Fragment Length
Polymorphism-based characterization of xanthomonads associated with
bacterial spot of tomato and pepper and their relatedness to
Xanthomonas species. Syst Appl Microbiol. 35:183–190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nei M and Gojobori T: Simple methods for
estimating the numbers of synonymous and nonsynonymous nucleotide
substitutions. Mol Biol Evol. 3:418–426. 1986.PubMed/NCBI
|
|
21
|
Yamada T, Wheeler CM, Halpern AL, Stewart
AC, Hildesheim A and Jenison SA: Human papillomavirus type 16
variant lineages in United States populations characterized by
nucleotide sequence analysis of the E6, L2, and L1 coding segments.
J Virol. 69:7743–7753. 1995.PubMed/NCBI
|
|
22
|
Yang Z: PAML 4: Phylogenetic analysis by
maximum likelihood. Mol Biol Evol. 24:1586–1591. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z, Schiffman M, Herrero R, Desalle R,
Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW and
Burk RD: Evolution and taxonomic classification of human
papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33,
HPV35, HPV52, HPV58 and HPV67. PLoS One. 6:e201832011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bosch FX and De Sanjosé S: Chapter 1:
Human papillomavirus and cervical cancer-burden and assessment of
causality. J Natl Cancer Inst Monogr 3–13. 2003. View Article : Google Scholar
|
|
25
|
Kasap B, Yetimalar H, Keklik A, Yildiz A,
Cukurova K and Soylu F: Prevalence and risk factors for human
papillomavirus DNA in cervical cytology. Eur J Obstet Gynecol
Reprod Biol. 159:168–171. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sichero L, Ferreira S, Trottier H,
Duarte-Franco E, Ferenczy A, Franco EL and Villa LL: High grade
cervical lesions are caused preferentially by non-European variants
of HPVs 16 and 18. Int J Cancer. 120:1763–1768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Z, Wang Q, Ding X, Li Q, Zhong R and
Ren H: Characteristics of HPV prevalence in Sichuan Province,
China. Int J Gynaecol Obstet. 131:277–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bao YP, Li N, Smith JS and Qiao YL; ACCPAB
members, : Human papillomavirus type distribution in women from
Asia: A meta-analysis. Int J Gynecol Cancer. 18:71–79. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bruni L, Diaz M, Castellsagué X, Ferrer E,
Bosch FX and de Sanjosé S: Cervical human papillomavirus prevalence
in 5 continents: Meta-analysis of 1 million women with normal
cytological findings. J Infect Dis. 202:1789–1799. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu M, He Z, Xi L, Li J, Liu F, Liu Y, Pan
Y, Ning T, Guo C, Xu R, et al: The distribution and common amino
acid polymorphisms of human papillomavirus (HPV)-31 variants in
2700 women from Northern China. PLoS One. 9:e991412014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xi LF, Koutsky LA, Galloway DA, Kuypers J,
Hughes JP, Wheeler CM, Holmes KK and Kiviat NB: Genomic variation
of human papillomavirus type 16 and risk for high grade cervical
intraepithelial neoplasia. J Natl Cancer Inst. 89:796–802. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zehbe I, Tachezy R, Mytilineos J, Voglino
G, Mikyskova I, Delius H, Marongiu A, Gissmann L, Wilander E and
Tommasino M: Human papillomavirus 16 E6 polymorphisms in cervical
lesions from different European populations and their correlation
with human leukocyte antigen class II haplotypes. Int J Cancer.
94:711–716. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee K, Magalhaes I, Clavel C, Briolat J,
Birembaut P, Tommasino M and Zehbe I: Human papillomavirus 16 E6,
L1, L2 and E2 gene variants in cervical lesion progression. Virus
Res. 131:106–110. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chagas BS, Batista MV, Guimarães V,
Balbino VQ, Crovella S and Freitas AC: New variants of E6 and E7
oncogenes of human papillomavirus type 31 identified in
Northeastern Brazil. Gynecol Oncol. 123:284–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chagas BS, Batista MV, Crovella S, Gurgel
AP, Silva Neto Jda C, Serra IG, Amaral CM, Balbino VQ, Muniz MT and
Freitas AC: Novel E6 and E7 oncogenes variants of human
papillomavirus type 31 in Brazilian women with abnormal cervical
cytology. Infect Genet Evol. 16:13–18. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ferenczi A, Gyöngyösi E, Szalmás A,
Hernádi Z, Tóth Z, Kónya J and Veress G: Sequence variation of
human papillomavirus type 31 long control region: Phylogenetic and
functional implications. J Med Virol. 85:852–859. 2013. View Article : Google Scholar : PubMed/NCBI
|