Introduction
Gliomas are the most common intrinsic brain tumors,
which make up ~30% of all brain and central nervous system tumors
and 80% of all malignant brain tumors (1). They are characterized by high
morbidity, high recurrence rates, high mortality and low cure rate
(2). In recent years with the
development of imaging diagnosis and microneurosurgery, diagnosis
and treatment of gliomas has made some progress but little
breakthrough in the treatment of malignant gliomas, most of
patients die within 1 year of diagnosis (3,4).
Despite the frequency of gliomas, the etiology that contributes to
tumor initiation remains to be elucidated. Previous studies
revealed that aberrant regulation of the Wnt signaling pathway has
a vital role in the process of glioma and malignant progression
(5–7).
Wnt/β-catenin signaling pathway has been reported to
be involved in the regulation of life processes, including cell
proliferation, cell apoptosis, organ growth and development, cancer
and inflammatory diseases (8,9). The
canonical Wnt pathway consists of Wnt protein, frizzled family
receptor, dishevelled, glycogen synthase kinase 3β, adenomatous
polyposis coli, axin, β-catenin and T cell transcriptional
regulation factors (TCF). It is activated by binding a Wnt-protein
ligand to a frizzled family receptor, which passes the biological
signal to the downstream pathway inside the cells. Following
activation by external stimulation, β-catenin is translocated into
nucleus to combine with TCF, followed by initiation of
transcription regulation of target genes such as Myc
proto-oncogene, bHLH transcription factor, cyclin D1 and cytochrome
c oxidase subunit II, which are involved in cell apoptosis
and may have carcinogenic effects (9,10).
TCF4 and β-catenin are upregulated in gliomas
compared with the normal brain, inhibiting cell proliferation and
inducing cell apoptosis (11).
TCF4, one of TCFs factors, including lymphoid enhancer-binding
factor (LEF1), TCF1, TCF3 and TCF4, has been previously reported to
exhibit high expression in renal carcinoma, colorectal cancer,
hepatocellular carcinoma and brain tumors (11–16).
Abnormal upregulation of TCF4 stimulating downstream target genes
is a common early event in tumorigenesis.
The human TCF4 gene consists of 17 exons with
several alternatively splicing sites, including a C-terminal tail
(exon 13–17) and exon 4 (16).
Previous studies revealed that TCF mRNAs were subject to
alternative splicing to form different isoforms, which are also
important in regulating transcription in the Wnt signaling pathway
and associated with tumorigenesis. Different TCF4 isoforms have
been identified in various types of cancer including renal
carcinoma (14), colorectal cancer
(16) and brain tumor (17). However, the importance of TCF4
isoforms in the process of gliomas remains to be elucidated. The
present study analyzed the alterative splicing forms of TCF4 and
characterized their biological functions in three different glioma
cell lines. The findings of the present study provide evidence that
TCF4 isoforms may contribute to human gliomas, which lay the
foundation for molecular mechanisms of regulation of malignant
biological behavior of glioma.
Materials and methods
Cell culture
U251, A172 and U-87MG human glioma cell lines (Type
Culture Collection of the Chinese Academy of Sciences, Shanghai,
China) were maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS) (both from Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin and were maintained at 37°C
and a 5% CO2 humidified environment.
RNA extraction, reverse
transcription-polymerase chain reaction (RT-PCR) and sequence
alignment
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was
synthesized using PrimeScript II First-Strand cDNA Synthesis kit
(Takara Bio, Inc., Otsu, Japan) at 42°C for 60 min and 95°C for 5
min. Subsequently, cDNA sequences were amplified by Premix Taq
(Takara Bio, Inc.) with specific primers presented in Table I at 98°C for 10 sec, 58°C for 30
sec, 72°C for 3 min and 72°C for 5 min. DNA fragments were isolated
from the agarose gels with Minibest Agarose Gel Extraction kit
(Takara Bio, Inc.). PCR products were then subcloned into T-vector
PMD19-T (Takara Bio, Inc.) and an individual clone was selected for
sequencing by Sangon Biotech Co., Ltd., (Shanghai, China). Sequence
alignments were performed with TCF4 genome (gene ID: 6934)
deposited in Genbank by BLAST (blast.ncbi.nlm.nhi.gov/Blast.cgi).
 | Table I.Sequences of primers used in the
present study. |
Table I.
Sequences of primers used in the
present study.
| Primer | Exon | Direction | Sequence (5′-3′) |
|---|
| H251 TCF4 | 10 | Forward |
AGTGCACGTTGAAAGAAAGCGCG |
| H252 TCF4 | 17SR | Reverse |
CTGCCTTCACCTTGTATGTAGAG |
| H557 TCF4 | 1 | Forward |
CTTCCAAAATTGCTGCTGGTG |
| H558 TCF4 | 11 | Reverse |
TCTCTGGACAGTGCATGCC |
| H559 TCF4 | 12 | Forward |
CAAGCAGCCGGGAGAGAC |
| H562 TCF4 | 15 | Forward |
ATGCAAATACTCCAAAGAAG |
| H563 TCF4 | 17LR | Reverse | TCAGCGAGCAGGAGGC |
| H580 TCF4 | 15 | Reverse |
CTGCACGGTTTGCACCAT |
Construction of expression vectors and
cell transfection
Three TCF4 isoforms (TCF4VX35, TCF4VX18 and TCF4V7)
were inserted into the corresponding site of the pVITRO2-neo-mcs
vector (Takara Bio, Inc.) site. Cells (2×105 cells/ml)
were seeded in 24-well plates and cultured until the cells were at
70–80% confluence, subsequently the cells were transfected with 800
ng/well of expression plasmids using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) as specified by the
manufacturer's protocol. After 48 h, green fluorescence was
examined under a fluorescence microscope (Olympus Corporation,
Tokyo, Japan).
MTT assay
Cell proliferation was determined using an MTT
assay. U251 cells (4×104 cells/ml) were seeded in
96-well plates and transfected with expression vectors or empty
vector (Takara Bio, Inc.). At 48 h following transfection, 20 µl
MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was
added to each culture and incubated for 4–6 h. At the end of the
incubation period the medium were removed and 150 µl DMSO was added
to each well. After agitation at a low speed for 10 min, absorbance
of converted dye was measured at a wavelength of 490 nm.
Flow cytometry
Apoptosis analysis was performed with the Annexin
V-PE/7-AAD flow cytometry kit (BD Biosciences, Franklin Lakes, NJ,
USA) according to the manufacturer's protocol. U251 cells
(1.5×106 cells/ml) in 6-well plate were transfected with
expression vectors or empty vector of TCF4 isoforms using
Lipofectamine 2000. After 48 h, cells were washed twice with
ice-cold PBS and resuspended in 1X binding buffer at a
concentration of 1×106 cells/ml. The solution (100 µl)
was transferred to a 5 ml culture tube. PE Annexin V (5 µl) and 5
µl 7-AAD was added and cells were incubated for 15 min at 25°C in
the dark. Finally, 400 µl 1X binding buffer was added to each tube
and was analyzed by FC500MCL flow cytometry using CXP (version 2.2;
Beckman Coulter, Inc., Brea, CA, USA) within 1 h.
Wound healing assay
Cell migration was assessed using a scratch wound
assay. U251 cells (1×106 cells/ml) were grown to 60–70%
confluence and transfected with expression vectors of TCF4 isoforms
using Lipofectamine 2000. After 24 h, a linear wound was made by
scraping a sterile micropipette tip across the confluent cell
layer. Cells were washed three times with PBS to remove detached
cells and debris. Subsequently the size of wounds was observed and
measured on a TS100-F inverted-phase contrast microscope (Nikon
Corporation, Tokyo, Japan) at 0 and 48 h.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean. Statistical analysis was performed using a one-way
analysis of variance followed by Duncan's multiple range test as a
post-hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Determination of TCF4 isoforms in
human glioma cell lines
TCF4 isoforms have been identified in various tumor
cells in the majority of body systems and organs (14,15,18).
In order to determine whether there are TCF4 isoforms in human
glioma cells, nest PCR analysis using specific primers of human
TCF4 in three cell lines (U251, A172 and U-87MG) was performed. PCR
products were inserted into a PMD19-T vector and an individual
clone was selected to be sequenced. A total of 13 different TCF
isoforms were expressed in U251 cells in the present study,
including 3 isoforms (vx35, vx18 and v7), which had high scores
after blasting the TCF genome in the NCBI database. A total of 19
different TCF isoforms were expressed in A172 cells and 3 isoforms
(18, 27 and vx21) gained top scores. In the U-87MG cells 12
different TCF isoforms were expressed and 3 isoforms (27, 18 and
vx21) had the highest scores. They had several distinct features:
v7 was found to lack exon 4, 6, whereas vx18 lacked exon 4, 6, 14,
15, vx21 lacked part of exon 17 and had an additional amino acid,
vx27 lacked exon 14 and part of exon 17 and vx35 lacked exon 4, 6,
15 and 16. The present study selected vx35, vx18 and v7 found in
U251 cells for further study. The three TCF4 isoforms (TCF4VX35,
TCF4VX18 and TCF4V7) were inserted into the pVITRO2-neo-mcs
vectors. The eukaryotic expression plasmids pVITRO-neo-TCF4vx35,
pVITRO-neo-TCF4vx18 and pVITRO-neo-TCF4vx7 were successfully
expressed in U215 cells.
Effect of TCF4 isoforms on cell
proliferation
TCF4 may serve different roles in different tumor
cells. Overexpression of the TCF4 isoforms has been previously
associated with oncogenesis and development of malignant tumors
(11,18,19).
In order to investigate the effect of TCF4 isoforms on U251 cell
proliferation, each TCF4 isoform expression plasmid was transiently
transfected into cells and growth rate was measured from 12 to 60
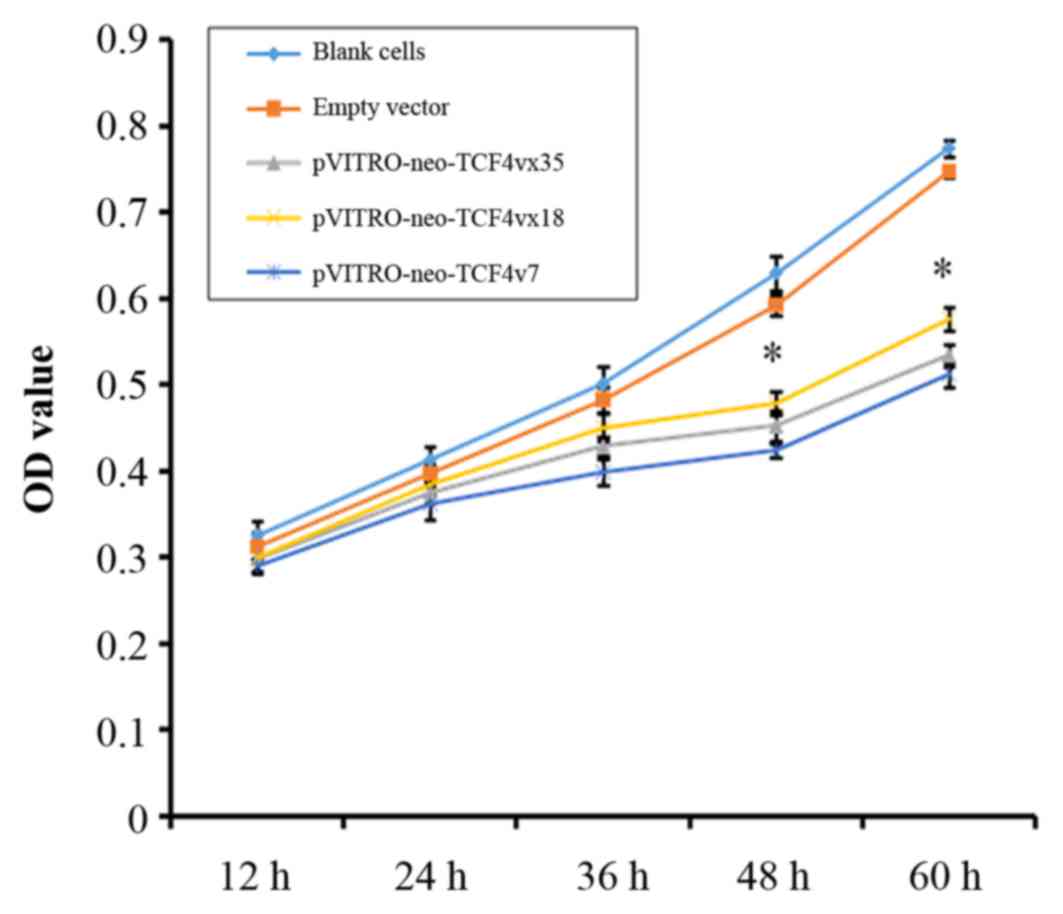
h. As presented in Fig. 1,
pVITRO-neo-TCF4vx35, pVITRO-neo-TCF4vx18 and pVITRO-neo-TCF4vx7
groups had reduced cell proliferation compared with the empty
vector control at 48 and 60 h (P<0.001).
Effect of TCF4 isoforms on cell
apoptosis
Following the observation that TCF4 isoforms
inhibited cell growth rate, the present study aimed to determine
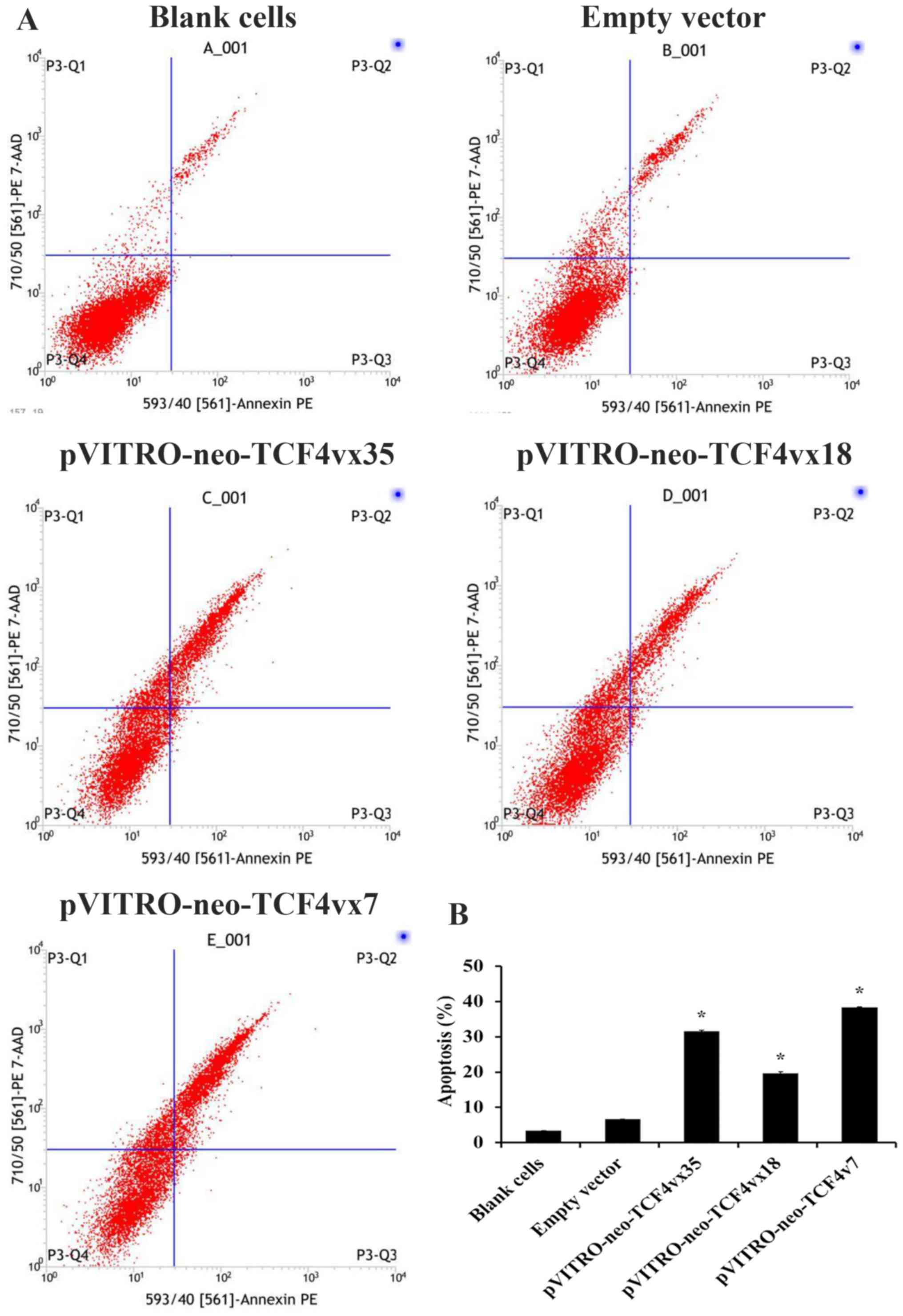
whether TCF4 isoforms induced cell apoptosis. The Annexin
V-PE/7-AAD flow cytometry was performed for apoptosis analysis. As
presented in Fig. 2, U251 cells
transfected with pVITRO-neo-TCF4vx35, pVITRO-neo-TCF4vx18 and
pVITRO-neo-TCF4vx7 had 31.59, 19.60 and 38.43% more apoptotic cells
compared with the empty vector control (6.67%; P<0.001; Fig. 2B).
Effect of TCF4 isoforms on cell
migration
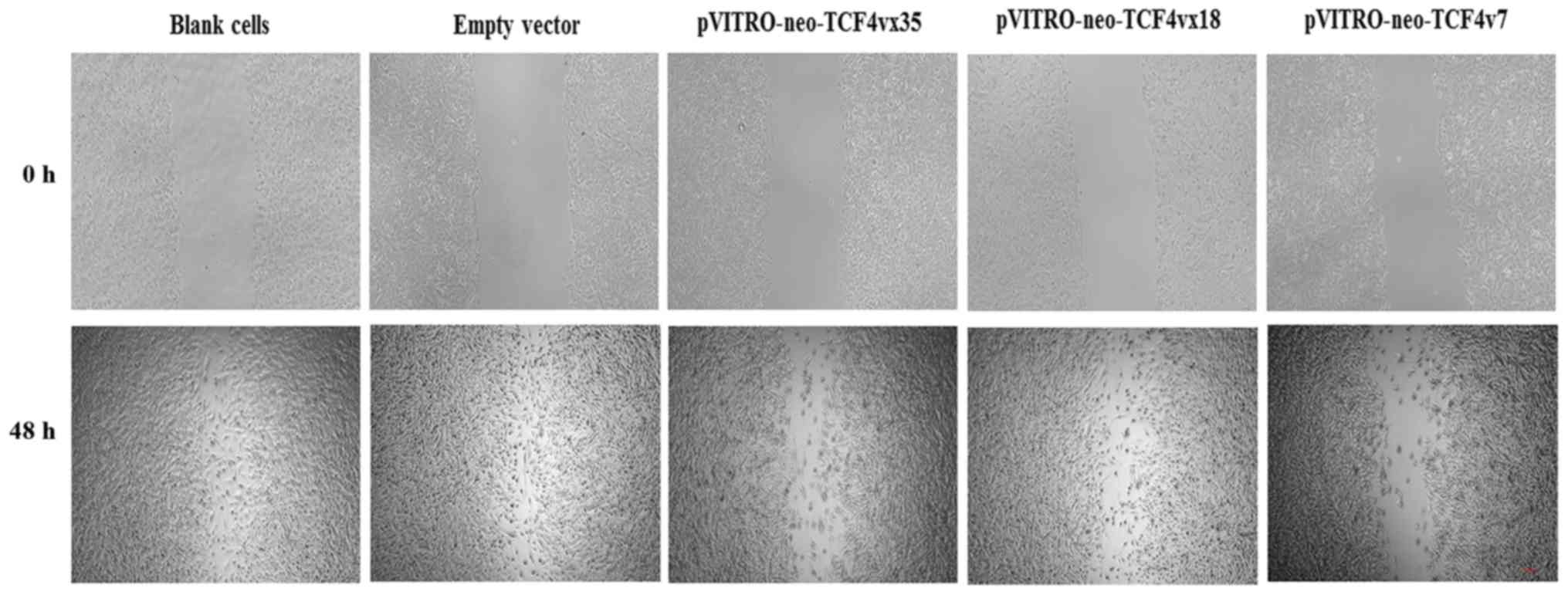
To determine if the TCF isoforms have a function in
cell migration, a wound healing assay was performed. As presented
in Fig. 3, U251 cells
overexpressing pVITRO-neo-TCF4vx35, pVITRO-neo-TCF4vx18 and
pVITRO-neo-TCF4vx7 had increased cell migration; however, this was
decreased compared with the control.
Discussion
The Wnt signaling pathway is involved in the
development and homeostasis of cells and tissues, and aberrant
regulation Wnt signal frequently occurs during the initiation of
various tumor types (8). TCF
proteins may act as transcriptional activators or repressors in
nucleus in the downstream in Wnt signaling. It has been previously
reported that TCF4 and its splice variants are critical
determinants of context-dependent Wnt signaling responses,
including physiological development or oncogenesis (20,21).
Previous studies revealed that human TCF4 gene was alternatively
spliced in various types of cancers, including brain tumors
(17). The present study aimed to
determine the TCF4 isoforms that may be associated with biological
properties of glioma cells and the development of the malignant
phenotype.
Differential expression and splicing isoform
analysis of TCF4 has been previously reported in the tissue of
brain tumors (17). However, to
the best of our knowledge, there is no report on gliomas regarding
the TCF4 isoform and its significance in tumorigenesis. The present
study analyzed TCF4 isoforms in three human glioma cell lines. In
the TCF4 gene, three major domains are present: β-catenin-binding
domain in exon 1, the DNA-binding HMG boxes in exons 10 and 11, and
the COOH-terminal-binding domain in exon 17 (22,23).
The current study identified 13 different TCF4 isoforms in U251
cells, 19 different TCF isoforms in A172 cells and 12 different TCF
isoforms in U-87MG cells. It was previously demonstrated that the
majority of alternative splicing occurred within exon 12–17 in
brain tumor (17). The present
study revealed that the majority of alternative spliced regions
were exon 4, 5, 14, 15 and 16 in glioma cells. However, in
colorectal cancer cell lines various splicing isoforms of TCF4 mRNA
were present in the COOH-terminal region (exon 17) (16). In renal cell carcinomas, four
splicing isoforms of the TCF4 were present in the region between
exon 12 and exon 17 (14). All the
isoforms the current study identified contained the N-terminal part
of TCF4 including the β-catenin binding domain; therefore, they may
not lack the transcriptional activity and Wnt/β-catenin was
activated in glioma cells to contribute to tumorigenesis. However,
the specific molecular mechanism remains to be elucidated.
To evaluate the functional properties of TCF4
isoforms in U251 cells, TCF4 isoform expression plasmid was
transfected and cell proliferation, apoptosis and migration were
quantified. The findings of the current study revealed that the
isoforms may inhibit the proliferation and induce the apoptosis and
migration of U251 cells. After Wnt signaling transduction cascade
is triggered, cytoplasmic β-catenin translocates to nucleus to bind
TCF and modulates expression of target genes critical for cell
proliferation, differentiation, survival and apoptosis (24). It is possible that overexpression
of TCF isoforms may lead to activation of β-catenin/TCF
transcription for target genes associated with cell proliferation
and apoptosis. Conversely, cells lose cell polarity and cell-cell
adhesion, and gain migratory and invasive properties during the
epithelial-mesenchymal transition, which is induced by
Wnt/β-catenin (25,26). Overexpression of TCF isoforms
appears to be sufficient to induce cell migration via the
Wnt/β-catenin pathway. However, the function and malignant
biological behaviors of TCF4 isoforms require further
investigation.
In conclusion, the TCF4 isoforms in three human
glioma cells lines were cloned and identified in the present study.
The functions and properties of TCF4 isoforms were analyzed in U251
cells. The findings of the present study revealed that TCF4
isoforms inhibited cell proliferation and induced the cell
apoptosis and migration. Therefore, this mechanism of
Wnt/β-catenin-mediated TCF isoforms transcription may regulate the
target genes that contribute to the tumorigenesis of glioma. Due to
the carcinogenic properties of the TCF isoforms, the interaction
between Wnt/β-catenin and the TCF isoforms should be assessed in
future studies.
Acknowledgements
The current study was supported by grants from Hubei
Province Health and Family Planning Scientific Research Project
(grant no. WJ2015MB118) and Medical Scientific Research Foundation
of Hubei Province, China (grant no. 2016CFC745).
References
|
1
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reuss D and von Deimling A: Hereditary
tumor syndromes and gliomas. Recent Results Cancer Res. 171:83–102.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Riezzo I, Zamparese R, Neri M, De Stefano
F, Parente R, Pomara C, Turillazzi E, Ventura F and Fineschi V:
Sudden, unexpected death due to glioblastoma: Report of three fatal
cases and review of the literature. Diagn Pathol. 8:732013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Swanson KD, Lok E and Wong ET: An overview
of alternating electric fields therapy (NovoTTF Therapy) for the
treatment of malignant glioma. Curr Neurol Neurosci Rep. 16:82016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sareddy GR, Panigrahi M, Challa S,
Mahadevan A and Babu PP: Activation of Wnt/beta-catenin/Tcf
signaling pathway in human astrocytomas. Neurochem Int. 55:307–317.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schüle R, Dictus C, Campos B, Wan F,
Felsberg J, Ahmadi R, Centner FS, Grabe N, Reifenberger G, Bermejo
JL, et al: Potential canonical wnt pathway activation in high-grade
astrocytomas. ScientificWorldJournal. 2012:6973132012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Huang K, Shi Z, Zou J, Wang Y,
Jia Z, Zhang A, Han L, Yue X, Liu N, et al: High β-catenin/Tcf-4
activity confers glioma progression via direct regulation of AKT2
gene expression. Neuro Oncol. 13:600–609. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duchartre Y, Kim YM and Kahn M: The Wnt
signaling pathway in cancer. Crit Rev Oncol Hematol. 99:141–149.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kikuchi A: Canonical Wnt signaling pathway
and cellular responses. Clin Calcium. 23:799–807. 2013.(In
Japanese). PubMed/NCBI
|
|
11
|
Chen L, Huang K, Han L, Shi Z, Zhang K, Pu
P, Jiang C and Kang C: β-catenin/Tcf-4 complex transcriptionally
regulates AKT1 in glioma. Int J Oncol. 39:883–890. 2011.PubMed/NCBI
|
|
12
|
Travis A, Amsterdam A, Belanger C and
Grosschedl R: LEF-1, a gene encoding a lymphoid-specific protein
with an HMG domain, regulates T-cell receptor alpha enhancer
function [corrected]. Genes Dev. 5:880–894. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van de Wetering M, Oosterwegel M, Dooijes
D and Clevers H: Identification and cloning of TCF-1, a T
lymphocyte-specific transcription factor containing a
sequence-specific HMG box. EMBO J. 10:123–132. 1991.PubMed/NCBI
|
|
14
|
Shiina H, Igawa M, Breault J,
Ribeiro-Filho L, Pookot D, Urakami S, Terashima M, Deguchi M,
Yamanaka M, Shirai M, et al: The human T-cell factor-4 gene
splicing isoforms, Wnt signal pathway, and apoptosis in renal cell
carcinoma. Clin Cancer Res. 9:2121–2132. 2003.PubMed/NCBI
|
|
15
|
Tsedensodnom O, Koga H, Rosenberg SA,
Nambotin SB, Carroll JJ, Wands JR and Kim M: Identification of
T-cell factor-4 isoforms that contribute to the malignant phenotype
of hepatocellular carcinoma cells. Exp Cell Res. 317:920–931. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duval A, Rolland S, Tubacher E, Bui H,
Thomas G and Hamelin R: The human T-cell transcription factor-4
gene: Structure, extensive characterization of alternative
splicings, and mutational analysis in colorectal cancer cell lines.
Cancer Res. 60:3872–3879. 2000.PubMed/NCBI
|
|
17
|
Howng SL, Huang FH, Hwang SL, Lieu AS, Sy
WD, Wang C and Hong YR: Differential expression and splicing
isoform analysis of human Tcf-4 transcription factor in brain
tumors. Int J Oncol. 25:1685–1692. 2004.PubMed/NCBI
|
|
18
|
He G, Guan X, Chen X, Wang Y, Luo C and
Zhang B: Expression and splice variant analysis of human TCF4
transcription factor in esophageal cancer. J Cancer. 6:333–341.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gu X, Yao L, Ma G, Cui L, Li Y, Liang W,
Zhao B and Li K: TCTP promotes glioma cell proliferation in vitro
and in vivo via enhanced β-catenin/TCF-4 transcription. Neuro
Oncol. 16:217–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wallmen B, Schrempp M and Hecht A:
Intrinsic properties of Tcf1 and Tcf4 splice variants determine
cell-type-specific Wnt/β-catenin target gene expression. Nucleic
Acids Res. 40:9455–9469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tomimaru Y, Koga H, Yano H, de la Monte S,
Wands JR and Kim M: Upregulation of T-cell factor-4
isoform-responsive target genes in hepatocellular carcinoma. Liver
Int. 33:1100–1112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clevers H and van de Wetering M: TCF/LEF
factor earn their wings. Trends Genet. 13:485–489. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Duval A, Gayet J, Zhou XP, Iacopetta B,
Thomas G and Hamelin R: Frequent frameshift mutations of the TCF-4
gene in colorectal cancers with microsatellite instability. Cancer
Res. 59:4213–4215. 1999.PubMed/NCBI
|
|
24
|
Barker N: The canonical Wnt/beta-catenin
signalling pathway. Methods Mol Biol. 468:5–15. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Micalizzi DS and Ford HL:
Epithelial-mesenchymal transition in development and cancer. Future
Oncol. 5:1129–1143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|