Introduction
Duchenne muscular dystrophy (DMD) is an X-linked
recessive disorder characterized by severe and progressive muscle
wasting and weakness due to degeneration of skeletal muscle. DMD
primarily affects males with an estimated incidence of 1/3,500 male
births (1). Females are usually
asymptomatic but some female carriers manifest milder forms of the
disease. This disorder is caused by defective expression of
dystrophin, a 427-kDa structural protein that is encoded by a 14-kb
mRNA transcribed from the DMD gene (2). Several studies of the DMD gene have
led to the identification of dystrophin isoforms that exhibit
tissue-specific expression and temporal regulation (3–6).
These isoforms are named according their molecular weight as Dp260,
Dp140, Dp116, and Dp71. The presence of at least seven independent
promoters in the DMD gene accounts for the complexity of its
transcriptional regulation.
Dp71, the smallest and the first expressed product
of DMD gene during embryogenesis (7), is ubiquitously present in all tissues
except in adult muscle cells (8).
The N-terminal of Dp71 has seven unique residues but retains the
cystein-rich and C-terminal domains of full-length dystrophin.
Despite homologies between Dp71 and 427-kDa dystrophin, many
studies have revealed different functions for both proteins
(9–11). Dp71 shows high levels of expression
in liver and brain (12). In
neuronal cells, Dp71 has been involved in differentiation, cell
cycle and adhesion processes (13–16).
Other studies have associated the Dp71 expression to mental
retardation, short stature in DMD patients and gastric
adenocarcinoma prognosis (17–19).
Despite functional studies of Dp71, it has been
necessary to identify the transcription factors and gene elements
involved in its regulation in order to elucidate fully the pathways
by which Dp71 expression is regulated in tissues. It has been
established that the Dp71 promoter, which lacks a TATA box, can be
transactivated by several transcription factors, including AP2α,
YY1, and members of the Sp family. For example, in mouse myoblasts,
the Dp71 promoter is consistently transactivated by Sp1 and Sp3,
but during differentiation these factors disappear, resulting in
downregulation of Dp71 (17). YY1,
Sp1 and Sp3 also transactivate the Dp71 promoter in hepatic cells
(18), while in neuronal cells the
transactivation is mediated by Sp1 and AP2α (19). Sp binding sites within the Dp71
promoter are highly conserved, which implies that the Sp proteins
(particularly Sp1) can exert similar effects on Dp71 expression in
different tissues and species.
The synthetic polyaromatic hydrocarbon,
β-naphthoflavone (β-NF), has been extensively used to analyze the
effect of xenobiotics on a large number of genes involved in
metabolic and adaptive processes (20,21).
In previous studies, we showed that both in vitro and in
vivo expression of Dp71 in hepatic cells is repressed by β-NF
(22). More recently, we
identified different DNA elements on the Dp71 promoter that are
crucial for Dp71 expression in hepatic cells, including binding
sites for YY1 and the Sp family members. The functionality of these
DNA elements and proteins was confirmed by EMSA, chromatin
immunoprecipitation, and site-directed mutagenesis analysis
(18). However, the underlying
molecular mechanisms by which β-NF inhibits Dp71 expression remain
poorly studied. The aim of the present study was to determine
whether β-NF represses Dp71 expression at the level of messenger
RNA stability or promoter activity.
Materials and methods
Cell cultures and treatments
Human HepG2 cells [American Type Culture Collection
(ATTC) Manassas, VA, USA; HB-8065], derived from hepatoblastoma
(23), were cultured in Minimum
Essential Media (Invitrogen, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum, 2 m M L-Glutamine, 1.5 g/l sodium
bicarbonate, 1 mM sodium pyruvate, 0.1 mM non-essential aminoacids,
penicillin (100 U/ml) and streptomycin (100 µg/ml). Mouse Hepa-1
cells (ATCC; CRL-1830), derived from hepatome, were cultured in
Dulbecco's modified Eagles medium (Invitrogen) supplemented with
10% fetal bovine serum, 2 mM L-Glutamine, 4.5 g/l D-Glucose,
penicillin (100 U/ml) and streptomycin (100 µg/ml). Both cell lines
were incubated at 37°C in a humidified atmosphere with 95% air and
5% CO2. Cells were seeded on 6-well culture plates
(1.5×105 cells per well) and treated for 24 h with 50 µM
of β-naphthoflavone (cat. no., N3633; Sigma, St. Louis, MO, USA)
diluted in dimethyl sulfoxide (DMSO) or with DMSO alone as control
(22). For all cell treatments,
the final DMSO concentration was adjusted to 0.1%. To inhibit
transcription, both β-NF-treated or DMSO-treated cells were exposed
to actinomycin D (50 µg/ml) for 0, 4, 8, 12 and 16 h.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction
Total RNA was extracted from β-NF- or DMSO-treated
Hepa-1 cells using the TRIzol reagent (Invitrogen) according to the
manufacturer's instructions. RNA concentration and purity were
estimated by optical density at 260 and 280 nm wavelength, and its
integrity was corroborated by electrophoresis on 1% agarose gels
stained with ethidium bromide. RNA was reversed transcribed with
the M-MLV reverse transcriptase (Invitrogen) and subjected to
real-time qPCR for Dp71, r18S and cytochrome P450 1A1 gene
expression analysis, as previously described (22). RT-qPCR was performed following the
MIQE guidelines (22) with the
next conditions: Each 25-µl reaction mixture consisted of 12.5 µl
of 2X TaqMan Master Mix (Applied Biosystems, Carlsbad, CA, USA),
1.25 µl of forward/reverse primers (25 µM each primer) and
hydrolysis probe (10 µM), and 3 µl of cDNA. Amplification was
performed under the following conditions: pre-denaturation at 50°C
for 2 min and 95°C for 10 min; denaturation at 95°C for 15 sec;
annealing and extension at 60°C for 1 min. mRNA levels were
normalized to the expression of the 18S rRNA housekeeping gene
(cat. 4310893E; Applied Biosystems). Samples were processed and
detected in a real-time PCR 7500 Fast System (Applied Biosystems).
Assays were performed in technical replicates and negative controls
where included in the same plate. Quantitative analyses of gene
expression were conducted using the 2−ΔΔCq formula
(24), where the first ∆Cq is the
difference between Cq values for Dp71 gene and r18S gene, and the
∆∆Cq is the difference between ∆Cq values of the
β-naphtopflavone-treated and control samples. Finally, 2 to the
power of negative ∆∆Ct gets the fold gene expression.
Transient cell transfections and
luciferase assays
The Dp71 promoter fragment (from −224 to +65) fused
to luciferase gene (18) was
transfected in human HepG2 and mouse Hepa-1 cells with
Lipofectamine 2000 reagent (Invitrogen), according to
manufacturer's instructions. Briefly, 3.6 µg of p224-Luc and 400 ng
of phRL-CMV plasmids (the latter used as a control for normalizing
transfection efficiency) were incubated with 250 µl of DMEM without
serum for 5 min. In a separate microtube, the plasmids were mixed
with 10 µl Lipofectamine 2000 previously diluted in 250 µl of DMEM
without serum. After 20 min of incubation at room temperature,
DNA-Lipofectamine complexes were added to 1×105 human or
mouse cells. In each assay, pGL3 Basic Vector and pGL3 Control
Vector (Promega, Madison, WI, USA) were transfected in parallel as
negative and positive controls, respectively. After 5 h, medium was
replaced with DMEM supplemented with 10% fetal bovine serum.
Twenty-four hours after transfection the cells were
exposed to 1, 5, 10 or 50 µM β-NF or 0.1% DMSO (control) for 24 h.
Before luciferase activity determination cells were washed with 1X
phosphate-buffered saline (PBS) solution, and then homogenized with
1X Passive Lysis Buffer (Promega) for 15 min on an oscillatory
shaker. Firefly and Renilla luciferase activity was measured
with the Dual-Luciferase Assay System (Promega) and the Modulus
Luminometer (Turner BioSystems, Sunnyvale, CA, USA). Luciferase
activity of DMSO-treated cells was set as 100%. Blanks were
analyzed by conducting luciferase activity assays in untransfected
cells. Luciferase activity levels were normalized to the
Renilla luciferase activity levels of the phRL-CMV vector
from the same cell culture.
Preparation of nuclear extracts and
electrophoretic mobility shift assays (EMSAs)
Nuclear extracts were prepared according to
Schreiber et al (1989) (25). Briefly, Hepa-1 cells were either
untreated or exposed to 50 µM β-NF or 0.1% DMSO (vehicle control)
for 24 h. The cells were then washed with cold 1X PBS, resuspended
in 400 µl of cold buffer A [10 mM HEPES, (pH 7.9), 10 mM KCl, 0.1
mM EDTA, 0.1 mM EGTA, 1 mM DTT and 0.5 mM PMSF], and incubated for
15 min on ice. Afterwards, 25 µl of 10% Igepal CA-630 solution
(Sigma-Aldrich, St. Louis, MO, USA) was added to each sample, and
cell disruption was performed by aspirating the contents several
times through a 22-gauge needle. The samples were centrifuged at
2,000 × g for 5 min at 4°C. Supernatants were removed and the
nuclear pellets resuspended in 50 µl of buffer C [20 mM HEPES (pH
7.9), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF) with
vigorous vortexing for 30 min at 4°C. Samples were then centrifuged
and the nuclear extracts isolated.
Nuclear extracts from untreated, DMSO-treated and
β-NF-treated hepatic cells were subjected electrophoretic
mobility-shift assays (EMSAs) using double-stranded oligonucleotide
probes (YY1 and Sp1/Sp3) (18).
These probes were end-labelled with [γ-32P]-ATP
(Amersham Pharmacia, GE Healthcare, Buckinghamshire, UK) using 10 U
of T4 polynucleotide kinase (Invitrogen), according to the
manufacturer's instructions. EMSAs were carried out by two
independent experiments on ice for 20 min in a 20-µl reaction
mixture containing 10 mM Tris-HCl (pH 8.0), 1 mM MgCl2,
5 mM NaCl, 0.5 mM EDTA (pH 8.0), 0.5 mM DTT, 4% glycerol, 15 µg
nuclear extract, 20 mM spermidine, 50 ng/µl poly(dI:dC), and 0.2
pmol of probe. Samples were separated on native polyacrylamide gels
(6%) and visualized by autoradiography.
Statistics
Data are expressed as the mean ± standard deviation
(SD). Statistical analyses were performed using the Mann Whitney U
test with STATA version 8.0 program (Stata Corporation, College
Station, TX, USA), and significant differences were considered at
P<0.05.
Results
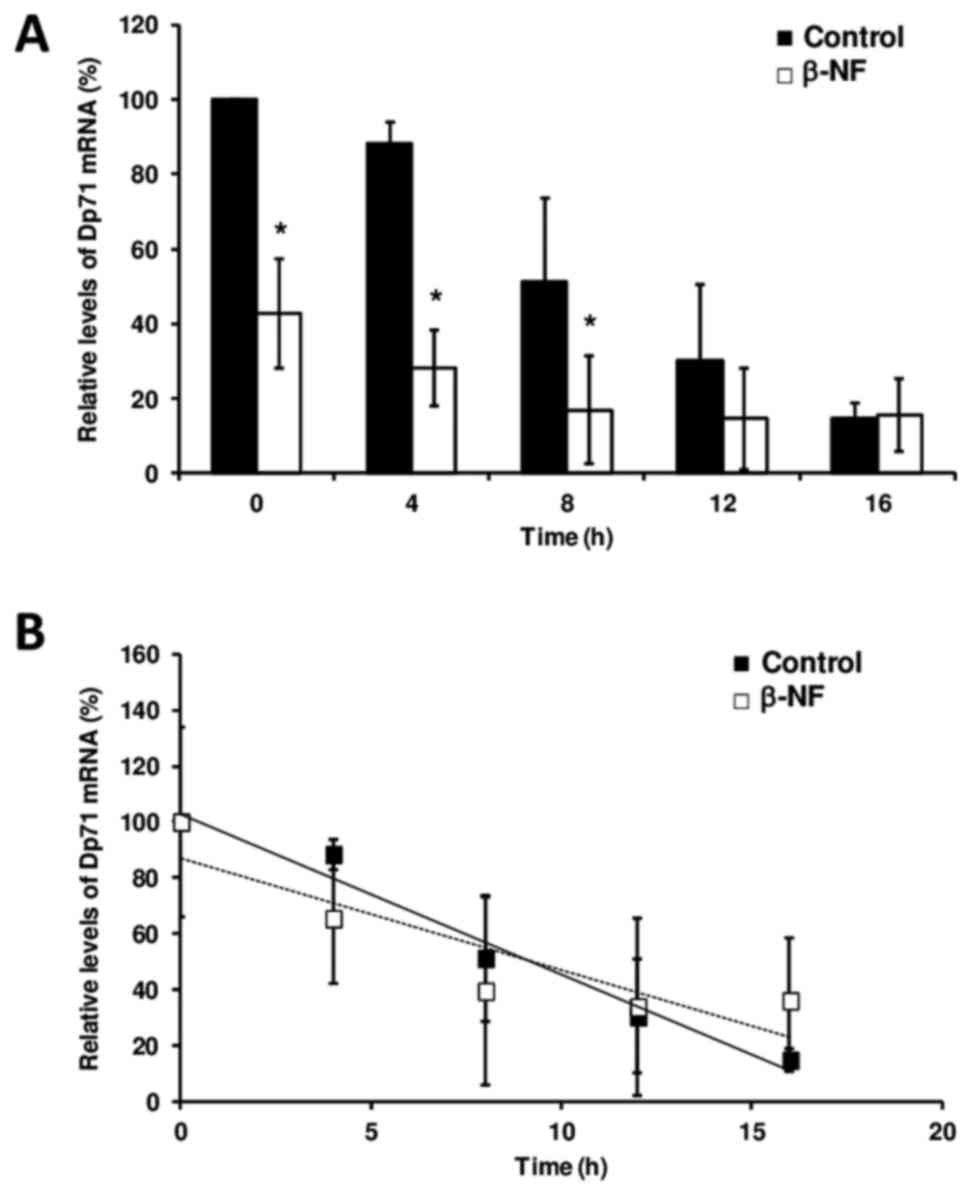
To ascertain whether β-NF affects mRNA stability,
Dp71 mRNA levels in Hepa-1 cells were measured by real-time
RT-qPCR. Our analysis confirms the transcriptional repression
exerted by β-NF in a 60% decrease in the mean Dp71 mRNA level
(P<0.05) that we previously observed, and it demonstrates that
this repression occurs in a time-dependent manner and was
maintained during transcription inhibition and subsequent mRNA
decay in response to actinomycin D treatment (Fig. 1A). However, β-NF did not alter the
mean half-life of Dp71 mRNA in hepatic cells compared to that in
untreated cells The Dp71 mRNA half-life in both DMSO-treated
(9.11±2.9 h) and β-NF-treated hepatic cells (9.36±1.6 h) determined
by our linear regression was not different (Fig. 1B). As expected, CYP1A1 expression
in β-NF-treated cells was increased compared to that in
DMSO-treated cells (data not shown).
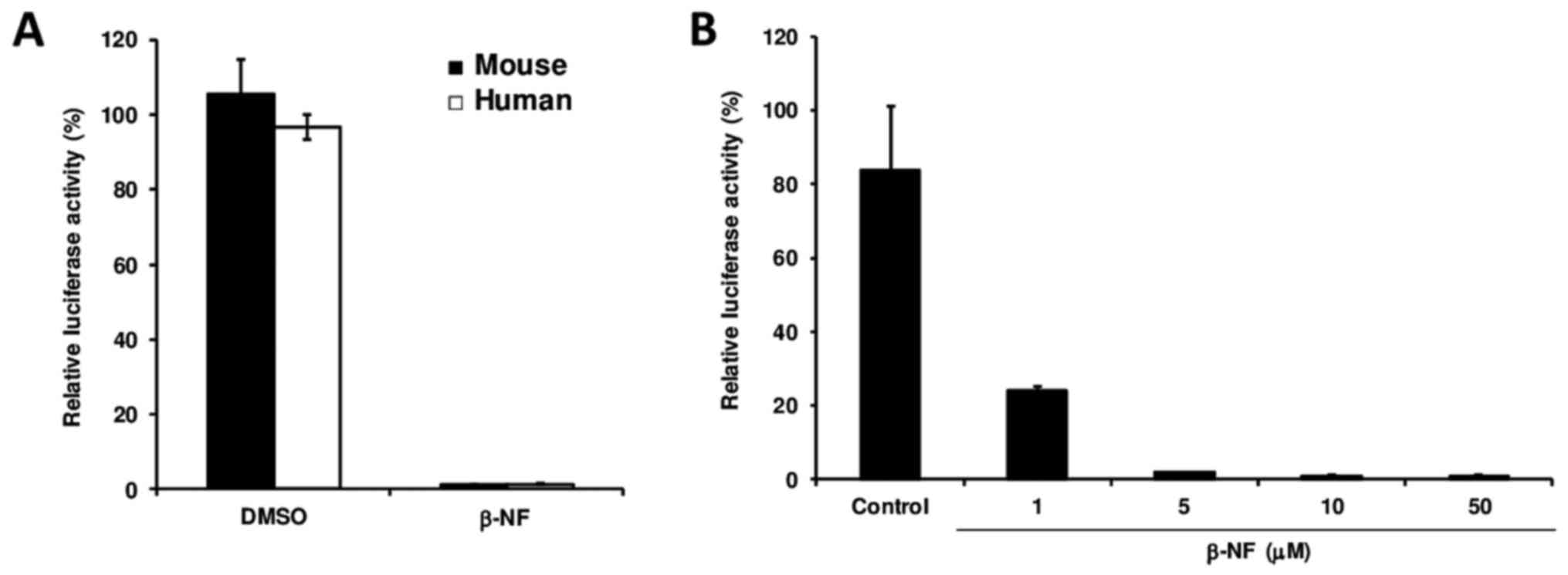
To determine whether the β-NF-induced downregulation
of Dp71 transcription occurs at the promoter level, mouse and human
hepatic cells were transfected with a vector carrying the Dp71
proximal promoter prior to β-NF treatment. Dp71 promoter full
repression in both cell lines upon β-NF treatment demonstrated that
this xenobiotic interferes with Dp71 promoter activity (Fig. 2A), and β-NF downregulates Dp71
expression in a dose-dependent manner via suppressing Dp71 promoter
activity rather than reducing Dp71 mRNA stability (Fig. 2B). Hepa-1 cells transfected with
pGL3 control vector (harboring CMV promoter) and exposed to β-NF
did not exhibit significant suppression (data not shown).
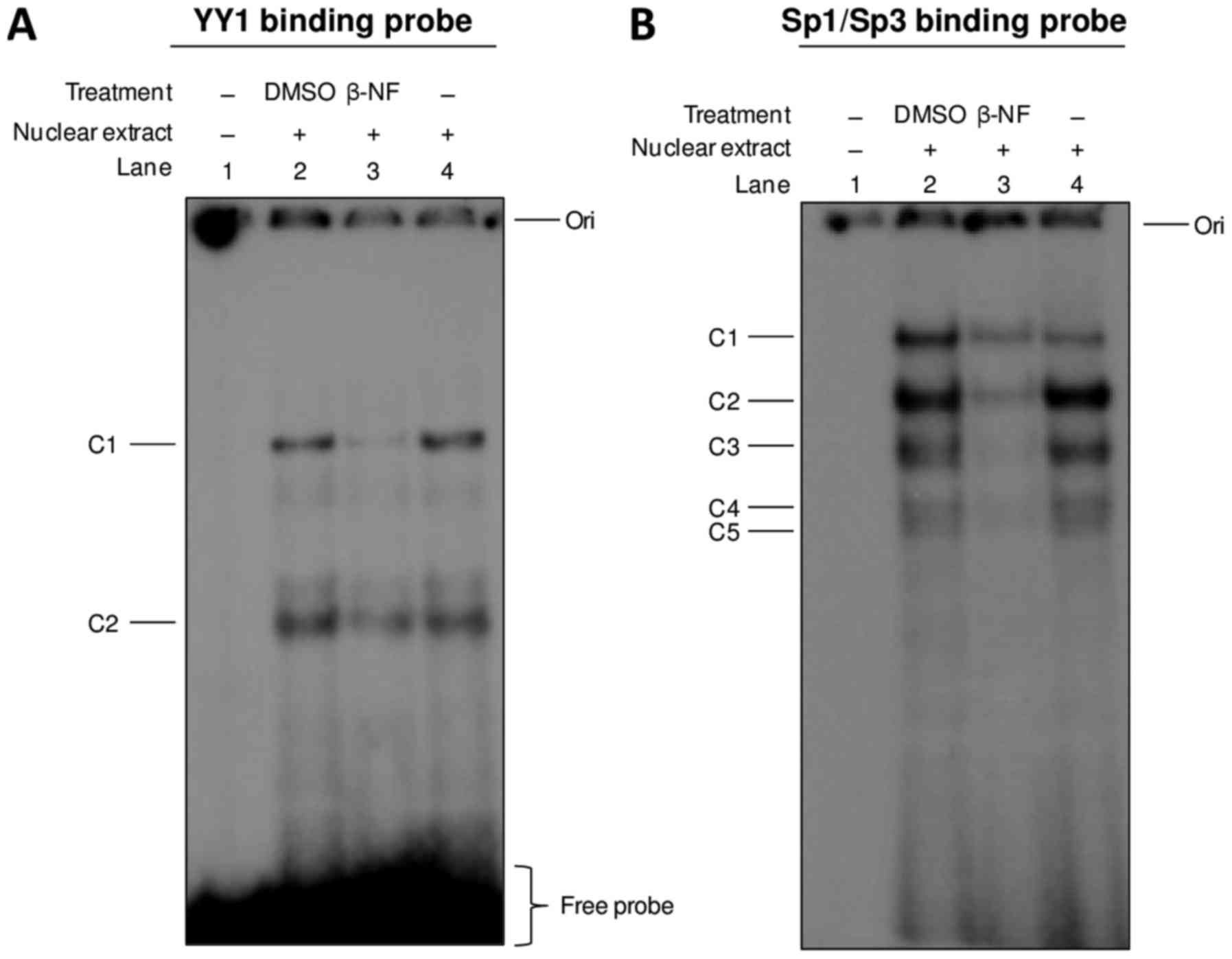
We previously demonstrated, by supershift assays,
the binding of YY1, Sp1, and Sp3 to Dp71 proximal promoter
(18); then we examined whether
β-NF alters the interaction of these transcription factors. Nuclear
extracts from DMSO-treated and β-NF-treated hepatic cells were
subjected to EMSA using YY1 and Sp1/Sp3 binding probes. As shown in
Fig. 3, β-NF reduced binding of
YY1 and Sp1/Sp3 to their respective DNA elements (lanes 3, Fig. 3A and B, respectively), which has
the clear implication that β-NF or its metabolites downregulate
Dp71 expression in hepatic cell by inhibiting binding of these
transcription factors to the Dp71 proximal promoter. By other hand,
the probe bearing the XRE element (22) did not form any specific complex
(data not shown).
Discussion
Dp71 is widely expressed in non-muscle tissues and
displays diverse functions in different tissues and cell types
(15), however the molecular
mechanisms underlying its expression remain poorly studied.
Previously, we demonstrated that Dp71 expression is negatively
regulated by the polyaromatic hydrocarbon β-NF as in vitro
as in vivo in hepatic cells (22). Moreover, we demonstrated different
DNA elements on Dp71 promoter that are crucial for Dp71 expression
in hepatic cells, including binding sites for YY1 and the Sp
family. The functionality of these DNA elements were confirmed by
EMSA, chromatin immunoprecipitation and site-directed mutagenesis
analysis (18). In the present
study, we explored the mechanisms underlying the repressive effect
of β-NF on Dp71 expression.
To ascertain whether β-NF affects mRNA stability,
Dp71 mRNA levels in Hepa-1 cells were measured by quantitative
real-time RT-qPCR. Our analysis confirms the transcriptional
repression exerted by β-NF that we previously observed (22) and demonstrates that this repression
occurs in a time-dependent manner. Despite this reduction in Dp71
expression, β-NF did not change the stability of the mRNA
transcript. The Dp71 mRNA half-life in both DMSO-treated and
β-NF-treated hepatic cells determined by our linear regression
analysis (9 h) is markedly lower than that measured in myogenic
cells (20 h) by Tennyson et al (26). This difference could be due to
differential transcriptional mechanisms operating in each cell
type.
We also determined whether the β-NF-induced
downregulation of Dp71 transcription occurs at the promoter level
by transfecting mouse and human hepatic cells with a vector
carrying the Dp71 proximal promoter prior to β-NF treatment. Our
data indicate that β-NF downregulates Dp71 expression in a
dose-dependent manner via suppressing Dp71 promoter activity rather
than reducing Dp71 mRNA stability. Furthermore, we observed this
β-NF-induced reduction of Dp71 promoter activity in both HepG2 and
Hepa-1 cell lines, indicating that this mechanism is conserved
between human and mouse hepatic cells.
In functional studies, we have previously shown that
mutations of YY1- and Sp-binding sites in Dp71 promoter
significantly reduced its activity, and because the binding of YY1,
Sp1, and Sp3 is relevant to Dp71 proximal promoter activity
(18), we examined whether β-NF
alters this interaction. β-NF remarkably decreased the binding of
YY1, Sp1, and Sp3 to the Dp71 proximal promoter, which implies that
β-NF and/or its metabolites may inhibit the expression of these
transcription factors. Alternatively, this xenobiotic may alter
post-translational modifications of these transcription factors,
such as glycosylation, phosphorylation, ubiquitination, or
acetylation, thereby reducing the affinity of these nuclear
proteins for their respective DNA elements (27,28).
Further studies are required to determine how β-NF modifies YY1,
Sp1, and Sp3 binding to the Dp71 promoter region.
Dp71 promoter sequence contains a single xenobiotic
response element (XRE) at the position −63/-59 (22). This kind of element is recognized
by the AhR/ARNT complex to regulate positively numerous genes
involved in cellular metabolism, detoxification process or
inflammatory process (29,30). Nevertheless, we failed to observe
interaction between of XRE and nuclear proteins from β-NF-treated
hepatic cells, which indicate that the repressive effect of β-NF on
Dp71 promoter activity is independent of the Aryl hydrocarbon
receptor.
In conclusion, our study demonstrates that
β-NF-induced repression of Dp71 expression in hepatic cells take
place at the promoter level, via inhibition of YY1, Sp1, and Sp3
binding to the Dp71 promoter. Further studies are warranted to
determine whether β-NF can alter the expression of other genes
regulated by these transcription factors.
Acknowledgements
The present study was supported by Consejo Nacional
de Ciencia y Tecnología (CONACyT)-Mexico (grant number 78764-M) for
MBL.
Glossary
Abbreviations
Abbreviations:
|
AhR
|
aryl hydrocarbon receptor
|
|
β-NF
|
β-naphthoflavone
|
|
DMD
|
Duchenne muscular dystrophy
|
|
DMSO
|
dimethylsulfoxide
|
|
Dp71
|
dystrophin Dp71
|
|
EMSA
|
Electrophoretic Mobility Shift
Assay
|
|
siRNA
|
small interfering RNA
|
|
Sp1
|
Stimulating factor 1
|
|
Sp3
|
stimulating protein 3
|
|
XRE
|
xenobiotic response element
|
|
YY1
|
Yin Yang 1
|
References
|
1
|
Ahn AH and Kunkel LM: The structural and
functional diversity of dystrophin. Nat Genet. 3:283–291. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Koenig M, Monaco AP and Kunkel LM: The
complete sequence of dystrophin predicts a rod-shaped cytoskeletal
protein. Cell. 53:219–228. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bar S, Barnea E, Levy Z, Neuman S, Yaffe D
and Nudel U: A novel product of the Duchenne muscular dystrophy
gene which greatly differs from the known isoforms in its structure
and tissue distribution. Biochem J. 272:557–560. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Byers TJ, Lidov HG and Kunkel LM: An
alternative dystrophin transcript specific to peripheral nerve. Nat
Genet. 4:77–81. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
D'Souza VN, Nguyen TM, Morris GE, Karges
W, Pillers DA and Ray PN: A novel dystrophin isoform is required
for normal retinal electrophysiology. Hum Mol Genet. 4:837–842.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lidov HG, Selig S and Kunkel LM: Dp140: A
novel 140 kDa CNS transcript from the dystrophin locus. Hum Mol
Genet. 4:329–335. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Greenberg DS, Schatz Y, Levy Z, Pizzo P,
Yaffe D and Nudel U: Reduced levels of dystrophin associated
proteins in the brains of mice deficient for Dp71. Hum Mol Genet.
5:1299–1303. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hugnot JP, Gilgenkrantz H, Vincent N,
Chafey P, Morris GE, Monaco AP, Berwald-Netter Y, Koulakoff A,
Kaplan JC, Kahn A, et al: Distal transcript of the dystrophin gene
initiated from an alternative first exon and encoding a 75-kDa
protein widely distributed in nonmuscle tissues. Proc Natl Acad Sci
USA. 89:pp. 7506–7510. 1992; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greenberg DS, Sunada Y, Campbell KP, Yaffe
D and Nudel U: Exogenous Dp71 restores the levels of dystrophin
associated proteins but does not alleviate muscle damage in mdx
mice. Nat Genet. 8:340–344. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cox GA, Sunada Y, Campbell KP and
Chamberlain JS: Dp71 can restore the dystrophin-associated
glycoprotein complex in muscle but fails to prevent dystrophy. Nat
Genet. 8:333–339. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sarig R, Mezger-Lallemand V, Gitelman I,
Davis C, Fuchs O, Yaffe D and Nudel U: Targeted inactivation of
Dp71, the major non-muscle product of the DMD gene: Differential
activity of the Dp71 promoter during development. Hum Mol Genet.
8:1–10. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lambert M, Chafey P, Hugnot JP, Koulakoff
A, Berwald-Netter Y, Billard C, Morris GE, Kahn A, Kaplan JC and
Gilgenkrantz H: Expression of the transcripts initiated in the 62nd
intron of the dystrophin gene. Neuromuscul Disord. 3:519–524. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Acosta R, Montanez C, Fuentes-Mera L,
Gonzalez E, Gómez P, Quintero-Mora L, Mornet D, Alvarez-Salas LM
and Cisneros B: Dystrophin Dp71 is required for neurite outgrowth
in PC12 cells. Exp Cell Res. 296:265–275. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Enríquez-Aragón JA, Cerna-Cortès J,
Bermúdez de León M, García-Sierra F, González E, Mornet D and
Cisneros B: Dystrophin Dp71 in PC12 cell adhesion. Neuroreport.
16:235–238. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tadayoni R, Rendon A, Soria-Jasso LE and
Cisneros B: Dystrophin Dp71: the smallest but multifunctional
product of the Duchenne muscular dystrophy gene. Mol Neurobiol.
45:43–60. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Villarreal-Silva M, Centeno-Cruz F,
Suàrez-Sànchez R, Garrido E and Cisneros B: Knockdown of dystrophin
Dp71 impairs PC12 cells cycle: Localization in the spindle and
cytokinesis structures implies a role for Dp71 in cell division.
PLoS One. 6:e235042011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de León MB, Montañez C, Gómez P,
Morales-Lázaro SL, Tapia-Ramírez V, Valadez-Graham V,
Recillas-Targa F, Yaffe D, Nudel U and Cisneros B: Dystrophin Dp71
expression is down-regulated during myogenesis: Role of Sp1 and Sp3
on the Dp71 promoter activity. J Biol Chem. 280:5290–5299. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peñuelas-Urquides K, Becerril-Esquivel C,
Mendoza-de-León LC, Silva-Ramírez B, Dávila-Velderrain J, Cisneros
B and de León MB: Transcription factors YY1, Sp1 and Sp3 modulate
dystrophin Dp71 gene expression in hepatic cells. Biochem J.
473:1967–1976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morales-Làzaro SL, Gonzàlez-Ramirez R,
Gòmez P, Tapia-Ramirez V, de León MB and Cisneros B: Induction of
dystrophin Dp71 expression during neuronal differentiation:
Opposite roles of Sp1 and AP2alpha in Dp71 promoter activity. J
Neurochem. 112:474–485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gerets HH, Tilmant K, Gerin B, Chanteux H,
Depelchin BO, Dhalluin S and Atienzar FA: Characterization of
primary human hepatocytes, HepG2 cells and HepaRG cells at the mRNA
level and CYP activity in response to inducers and their
predictivity for the detection of human hepatotoxins. Cell Biol
Toxicol. 28:69–87. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Volkov MS, Bolotina NA, Evteev VA and
Koblyakov VA: Ah-receptor-independent stimulation of hepatoma 27
culture cell proliferation by polycyclic aromatic hydrocarbons.
Biochemistry. 77:201–207. 2012.PubMed/NCBI
|
|
22
|
Bermúdez de Leòn M, Gómez P, Elizondo G,
Zatarain-Palacios R, García-Sierra F and Cisneros B:
Beta-naphthoflavone represses dystrophin Dp71 expression in hepatic
cells. Biochim Biophys Acta. 1759:152–158. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schreiber E, Matthias P, Müller MM and
Schaffner W: Rapid detection of octamer binding proteins with
‘mini-extracts’, prepared from a small number of cells. Nucleic
Acids Res. 17:64191989. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tennyson CN, Dally GY, Ray PN and Worton
RG: Expression of the dystrophin isoform Dp71 in differentiating
human fetal myogenic cultures. Hum Mol Genet. 5:1559–1566. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jokela TA, Makkonen KM, Oikari S, Kärnä R,
Koli E, Hart GW, Tammi RH, Carlberg C and Tammi MI: Cellular
content of UDP-N-acetylhexosamines controls hyaluronan synthase 2
expression and correlates with O-linked N-acetylglucosamine
modification of transcription factors YY1 and SP1. J Biol Chem.
286:33632–33640. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tan NY and Khachigian LM: Sp1
phosphorylation and its regulation of gene transcription. Mol Cell
Biol. 29:2483–2488. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chiba T, Chihara J and Furue M: Role of
the arylhydrocarbon receptor (AhR) in the pathology of asthma and
COPD. J Allergy. 2012:3723842012. View Article : Google Scholar
|
|
30
|
Nebert DW, Roe AL, Dieter MZ, Solis WA,
Yang Y and Dalton TP: Role of the aromatic hydrocarbon receptor and
[Ah] gene battery in the oxidative stress response, cell cycle
control, and apoptosis. Biochem Pharmacol. 59:65–85. 2000.
View Article : Google Scholar : PubMed/NCBI
|