Introduction
The term transcriptome encompasses the
complete set of RNA molecules applying to an entire organism or
specific cell type, making it more dynamic than the genome. It
comprises several types of transcripts which are responsible for
changes in gene expression. Two major classes of regulatory
non-coding RNAs (ncRNAs) which lack any transcriptive capability
include microRNAs and long non-coding RNASs (lncRNAs). They play a
significant role in diverse cellular processes from enabling normal
development to disease progression, as well as cell communication.
A valuable example of a biological carrier that can transport
abundant quantities of lncRNA is that of the exosome: A type of
extracellular vesicle involved in cell-to-cell communication and
disease transmission. Its role has been documented in both
physiological conditions and pathological changes such as cancer
development, where they are responsible for the regulation of
various processes including immunosuppression, proliferation and
induction of pre-metastatic niche formation. In this review, the
role of a number of exosomal lncRNA molecules will be
discussed.
Characteristics of exosomes
The first scientific report to examine the
possibility that intracellular structures could be associated with
the process of budding vesicles bearing receptors during
maturation, using ferritin-labelled proteins and anti-IgG, recorded
the externalization of transferrin receptors into the intercellular
space during sheep reticulocyte maturation (1). The term exosome was later
proposed in 1987 (2).
Exosomes are small spherical nanovesicles ranging
from 20 to 120 nm (3,4). They belong to the group of
membrane-derived vesicles known as extracellular vesicles (EVs),
together with oncosomes, ectosomes or apoptotic bodies (5). The classification of each vesicle to
its specific group depends on genesis, composition, size and
density (see Table I).
Extracellular vesicles can be secreted constitutively or as a
result of specific induction (6).
The origin of exosomes is linked to the creation of multivesicular
bodies (MVBs) in the endosomal pathway; MVBs are structures with a
lipid bilayer and a density of 1.12–1.19 g/Ml (7) and which display 5′ nucleotidase
activity (8).
 | Table I.Subdivision of extracellular vesicles
based on their physiological features [based on (5,14,75)]. |
Table I.
Subdivision of extracellular vesicles
based on their physiological features [based on (5,14,75)].
| Extracellular
vesicles | Size | Density | Origin of
vesicles |
|---|
| Exosomes | 20–120 nm | 1.12–1.19 g/ml | Multivesicular
body |
| Ectosomes | ~0.1–1 µm | ~1.16 g/ml | Cell membrane |
| Apoptotic
bodies | 0.05–5 µm | 1.24–1.28 g/ml | Cell membrane |
| Oncosomes | 1–10 µm | Not specified | Cell membrane |
Exosomes are released from many types of healthy or
tumour cells, although more abundantly in the latter (9), and can be found in the tumour
microenvironment (TME) and blood samples of cancer patients
(10). Physiologically, exosomes
can be found in urine (10), sperm
(3), amniotic fluid (9) and various other biological fluids.
The role of exosomes in metastasis, angiogenesis and
epithelial-to-mesenchymal transition (EMT) has been confirmed in
cancer development by various studies (9). Exosomes released by tumour cells were
shown to constitute a leading source of antigens for dendritic
cells inducing antitumour immune responses (11). The physiological functions of
exosomes include cellular communication, immunity, infections,
elimination of waste products and signalling (12). There is also evidence of their
participation in aging-related diseases i.e. neurodegeneration when
different types of mRNAs, miRNAs or lncRNA change their abundance
inside nanovesicles (13).
Biogenesis
Exosome secretions and their molecular mechanisms
are rather obscure, since their release is regulated by several
signal molecules and various processes including ceramide synthesis
and Ca2+signalling (3).
Proteins that interact with the cell membrane and perform lipid
bilayer deformation are epsin, amphiphysin, endofilin, syndapin and
dynamin (4). The biogenesis
follows the following sequence: The process begins with membrane
invagination and exosome release to cytoplasm, followed by early
endosome (EE) formation, and then the formation of intraluminal
vesicles (ILVs) inside the late endosome (LE).
Under both normal and pathological conditions,
exosomes have an endocytic origin and are released by different
types of cells. Although exosomes typically mimic the composition
of the parent cells, they may contain a few signature proteins.
Following the formation of exosomes within the endocytic pathway,
they are released from the plasma membrane via MVBs from early
endosome maturation and may further fuse with lysosomes or the
plasma membrane. The most common and well-known mechanism of cargo
sorting is based on the action of proteins from Endosomal Sorting
Complex Required for Transport (ESCRT) (6). When this pathway is inactivated,
there is a possibility to recruit sumoylated protein hnRNPA2B1 [it
binds microRNA (14)] or maintain
the signal through tetraspanin-enriched microdomains (TEMs)
composed of transmembrane glycoproteins (8,15).
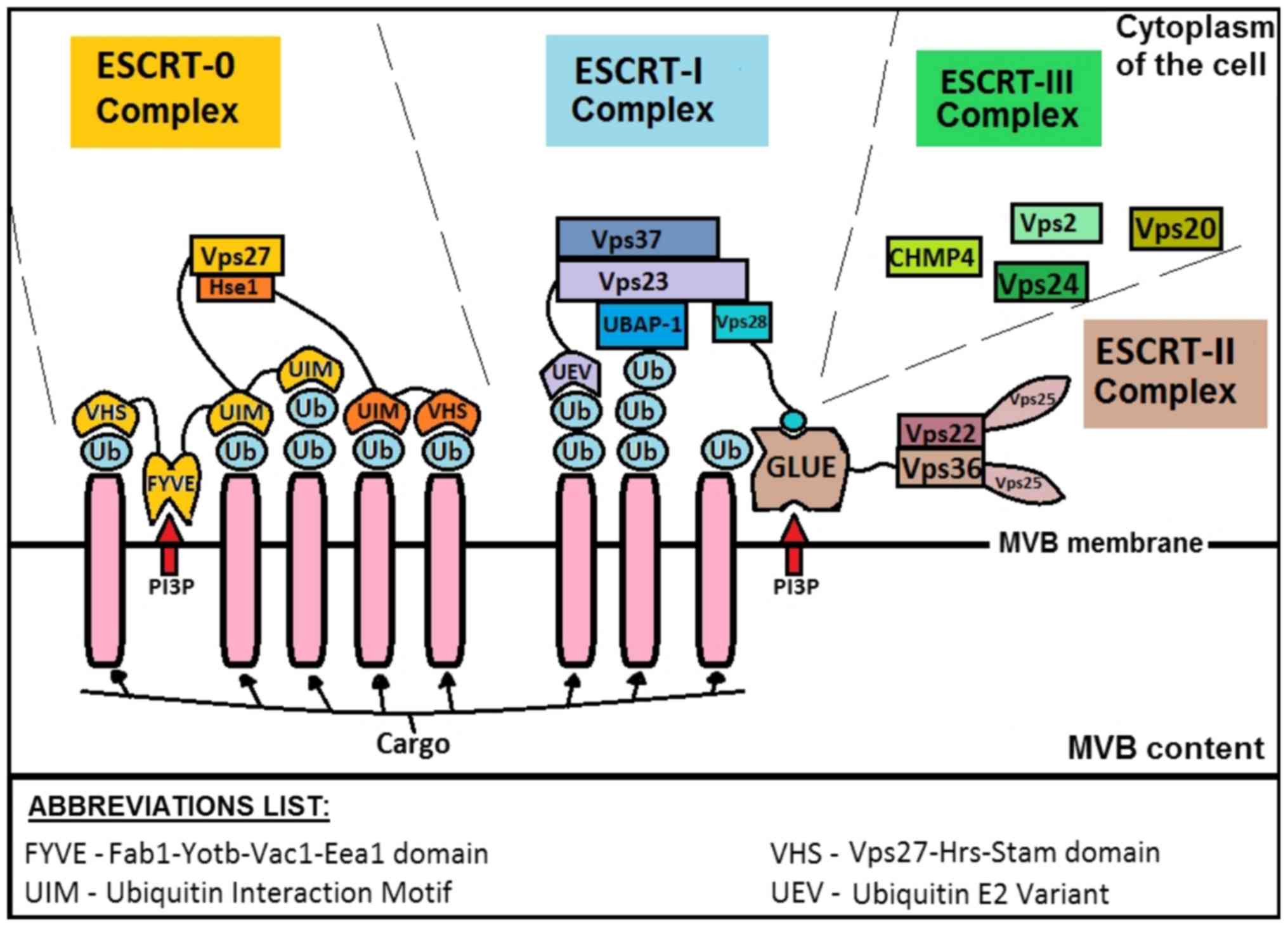
The schematic of ESCRT complex based on yeast, is shown in Fig. 1.
The most important proteins associated with ESCRT
are: ALIX (ALG-2-interacting protein X), syndecan and syntenin
(16). Syntenin ends the formation
process through syndecan sorting and binding to ALIX, which affects
ESCRT-III (4). Direct
ALIX-syntenin interaction is possible through LYPX(n)L motifs
(16), which are observed in the
viral proteins found in late assembly domains (L-domains), these
being responsible for recruiting the host ESCRT machinery directly
or via ESCRT-associated proteins. The final product of this ESCRT
and auxiliary protein activity is the creation of an MVB containing
exosomes.
Despite general syntenin-syndecan-ALIX axis, there
are some other molecular individuals involved in controlling ESCRT.
Examples are ADP-ribosylation factor 6 (ARF6) GTPase and
Phospholipase D2 (PLD2) enzyme which are considered as proteins
associated with syntenin (17) and
are responsible for the control of the budding of ILVs into MVBs
(18). LIP5 (LYST-interacting
protein 5), also known as vesicle trafficking 1 protein (VTA1), is
a cofactor for Vps4 ATPase that enhances Vps4-mediated disassembly
of ESCRT-III polymer (19). Cep55
(centrosomal protein 55) can mediate ALIX and ESCRT-I complex
recruitment; however, interactions with the components of ESCRT-II
and ESCRT-III have also been observed (20). Further examples consist of CD2AP
(CD2-associated protein), IQGAP1 (IQ-motif-containing
GTPase-activating protein 1) or ROCK1 (Rho-associated kinase 1)
which are thought to interact with ALIX or TSG-101 (Tumor
susceptibility gene 101). They participate in processes such as
cytokinesis (20), viral budding
(21) and cell polarity (22), respectively. Finally, enzyme
heparanase, able to internally cleave heparan sulfate chains on
syndecans, acts by increasing exosome production by stimulating
intraluminal budding of syndecan and syntenin (23).
After the ‘mature’ multivesicular body is formed, it
can be transported to lysosomes (degraded hydrolytic enzymes) or
interact with the lipid membrane, resulting in exosome release
outside the cell. In the latter case, the attachment of MVB to the
lipid bilayer takes place with the help of Rab-GTP and SNARE family
proteins. Examples of these groups are given in Table II.
 | Table II.Selected proteins involved in fusion
of MVB with cell membrane and in exosome secretion [based on
(3,4,7,87)]. |
Table II.
Selected proteins involved in fusion
of MVB with cell membrane and in exosome secretion [based on
(3,4,7,87)].
| Protein | Proteins
family | Function |
|---|
| Rab7; Rab11 | RabGTP | Promotes proper
protein attachment in pathways dependent on calcium ions |
| Rab27 (a, b) | RabGTP | Regulates the
stages of exosome secretion, controls the location of MVB |
| Rab35 | RabGTP | Regulates exosomes
secretion by interacting with GTP-activating protein-
TBC1(Tre-2/Bub2/Cdc16) |
| RAL-1 | RasGTP | Recruiting the
Syx-5 protein, thereby mediating in fusion of membranes |
| Syx-5
(Syntaxin5) | SNARE | Stimulates fusion
of MVB with cell membrane |
| VAMP7 | SNARE | Necessary for MVB
fusion with cell membrane (Vesicle-Associated Membrane
Protein) |
| YKT6 | SNARE | Mediates the
release of exosomes from the cell |
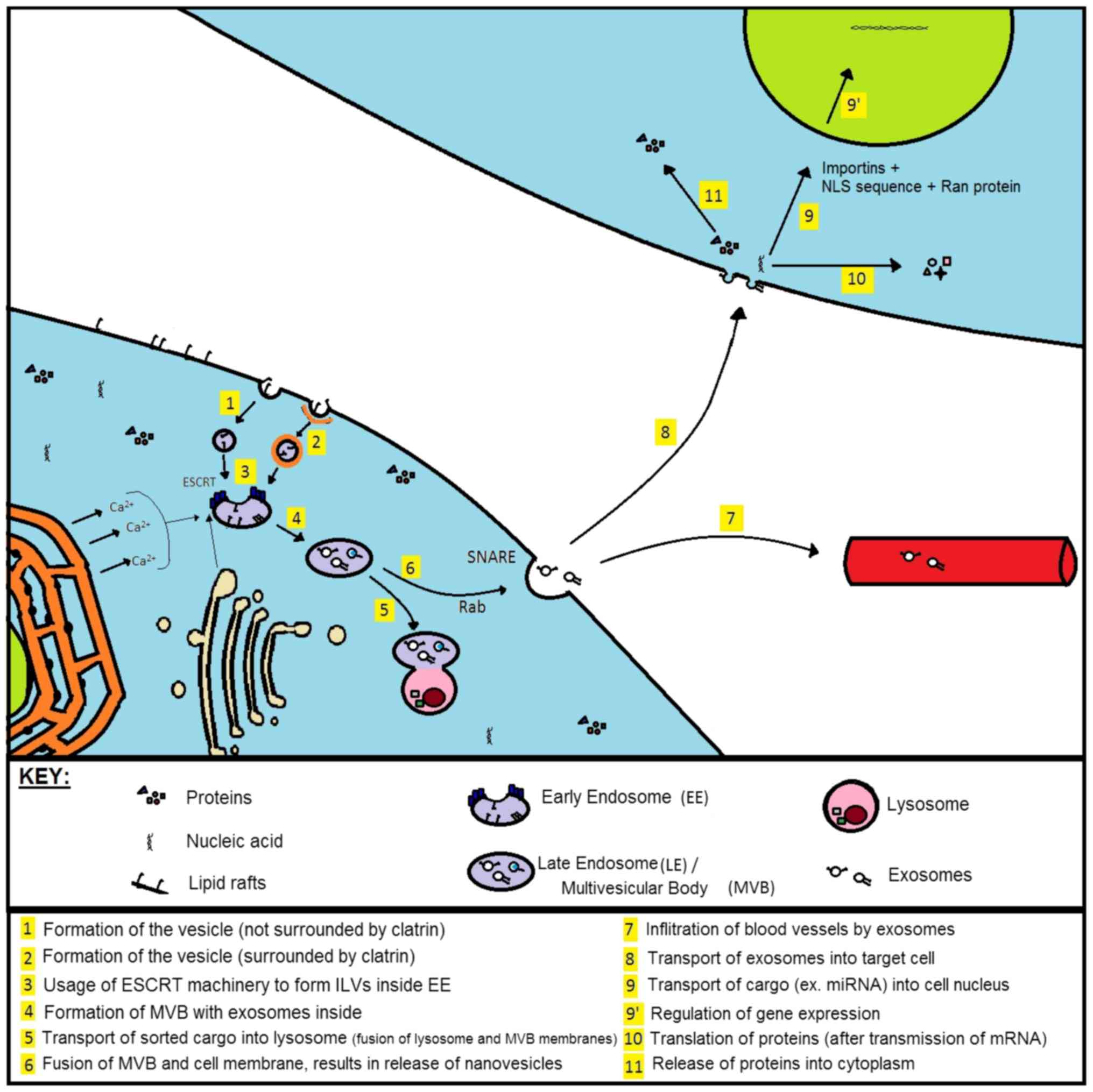
Following the fusion of MVB and cell membrane, the
exosomes can be transported to distant areas of the organism (via
blood vessels) or interact with neighbouring cells (4). ‘Targeted’ cells can internalize
nanovesicles in endocytic way where the absorption itself is
facilitated through phagocytosis, fusion with cell membrane or
ligand-receptor interaction. Fig.
2 presents the process of biogenesis, secretion and absorption
of exosomes by means of ESCRT machinery.
Molecular content
Nanovesicle formation exhibits some characteristic
stages of biogenesis and involves specific structures present in
the cell. The selective mechanism of transporting cargo to the
exosome interior is a more heterogenous process, as evidenced by
their varied content, even in different types and stages of cancer
development (8). The composition
of exosomes contains countless types of proteins, including
cytokines and growth factors, lipids or nucleic acids (3), including characteristic biomarkers
such as CD63, CD81, CD9, TSG-101, ALIX or HSP70 which allow direct
identification of exosomes (6).
Proteins
The profile of the proteins within the vesicles
consists of cytoplasmic proteins, cell membrane proteins or
proteins from endosomes; however, no proteins from organelles have
been detected (6). These proteins
include a number of exosomal molecules, such as those connected
with MVB biogenesis (e.g., ALIX, TSG-101) (6), ESCRT components (3), transport and fusion proteins
(annexin, Rab-GTP, flotillin) and heat shock cognate (HSC). Other
important ones include integrins, tetraspanins or subunits of G
proteins. The tumour exosomes (TEXs) include additional molecules
that participate in the EMT process including matrix
metalloproteinases (MMPs), IL-6, TGF-β and immunosupression factors
(FasL, TRAIL and GAL-9 that stimulates T cell apoptosis) (8). Some of them are presented in Table III.
 | Table III.Examples of exosomal molecules
involved in tumour progression, immunosuppression and apoptosis
[based on (3,4,6,10,88,89)]. |
Table III.
Examples of exosomal molecules
involved in tumour progression, immunosuppression and apoptosis
[based on (3,4,6,10,88,89)].
| Type of molecule in
exosome | Function | Type of cells |
|---|
| ITG α6β4; ITG
α6β1 | Connected with
metastasis to the lungs, promotes TEXs adhesion in the lungs | Lung cancer
cells |
| ITG αVβ5 | Connected with
metastasis to the liver, binding to Kupffer cells | Liver cancer
cells |
| TGF-β; IL-10; MCP-1
(Monocyte Chemoattractant Protein-1) | Promote cell
migration | Lung cancer cells,
melanoma cells |
| MHC-II (Major
Histocompatibility Complex-II) | Stimulation of
CD4+ cells | B lymphocytes, DC
cells |
| OVA
(Ovalbumin) EGFR (Epidermal Growth Factor
Receptor); | Inhibition of
immune response | Melanoma cells |
| KRAS (Kirsten
RAt Sarcoma viral oncogene homolog) | Proliferation,
resistance to treatment | Lung cancer
cells |
| EGFRvIII (EGFR
variant III) | Anti-apoptotic
abilities | Glioma cells |
| MDR-1
(Multi-Drug Resistance) | Resistance for
drug | Prostate cancer
cells |
| HER2 (Human
Epidermal growth factor Receptor 2) | Resistance to
treatment with Trastuzumab | Breast cancer
cells |
Lipids
The lipid composition of exosomes is distinct from
that of the cell membrane: it is rich in components such as
cholesterol, sphingomyelin, glycerophospholipids,
phosphatydylinositol, ceramide or phosphatidilserine, as well as
prostaglandins and leukotrienes (6). The lipid content in exosomes is
higher than that of the parent cell except phosphatidylcholine
which is ordinarily decreased (24). An example of exosomal lipid
activity is the differentiation of bone-marrow myeloid cells into
myeloid-derived suppressor cells (MDSCs) by prostaglandin E2. The
accumulation of MDSCs leads to immunosuppression, which provides
favourable conditions for metastasis (8). The participation of lipids in the
biogenesis of exosomes is summarized below (Table IV).
 | Table IV.The examples of lipids involved in
formation and secretion of exosomes [based on (9)]. |
Table IV.
The examples of lipids involved in
formation and secretion of exosomes [based on (9)].
| Lipid | Function |
|---|
|
Phosphatidylserine | Participation in
microautophagy connected with HSC70 protein (process of exosomes
synthesis) |
| LysoBisPhosphAtidic
(LBPA) | Change in membrane
dynamics (in collaboration with ALIX) |
|
BisMonoacylglycerolPhosphate (BMP) | Binding to ALIX
protein; (BMP formed by PhosphoLipase D2) |
| Ceramide | Involved in the
biosynthesis of exosomes and packaging miRNA into them (in a
process independent of the ESCRT machinery) |
| Cholesterol | Participation in
secretion of FLOT2-positive exosomes |
| Lipid rafts
(cholesterol-rich sphingolipid microdomains) | Participation in
the biogenesis of exosomes inside MSCs (Mesenchymal Stem
Cells) |
| Phosphatidylcholine
transporter ABCA3 | Participation in
the biogenesis of exosomes derived from B-cell lymphoma |
Nucleic acids
Exosomes also can include different types of RNA or
DNA such as mRNA, miRNA (6),
lncRNA, dsDNA, RNA-ROR (regulator of reprogramming) (3), and therefore play a vital role in
regulating signaling pathways and gene expression (refer to
Table V). These DNA and RNA
molecules may be transferred into recipient cell by the fusion of
exosome and cell membranes, aiding the regulation of large numbers
of signaling pathways or expression of genes (3). Many studies indicate elevated levels
of lncRNA in extracellular exosomes compared to parent cells
(25,26). This can be attributed to the action
of nucleotide patterns that are present in transcripts as
cis-acting elements responsible for targeting them into exosomes
(27).
 | Table V.The examples of nucleic acids that
occur in exosomes [based on (3,6,90)]. |
Table V.
The examples of nucleic acids that
occur in exosomes [based on (3,6,90)].
| Nucleic acid | Length in base
pairs (bp) | Function |
|---|
| mRNA | ~3,250 bp (for
TMPRSS2 in prostate cancer, TransMembrane PRotease Serine 2) | Participation in
metastasis, progression of cancer |
| lncRNA | More than 200
bp | Regulation of gene
expression at transcriptional, posttranscriptional and epigenetic
levels |
| dsDNA | More than 2,500
bp | Participation in
neoplasm (more frequent occurrences in tumour exosomes than
exosomes from normal cells) |
| RNA-ROR | ~2,600 bp | Resistance to
chemotherapy in HepatoCellular Carcinoma, promoting cancerogenesis
through specific histone methylation |
The purpose of this review is to highlight the
significance of lncRNA as a pivotal molecule inside exosomes.
Therefore, it will summarize the role of various exosomal lncRNAs,
together with their function in carcinogenesis and in other
biological processes.
lncRNA
The functional annotation of 60,770 full length
longer cDNAs and rare transcripts in the mouse transcriptome
resulted in the discovery of a novel class of non-coding cDNAs,
among which ~80% of the transcripts are not spliced (28). The lncRNAs constitute a class of
transcripts that does not encode proteins and whose length exceeds
200 bp (29). This group includes
several heterogenous molecules that are difficult to categorize:
LncRNAs can be divided into long intergenic non-coding RNAs
(lincRNAs), long intronic non-coding RNAs and half-STAU1-binding
site RNAs (1/2sbsRNAs) (30).
Depending on the DNA coding strand, the lncRNAs can be classified
as sense or antisense transcripts (31).
Some resemblance can be found between mRNAs and
lncRNAs in some biochemical processes, for example mechanisms of
polyadenylation and splicing or participation of RNA polymerase II
in the process of their transcription (32). The roles of these molecules are
connected with transduction signal, decoys, guides or scaffolds
(31); however, their main
function is associated with their action as transcription
regulators of protein-coding genes (32). There is also the probability that
lncRNAs act like a ‘sponge’-leading to miRNA recruitment into EVs,
or possibly to provide the mechanisms required for microRNA loading
(13).
These findings confirm both the diversity of lncRNAs
and their participation in carcinogenesis by demonstrating their
influence on the promotion of angiogenesis, protein stability or
miRNA inhibition by acting as a sponge. In addition, as lncRNAs are
detected in TEXs or apoptotic bodies, and have high stability
during circulation, they could be considered as potential cancer
biomarkers (33); however, some
studies indicate they may display tumour suppression behavior
through the blocking of the transcription of apoptosis-inhibiting
genes and stimulation of p53 expression (34) or the structural modification of
chromatin (35).
lncRNAs as protumorigenic
molecules
Numerous studies implicate lncRNAs as
cancer-associated molecules that are enriched in exosomes. A
classic feature of the tumour microenvironment is hypoxia, which
has a notable impact on cancer progression. Activation of its
pathway via the transcription factor hypoxia inducible factor (HIF)
plays a part in metastasis and a more aggressive phenotype. The
role of lncRNAs in hypoxia-driven cancer progression came into view
recently, where these hypoxia-responsive lncRNAs play a critical
part in regulating hypoxic gene expressions (36). The next subsections will present
the characteristics of the most frequent types.
HOTAIR: HOX Transcript Antisense RNA has been
associated with progression and poor prognosis in patients with
squamous cell carcinoma (SCC), hepatocellular carcinoma (HCC),
urothelial bladder cancer (UBC) or colorectal cancer (CRC)
(37). Yan et al (38) report the involvement of HOTAIR in
the repression of WIF-1-a protein implicated in the Wnt signaling
pathway. Moreover, epigenetic regulation is also achievable: It
participates in the migration, invasion or proliferation,
potentiating through silencing of miR-205 in bladder cancer. In
addition, UBC cells displayed increased invasion with migration
corresponding to regulation of the EMT pathway, which was confirmed
by knockdown of HOTAIR. This resulted in reduced invasiveness by
influencing genes connected with EMT such as MMP1 (Matrix
metalloproteinase-1), ZEB1 (Zinc finger E-box-binding
homeobox 1), TWIST1 (Twist-related protein 1), SNAI1
(Snail family zinc finger 1), ZO-1 (Zonula occludens 1),
LAMC2 (Laminin, gamma 2), LAMB3 (Laminin, beta 3) or
ABL2 (ABL proto-oncogene 2) (37). Studies by Xie et al, have
shown that salivary HOTAIR could be used as biomarker in
diagnostics: it had good discriminatory power for differentiating
pancreatic cancer from healthy patients and benign pancreatic
tumour (39, 40). In addition to its contribution in cancerous
diseases, HOTAIR has also been described to play a role in
rheumatoid arthritis, where its transcript activates MMP-2 and
MMP-13 in synoviocytes and osteoclasts. This could promote joint
and cartilage matrix cessation advancing joint destruction.
Furthermore, HOTAIR-loaded exosomes with enriched lncRNA activate
macrophages and influence immune response (41).
MALAT-1: Metastasis-Associated Lung
Adenocarcinoma Transcript 1 or Nuclear-Enriched Abundant Transcript
2 (NEAT-2) was the first lncRNA to be associated with metastasis in
Non-Small Cell Lung Cancer (NSCLC). This transcript, similar to the
HOTAIR lncRNA, is also connected with migration or tumour growth
induction. Zhang et al (42), indicated increased expression of
transcript compared to healthy controls, and an association with
higher TNM stage in patients with cancer. In vitro study
knockdown of MALAT-1 in cancer cell lines significantly inhibited
cell proliferation and colony formation, although it also induced
cell cycle arrest and cell apoptosis due to a decrease in S phase
progression. In addition, cyclin D1 and cyclin D2 downregulation
was also observed, suggesting that MALAT-1 was involved in
acceleration of the cell cycle. This indicates that MALAT-1 can be
an important therapeutic target when developing treatments for
NSCLC patients, because of its participation in the malignant
phenotype of cancer through gene expression regulation (43). Alongside NSCLC, MALAT-1 has been
proposed as a mediator of angiogenesis in Thyroid Cancer. Huang
et al (44), found it to
play a role in the secretion of the fibroblast growth factor 2
(FGF) protein from tumour-associated macrophages (TAMs), which led
to vascularization, promotion of proliferation, invasion and
migration of cancer cells. Nonetheless, MALAT-1 participation in
releasing inflammatory cytokines or FGF2 secretion was
predominantly blocked by overexpression of FGF2.
ZFAS1: Initial studies concerning the Zinc
Finger AntiSense 1 (ZFAS1) lncRNA refer to alveolar development in
the mammary gland, with a proposed role in the regulation of
alveolar development and epithelial cell differentiation in the
mammary gland (45). The
transcript has been found to promote cancer progression by cell
cycle regulation through cyclin-dependent kinase 1 (CDK1)
interaction or destabilization of the p53 protein. The level of
ZFAS1 was also upregulated in exosomes from HCC, CRC and gastric
cancer (GC). Pan et al (46), noticed that overexpression of this
transcript leads to proliferation and migration of GC cells and
ZFAS1-positive exosomes which can be internalized by MKN-28 cells.
Knockdown of ZFAS1 inhibited the migration, proliferation and EMT
process; however, it also induced cell cycle arrest and programmed
cell death, serving as a potential tumour suppressor. These
findings have been attributed to decreased ERK, Bcl-2 and increased
Bax protein levels. Additionally, ZFAS1 knockdown was observed to
suppress cell migration by decreasing the expression of EMT
transcription factors such as Slug or Twist. These findings suggest
that ZFAS1 could promote metastasis by cell cycle acceleration or
support EMT playing a pivotal role in GC diagnosis (47).
H19: H19 lncRNA is also involved in tumor
metastasis and has been described as a master factor in cancer
biology due to its frequent deregulation in almost all tumours,
both in the initiation and progression steps (48). H19 is connected with the
downregulation of E-Cadherin, which influences cell adhesion and
leads to spread of cancer cells. In addition, Conigliaro et
al (49) investigated H19 as a
pro-angiogenic molecule promoting cell-to-cell adhesion (CD90+
cells with endothelial cell monolayer) and association with
increased expression of VEGF and ICAM1 transcripts which in turn
leads to new vessel development. H19 may also act as an epigenetic
modulator or as miRNA sponge (50). In conclusion, this type of lncRNA
could have a range of impacts on target cells and some of its
functions still remain unknown; nevertheless, H19 has been proposed
as a potential therapeutic target (e.g., in HCC) (51).
POU3F3: In vitro studies indicate that this
transcript increases viability and cell proliferation in gliomas
and its overexpression is associated with tumour grading (52). Hu et al (53) investigated whether exosomal POU3F3
from glioma cells could promote angiogenesis and its effect on
endothelial cells. They indicate that lncRNA enriched the exosomes
regulating the migration, proliferation and angiogenesis of Human
Brain Microvascular Endothelial Cells (HBMECs) in both in
vitro and in vivo studies. Silencing POU3F3 using shRNA
decreased the migratory ability of endothelial cells but did not
affect their motility. POU3F3 overexpression in HBMECs caused by
lncRNA enriched exosome internalization, and significantly
upregulated the transcript level of bFGF, bFGFR, Angio and VEGFA.
These findings indicate that POU3F3 lncRNA can be an essential
molecule in cell-to-cell communication or a regulator of
angiogenesis in glioma cells (53).
TUC339: One of lncRNA transcribed from
ultraconserved elements (UCEs) is TUC339. UCEs are 100% maintained
genomic sequences that are greater than 200 bases in length and are
conserved across human, mouse and rat genomes (54). The localization of these elements
is connected with fragile sites and cancer-associated regions of
the genome. Overexpression of this type of lncRNA has been reported
in HCC (55). A study on the role
of exosomal ultraconserved lncRNA in modulating target cell
phenotype as a result of genomic changes found that TUC339
upregulation in HCC cell lines resulted in an enhancement of cell
growth and a reduction of adhesion to cell-extracellular matrix
(ECM). The knockdown of TUC339 decreased proliferation and altered
the expression of a total of 843 genes, 469 of which were inhibited
(56).
ROR: LncRNA ROR participates in liver cancer
through disruption in TGF-β pathway. Although TGF-β has been
related to cancer chemoresistance, the molecular actions involved
in this event remain unknown. Takahashi et al (57), identified a possibly mechanism by
which lncRNA may mediate chemoresistance dependent on TGF-β. They
indicate that TGF-β reduced the sensitivity of HCC cells to
sorafenib or doxorubicin, with a further reduction of
sorafenib-dependent caspase 3/7 activity, leading to inhibition of
the apoptosis process. Moreover, the release of extracellular
vesicles and packaging of lncRNA within vesicles were also
modulated by TGF-β. One of the selectively enriched exosomal
lncRNAs thought to be a potential mediator of chemoresistance is
RNA-ROR. Using a molecular pathway connected with p53 dependent
signaling, lincRNA can contribute to tumor development by
inhibiting apoptosis through the repression of p53. Additionally,
knockdown of this lncRNA results in increased expression of Caspase
8 and GADD45B, leading to apoptosis induction and growth arrest.
These data suggest that lincRNA ROR can be an important mediator in
cell-to-cell communication and its contribution in chemoresistance
has been confirmed in research. Inhibition of this transcript
resulted in reversion of drug resistance, leading to cell death
(58).
VLDLR: One of the long-intergenic non-coding
RNA transcripts that support HCC development is called VLDLR
(59). By showing that knockdown
of this RNA can result in cell cycle arrest in G1/S phase,
Takahashi et al (60),
confirmed that it regulates HCC cell proliferation. Moreover, the
quantity of lncRNA was increased in both HCC cells and
extracellular vesicles following chemotherapeutic stress caused by
sorafenib, doxorubicin and camptothecin, thus enhancing cell
viability. Furthermore, the extracellular transfer of VLDLR to
adjacent tumour cells could participate in chemoresistance through
the modulation of ABCG2 gene expression.
The summary of the other lncRNA molecules that
participate in tumour development is presented in Table VI.
 | Table VI.Functions of chosen lncRNA molecules
in numerous tumour types [based on (91–102)]. |
Table VI.
Functions of chosen lncRNA molecules
in numerous tumour types [based on (91–102)].
| lncRNA | Function | Tumor type | (Refs.) |
|---|
| ARSR | Resistance to
sunitinib; overexpression leads to poor response of renal cancer
patients; promoting c-MET expression | Renal cell
carcinoma (RCC) | (91) |
| MEG3 | Cell cycle arrest;
decreases apoptosis by regulation of miR-21 | Cervical cancer
(CC) | (92,93) |
| RMRP | Acts as a miR-206
sponge and modulate the cell cycle by regulating the Cyclin D2
expression | Gastric cancer
(GC) | (94) |
| NEAT1 | Promotes cancer
progression through regulating CDK6 mediated by influence on
miR-107 | Laryngeal squamous
Cell cancer (LSCC) | (95) |
| UCA1 | Enhances cell
proliferation, migration, invasion, EMT process | Bladder cancer
(BC) | (96) |
| HULC | Induces
angiogenesis and modulates VEGF expression; upregulates sphingosine
kinase 1 (SPHK1) by miR-107/E2F1 pathway | Liver cancer
(LC) | (97,98) |
| TUG1 | Promotes
radioresistance and EMT transition; enhance proliferation and
migration | Bladder cancer (BC)
Pancreatic cancer (PC) | (99,100) |
| HOTTIP | Increases the
chemoresistance by Wnt/β-catenin pathway activation | Osteosarcoma | (101) |
| CCAT2 | Upregulates VEGFA
and TGFβ, promotes angiogenesis, decreases apoptosis by elevated
Bcl-2 expression and inhibition of Bax and caspase-3 | Glioma cancer | (102) |
Suppressor function of lncRNA
In contrast to the lncRNAs with pro-cancerous
properties, there is also another group of transcripts that could
support normal cells rather than stimulating their conversion into
a neoplastic phenotype (61).
GAS5: Several studies have implied that the
Growth Arrest Specific 5 gene molecule has not been connected with
cancer progression, as its expression has been observed to be
reduced in various cancer types (62,63).
Pickard et al (64),
noticed that GAS5 participates in apoptosis regulation in prostate
cancer cells. They indicated that the high expression of GAS5 level
reduces tumour cell survival and hence induces pro-apoptotic
activity. On the other hand, when GAS5 was downregulated,
programmed cell death was diminished but growth arrest was not. In
prostate carcinoma GAS5 has been found to bind to the receptor
domain and inhibit transcriptional stimulation, leading to
increased death of tumour cells (65).
PARTICL: Some studies also confirmed the role
of lncRNAs in the methylation process in response to radiation. One
of this lncRNA is PARTICL (promoter of MAT2A antisense
radiation-induced circulating long non-coding RNA) a trans-acting
mediator of DNA, and histone lysine methylation, a clever component
for gene silencing (66). The
findings indicate that in low dose irradiation, PARTICL can enhance
EZH2 (enhancer of zeste homolog 2) expression, which in turn
catalyzes the addition of methyl groups to histone H3 at lysine 27.
Moreover, there was also an observed increase in the activity of
the DNMT1 enzyme (DNA methyltransferase 1), global methylome
enhancement methylome regulation e.g., CpG island methylation of
WWOX gene. Finally, speculation about PARTICL as an
epigenetic modifying platform targeting chromatin structure
modulation through recruitment of PRC2 (Polycomb Repressive Complex
2) has been proposed; this has been proposed to act by the binding
of lncRNA to a subunit of PRC2 called SUZ12 (Suppressor of Zeste
12). These data indicate that PARTICL interlinks regulators of
epigenetic modifications that may affect the suppression of
transcription (67).
CCND1: Another lncRNA found in exosomes
(68) and connected with ionizing
radiation-dependent transcription (69) is ncRNA CCND1 (cyclin D1). This
single strand transcript binds to the TLS (translocated in
liposarcoma) modulator, thus recruiting it to CCND1 promoter and
inducing negative regulation of gene transcription. In addition, it
also blocks the activity of CREB (cAMP response element binding)
and p300 proteins.
EXO1-4: The four molecules that occur
abundantly in exosomes in cervical cancer were defined as Exo1-4
(70). Those transcripts contain
DNase hypersensitive regions as well as RNAPII and CTCF binding
sites (71). Exo1-4 is a subset of
lncRNAs that modulate cell function through increasing recipient
cell viability by binding to various proteins: For example, Exo2
directly interacts with lactate dehydrogenase B (LDHB) while Exo4
reacts with high mobility group protein 17 (HMG-17). This
speculates that lncRNA Exo1-4 has an impact on processes like cell
metabolism or nucleosomal architecture.
lncRNAs as novel class of tumour
biomarkers
Because lncRNA has greater tissue specificity than
mRNA (72), these molecules may
act as specific diagnostic biomarkers (73). When included in extracellular
vesicles, circulation of RNA in body fluids is stable and able to
resist the activity of ribonucleases (74), which makes them potential cancer
prognostic markers (75). The
profiles of lncRNA varies depending on tumour types that may be
useful for clinical research, and moreover, these molecules could
replace biopsies owing to the reduced level of invasiveness
(76). Exosomal lncRNA can be
nominated as cancer biomarkers but some of them are responsible for
the development of other diseases (77). A set of differently expressed
lncRNA associated with non-cancerous or cancerous diseases is shown
in Table VII.
 | Table VII.Participation of lncRNAs in various
diseases [based on (77, 103–110)]. |
Table VII.
Participation of lncRNAs in various
diseases [based on (77, 103–110)].
| lncRNA | Disease | Function | (Refs.) |
|---|
| CRNDE-h | Colorectal cancer
(CRC) | Promotes metabolic
changes through insulin/IGF signaling | (103) |
| p21 | Prostate cancer
(PCa) | Alter gene
expression by modulating mRNA translation and suppressing the p53
or Wnt/β-catenin pathway | (104) |
| PCA3 | Prostate cancer
(PCa) | Modulates the
survival of cells downstream of androgen receptor signaling | (105) |
| BCAR4 | Breast cancer
(BC) | Convert phenotype
into an estrogen-independent, antiestrogen-resistant; tamoxifen
resistance via HER2 signaling | (106) |
| ATB | Hepatocellular
carcinoma (HCC) | Suppresses
E-cadherin, leading to progression of epithelial tumor cells via
inducing EMT | (107) |
| ANRIL | Non-small cell lung
carcinoma (NSCLC) | Affects
proliferation and apoptosis using suppression of KLF2 and P21
transcription | (108) |
| LINC00152 | Gastric cancer
(GCA) | Connects with depth
of invasion among gastric cancer patients | (109) |
| RP11-445H22.4 | Breast cancer
(BC) | Its expression
levels correlated with estrogen receptor (ER), progesterone
receptor (PR), and menopausal status of the breast cancer patients;
this molecule showed a remarkable improvement compared with
CEA | (110) |
| RP11-1382.1 | Chronic kidney
disease (CKD) | Unknown
function | (77) |
Potential usefulness and perspectives
As a result of maintaining full stability of lncRNA
in extracellular vesicles as mentioned earlier, these transcripts
can be taken up by target cells with their full functionality
preserved (78). This make EVs
effective nanocarriers that are capable of regulating lncRNAs
expression in cancer cells (79).
An attractive potential tumour therapy that requires further
development maintaining high levels of tumour suppressor lncRNAs or
destabilizing oncogenic ones (80). H19 is an example of the use of
pro-tumorigenic long non-coding transcripts in the specific
targeting of tumour cells. The regulatory sequence of this lncRNA
was used in the successful formation of the BC-819 plasmid. In
vivo this construct encodes the toxin Diphtheria Toxin A (DTA)
which provided encouraging results in the treatment of bladder or
colon cancer, as well as pancreas carcinoma or NSCLC (81). There are also potential approaches
to inducing overexpression of lncRNA MEG3 (Maternally Expressed
Gene 3) in lung adenocarcinoma in order to increase sensitivity to
cisplatin-based chemotherapy (82). Another potential type of
lncRNA-based therapy exploits the ‘sponge’ activity of those
transcripts, resulting in lowered miRNA activity in tumours
(81).
Looking prospectively, there are still many
challenges to be tackled. Even following the identification of
numerous lncRNAs connected with various diseases, there is still
the need to identify more transcripts and clarify their mechanism
of action. Unbiased genetic screening, lncRNA expression patterns
and characterization of associated proteins should provide
guidance. Moreover, the development of better technologies directed
to improve lncRNA-based therapies will effectively expand their
usage as therapeutic or diagnostic strategies. Finally,
investigation of new tools using the epigenetic functionality of
lncRNAs could help in the research and treatment of diseases that
depend on epigenetics (83). When
it comes to the development of exosomal lncRNA-based therapeutics,
a few problems were identified including uncertain functionality in
biological pathways, the pharmacokinetics and toxicity of lncRNAs
and the precise quantification of EV-bounded non-coding transcripts
which need to be addressed accordingly (84).
In conclusion, progressive studies of lncRNA
properties can lead to their use as cancer biomarkers with high
sensitivity and accuracy, resulting in early-stage detection, and
improved performance of existing clinical biomarkers. Long
non-coding RNA can either have a suppressive function in terms of
cancer development, or promote its progression. A good example of
transporters that carry lncRNAs are exosomes, participating in
cell-to-cell communication and thereafter affecting physiological
and pathological changes. There are still several challenges that
needs to be tackled in order to provide efficient EV-based lncRNAs
therapeutics for clinical application, but the current state of
research offers promise.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
DK drafted the manuscript, designed the figures, and
researched the literature. DK and EP designed the article. EP, AKB
and RH revised the article. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pan BT and Johnstone RM: Fate of the
transferrin receptor during maturation of sheep reticulocytes in
vitro: Selective externalization of the receptor. Cell. 33:967–978.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnstone RM, Adam M, Hammond JR, Orr L
and Turbide C: Vesicle formation during reticulocyte maturation.
Association of plasma membrane activities with released vesicles
(exosomes). J Boil Chem. 262:9412–9420. 1987.
|
|
3
|
Yu S, Cao H, Shen B and Feng J:
Tumor-derived exosomes in cancer progression and treatment failure.
Oncotarget. 6:37151–37168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Isola AL and Chen S: Exosomes: The link
between GPCR activation and metastatic potential? Front Genet.
7:562016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ciardiello C, Cavallini L, Spinelli C,
Yang J, Reis-Sobreiro M, de Candia P, Minciacchi VR and Di Vizio D:
Focus on extracellular vesicles: New frontiers of cell-to-cell
communication in cancer. Int J Mol Sci. 17:1752016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taverna S, Giallombardo M, Gil-Bazo I,
Carreca AP, Castiglia M, Chacartegui J, Araujo A, Alessandro R,
Pauwels P, Peeters M and Rolfo C: Exosomes isolation and
characterization in serum is feasible in non-small cell lung cancer
patients: Critical analysis of evidence and potential role in
clinical practice. Oncotarget. 7:28748–28760. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fu H, Yang H, Zhang X and Xu W: The
emerging roles of exosomes in tumor-stroma interaction. J Cancer
Res Clin Oncol. 142:1897–1907. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soung YH, Nguyen T, Cao H, Lee J and Chung
J: Emerging roles of exosomes in cancer invasion and metastasis.
BMB Rep. 49:18–25. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rahman MA, Barger JF, Lovat F, Gao M,
Otterson GA and Nana-Sinkam P: Lung cancer exosomes as drivers of
epithelial mesenchymal transition. Oncotarget. 7:54852–54866. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Yi J, Chen X, Zhang Y, Xu M and
Yang Z: The regulation of cancer cell migration by lung cancer
cell-derived exosomes through TGF-β and IL-10. Oncol Lett.
11:1527–1530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bobrie A and Thery C: Unraveling the
physiological functions of exosome secretion by tumors.
Oncoimmunology. 2:e225652013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Toro J, Herschlik L, Waldner C and
Mongini C: Emerging roles of exosomes in normal and pathological
conditions: New insights for diagnosis and therapeutic
applications. Front Immunol. 6:2032015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim KM, Abdelmohsen K, Mustapic M,
Kapogiannis D and Gorospe M: RNA in extracellular vesicles. Wiley
Interdiscip Rev RNA. 8:2017. View Article : Google Scholar
|
|
14
|
Neviani P and Fabbri M: Exosomic microRNAs
in the tumor microenvironment. Front Med (Lausanne).
2:472015.PubMed/NCBI
|
|
15
|
Martin F, Roth DM, Jans DA, Pouton CW,
Partridge LJ, Monk PN and Moseley GW: Tetraspanins in viral
infections: A fundamental role in viral biology? J Virol.
79:10839–10851. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baietti MF, Zhang Z, Mortier E, Melchior
A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C,
Vermeiren E, et al: Syndecan-syntenin-ALIX regulates the biogenesis
of exosomes. Nat Cell Biol. 14:677–685. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fares J, Kashyap R and Zimmermann P:
Syntenin: Key player in cancer exosome biogenesis and uptake? Cell
Adh Migr. 11:124–126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ghossoub R, Lembo F, Rubio A, Gaillard CB,
Bouchet J, Vitale N, Slavík J, Machala M and Zimmermann P:
Syntenin-ALIX exosome biogenesis and budding into multivesicular
bodies are controlled by ARF6 and PLD2. Nat Commun. 5:34772014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shim S: Role and regulation of ESCRT-III
In multivesiculr body biogenesis. All Theses and Dissertations
(ETDs). 3232009.
|
|
20
|
Roxrud I, Stenmark H and Malerod L: ESCRT
& Co. Biol Cell. 102:293–318. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Votteler J and Sundquist WI: Virus budding
and the ESCRT pathway. Cell Host Microbe. 14:232–241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amano M, Nakayama M and Kaibuchi K:
Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell
polarity. Cytoskeleton (Hoboken). 67:545–554. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roucourt B, Meeussen S, Bao J, Zimmermann
P and David G: Heparanase activates the syndecan-syntenin-ALIX
exosome pathway. Cell Res. 25:412–428. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roma-Rodrigues C, Fernandes AR and
Baptista PV: Exosome in tumour microenvironment: Overview of the
crosstalk between normal and cancer cells. Biomed Res Int.
2014:1794862014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gezer U, Ozgur E, Cetinkaya M, Isin M and
Dalay N: Long non-coding RNAs with low expression levels in cells
are enriched in secreted exosomes. Cell Biol Int. 38:1076–1079.
2014.PubMed/NCBI
|
|
26
|
Schageman J, Zeringer E, Li M, Barta T,
Lea K, Gu J, Magdaleno S, Setterquist R and Vlassov AV: The
complete exosome workflow solution: From isolation to
characterization of RNA cargo. Biomed Res Int. 2013:2539572013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Batagov AO, Kuznetsov VA and Kurochkin IV:
Identification of nucleotide patterns enriched in secreted RNAs as
putative cis-acting elements targeting them to exosome
nano-vesicles. BMC Genomics. 3 (Suppl 12):S182011. View Article : Google Scholar
|
|
28
|
Okazaki Y, Furuno M, Kasukawa T, Adachi J,
Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al:
Analysis of the mouse transcriptome based on functional annotation
of 60,770 full-length cDNAs. Nature. 420:563–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Perkel JM: Visiting ‘noncodarnia’.
Biotechniques. 54:301 303–304. 2013. View Article : Google Scholar
|
|
31
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sato-Kuwabara Y, Melo SA, Soares FA and
Calin GA: The fusion of two worlds: Non-coding RNAs and
extracellular vesicles-diagnostic and therapeutic implications
(Review). Int J Oncol. 46:17–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bolha L, Ravnik-Glavac M and Glavac D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:72439682017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hewson C and Morris KV: Form and function
of exosome-associated long non-coding RNAs in cancer. Curr Top
Microbiol Immunol. 394:41–56. 2016.PubMed/NCBI
|
|
35
|
Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau
DJ, Sarma K, Song JJ, Kingston RE, Borowsky M and Lee JT:
Genome-wide identification of polycomb-associated RNAs by RIP-seq.
Mol Cell. 40:939–953. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shih JW and Kung HJ: Long non-coding RNA
and tumor hypoxia: New players ushered toward an old arena. J
Biomed Sci. 24:532017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Berrondo C, Flax J, Kucherov V, Siebert A,
Osinski T, Rosenberg A, Fucile C, Richheimer S and Beckham CJ:
Expression of the long non-coding RNA HOTAIR correlates with
disease progression in bladder cancer and is contained in bladder
cancer patient urinary exosomes. PLoS One. 11:e01472362016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yan TH, Lu SW, Huang YQ, Que GB, Chen JH,
Chen YP, et al: Upregulation of the long noncoding RNA HOTAIR
predicts recurrence in stage Ta/T1 bladder cancer. Tumour biology:
the journal of the International Society for Oncodevelopmental
Biology and Medicine. 2014.35(10): 10249–57. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xie Z, Chen X, Li J, Guo Y, Li H, Pan X,
Jiang J, Liu H and Wu B: Salivary HOTAIR and PVT1 as novel
biomarkers for early pancreatic cancer. Oncotarget. 7:25408–25419.
2016.PubMed/NCBI
|
|
40
|
Wang J, Zhang X, Ji C, Zhang L, Di Y, Lou
W, Zhang X and X J: Novel implications of exosomes and lncRNAs in
the diagnosis and treatment of pancreatic cancer. Novel
Implications of Exosomes in Diagnosis and Treatment of Cancer and
Infectious Diseases. Wang J; IntechOpen, : 2017, https://www.intechopen.com/books/novel-implications-of-exosomes-in-diagnosis-and-treatment-of-cancer-and-infectious-diseases/novel-implications-of-exosomes-and-lncrnas-in-the-diagnosis-and-treatment-of-pancreatic-cancer09th–May.
2019 View Article : Google Scholar
|
|
41
|
Song J, Kim D, Han J, Kim Y, Lee M and Jin
EJ: PBMC and exosome-derived Hotair is a critical regulator and
potent marker for rheumatoid arthritis. Clin Exp Med. 15:121–126.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y,
Li X, Wang Y and Ming H: Serum long non coding RNA MALAT-1
protected by exosomes is up-regulated and promotes cell
proliferation and migration in non-small cell lung cancer. Biochem
Biophys Res Commun. 490:406–414. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gutschner T, Hammerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang JK, Ma L, Song WH, Lu BY, Huang YB,
Dong HM, Ma XK, Zhu ZZ and Zhou R: LncRNA-MALAT1 promotes
angiogenesis of thyroid cancer by modulating tumor-associated
macrophage FGF2 protein secretion. J Cell Biochem. 118:4821–4830.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Askarian-Amiri ME, Crawford J, French JD,
Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani
SR, et al: SNORD-host RNA Zfas1 is a regulator of mammary
development and a potential marker for breast cancer. RNA.
17:878–891. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pan L, Liang W, Fu M, Huang ZH, Li X,
Zhang W, Zhang P, Qian H, Jiang PC, Xu WR and Zhang X:
Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes
gastric cancer progression. J Cancer Res Clin Oncol. 143:991–1004.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou H, Wang F, Chen H, Tan Q, Qiu S, Chen
S, Jing W, Yu M, Liang C, Ye S and Tu J: Increased expression of
long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal
transition of gastric cancer. Aging (Albany NY). 8:2023–2038. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Matouk IJ, Halle D, Raveh E, Gilon M,
Sorin V and Hochberg A: The role of the oncofetal H19 lncRNA in
tumor metastasis: Orchestrating the EMT-MET decision. Oncotarget.
7:3748–3765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Conigliaro A, Costa V, Lo Dico A, Saieva
L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M,
et al: CD90+ liver cancer cells modulate endothelial cell phenotype
through the release of exosomes containing H19 lncRNA. Mol Cancer.
14:1552015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
George J and Patel T: Noncoding RNA as
therapeutic targets for hepatocellular carcinoma. Semin Liver Dis.
35:63–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lang HL, Hu GW, Chen Y, Liu Y, Tu W, Lu
YM, Wu L and Xu GH: Glioma cells promote angiogenesis through the
release of exosomes containing long non-coding RNA POU3F3. Eur Rev
Med Pharmacol Sci. 21:959–972. 2017.PubMed/NCBI
|
|
53
|
Hu C, Chen M, Jiang R, Guo Y, Wu M and
Zhang X: Exosome-related tumor microenvironment. Journal of Cancer.
2018.9(17): 3084–92. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mitra SA, Mitra AP and Triche TJ: A
central role for long non-coding RNA in cancer. Front Genet.
3:172012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Braconi C, Valeri N, Kogure T, Gasparini
P, Huang N, Nuovo GJ, Terracciano L, Croce CM and Patel T:
Expression and functional role of a transcribed noncoding RNA with
an ultraconserved element in hepatocellular carcinoma. Proc Natl
Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kogure T, Yan IK, Lin WL and Patel T:
Extracellular vesicle-mediated transfer of a novel long noncoding
RNA TUC339: A mechanism of intercellular signaling in human
hepatocellular cancer. Genes Cancer. 4:261–272. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Takahashi K, Yan IK, Kogure T, Haga H and
Patel T: Extracellular vesicle-mediated transfer of long non-coding
RNA ROR modulates chemosensitivity in human hepatocellular cancer.
FEBS Open Bio. 4:458–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li Y, Jiang B, Zhu H, Qu X, Zhao L, Tan Y,
Jiang Y, Liao M and Wu X: Inhibition of long non-coding RNA ROR
reverses resistance to Tamoxifen by inducing autophagy in breast
cancer. Tumour Biol. 39:10104283177057902017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang N, Li S, Li G, Zhang S, Tang X, Ni S,
Jian X, Xu C, Zhu J and Lu M: The role of extracellular vesicles in
mediating progression, metastasis and potential treatment of
hepatocellular carcinoma. Oncotarget. 8:3683–3695. 2017.PubMed/NCBI
|
|
60
|
Takahashi K, Yan IK, Wood J, Haga H and
Patel T: Involvement of extracellular vesicle long noncoding RNA
(linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer
Res. 12:1377–1387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fritah S, Niclou SP and Azuaje F:
Databases for lncRNAs: A comparative evaluation of emerging tools.
RNA. 20:1655–1665. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lucafò M, Stocco G and Decorti GJTCR:
Emerging molecular mechanisms underlying cancer metastasis: The
rising role of the long non-coding RNA GAS5. Transl Cancer Res. 5
(Suppl 4):S827–S830. 2016. View Article : Google Scholar
|
|
64
|
Pickard MR, Mourtada-Maarabouni M and
Williams GT: Long non-coding RNA GAS5 regulates apoptosis in
prostate cancer cell lines. Biochim Biophys Acta. 1832:1613–1623.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
O'Leary VB, Hain S, Maugg D, Smida J,
Azimzadeh O, Tapio S, Ovsepian SV and Atkinson MJ: Long non-coding
RNA PARTICLE bridges histone and DNA methylation. Sci Rep.
7:17902017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
O'Leary VB, Ovsepian SV, Carrascosa LG,
Buske FA, Radulovic V, Niyazi M, Moertl S, Trau M, Atkinson MJ and
Anastasov N: PARTICLE, a triplex-forming long ncrna, regulates
locus-specific methylation in response to low-dose irradiation.
Cell Rep. 11:474–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Deng J, Tang J, Wang G and Zhu YS: Long
non-coding RNA as potential biomarker for prostate cancer: Is it
making a difference? Int J Environ Res Public Health. 14:E2702017.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Di Liegro CM, Schiera G and Di Liegro I:
Extracellular vesicle-associated RNA as a carrier of epigenetic
information. Genes (Basel). 8:E2402017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hewson C, Capraro D, Burdach J, Whitaker N
and Morris KV: Extracellular vesicle associated long non-coding
RNAs functionally enhance cell viability. Noncoding RNA Res.
1:3–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yarmishyn AA and Kurochkin IV: Long
noncoding RNAs: A potential novel class of cancer biomarkers. Front
Genet. 6:1452015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Shi T, Gao G and Cao Y: Long noncoding
RNAs as novel biomarkers have a promising future in cancer
diagnostics. Dis Markers. 2016:90851952016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Akers JC, Gonda D, Kim R, Carter BS and
Chen CC: Biogenesis of extracellular vesicles (EV): Exosomes,
microvesicles, retrovirus-like vesicles, and apoptotic bodies. J
Neurooncol. 113:1–11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Silva A, Bullock M and Calin G: The
clinical relevance of long non-coding RNAs in cancer. Cancers
(Basel). 7:2169–2182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Khurana R, Ranches G, Schafferer S,
Lukasser M, Rudnicki M, Mayer G and Hüttenhofer A: Identification
of urinary exosomal noncoding RNAs as novel biomarkers in chronic
kidney disease. RNA. 23:142–152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Takahashi K, Yan IK, Haga H and Patel T:
Modulation of hypoxia-signaling pathways by extracellular linc-RoR.
J Cell Sci. 127:1585–1594. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Gutschner T, Richtig G, Haemmerle M and
Pichler M: From biomarkers to therapeutic targets-the promises and
perils of long non-coding RNAs in cancer. Cancer Metastasis Rev.
37:83–105. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Rao AKDM, Rajkumar T and Mani S:
Perspectives of long non-coding RNAs in cancer. Mol Boil Rep.
44:203–18. 2017. View Article : Google Scholar
|
|
81
|
Smaldone MC and Davies BJ: BC-819, a
plasmid comprising the H19 gene regulatory sequences and diphtheria
toxin A, for the potential targeted therapy of cancers. Curr Opin
Mol Ther. 12:607–616. 2010.PubMed/NCBI
|
|
82
|
Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P,
Lu B, Liu G and Wang Z: The long noncoding RNA MEG3 contributes to
cisplatin resistance of human lung adenocarcinoma. PLoS One.
10:e01145862015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wu R, Su Y, Wu H, Dai Y, Zhao M and Lu Q:
Characters, functions and clinical perspectives of long non-coding
RNAs. Mol Genet Genomics. 291:1013–1033. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Qi P, Zhou XY and Du X: Circulating long
non-coding RNAs in cancer: Current status and future perspectives.
Mol Cancer. 15:392016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Schmidt O and Teis D: The ESCRT machinery.
Curr Biol. 22:R116–R120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Keller S, Sanderson MP, Stoeck A and
Altevogt P: Exosomes: From biogenesis and secretion to biological
function. Immunol Lett. 107:102–108. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ruiz-Martinez M, Navarro A, Marrades RM,
Vinolas N, Santasusagna S, Munoz C, Ramírez J, Molins L and Monzo
M: YKT6 expression, exosome release, and survival in non-small cell
lung cancer. Oncotarget. 7:51515–51524. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang C and Robbins PD: The roles of
tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol.
2011:8428492011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang A, Zhou N, Huang J, Liu Q, Fukuda K,
Ma D, Lu Z, Bai C, Watabe K and Mo YY: The human long non-coding
RNA-RoR is a p53 repressor in response to DNA damage. Cell Res.
23:340–350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-transmitted lncARSR
promotes sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang J, Liu SC, Luo XH, Tao GX, Guan M,
Yuan H and Hu DK: Exosomal long noncoding RNAs are differentially
expressed in the cervicovaginal lavage samples of cervical cancer
patients. J Clin Lab Anal. 30:1116–1121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang J, Yao T, Wang Y, Yu J, Liu Y and
Lin Z: Long noncoding RNA MEG3 is downregulated in cervical cancer
and affects cell proliferation and apoptosis by regulating miR-21.
Cancer Boil Ther. 17:104–113. 2016. View Article : Google Scholar
|
|
94
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Xue M, Chen W, Xiang A, Wang R, Chen H,
Pan J, Pang H, An H, Wang X, Hou H and Li X: Hypoxic exosomes
facilitate bladder tumor growth and development through
transferring long non-coding RNA-UCA1. Mol Cancer. 16:1432017.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Juni RP, Abreu RC and da Costa Martins PA:
Regulation of microvascularization in heart failure-an endothelial
cell, non-coding RNAs and exosome liaison. Noncoding RNA Res.
2:45–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z,
Li J, Ye L and Zhang X: Long non-coding RNA HULC promotes tumor
angiogenesis in liver cancer by up-regulating sphingosine kinase 1
(SPHK1). Oncotarget. 7:241–254. 2016.PubMed/NCBI
|
|
99
|
Tan J, Qiu K, Li M and Liang Y:
Double-negative feedback loop between long non-coding RNA TUG1 and
miR-145 promotes epithelial to mesenchymal transition and
radioresistance in human bladder cancer cells. FEBS Lett.
589:3175–3181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Qin CF and Zhao FL: Long non-coding RNA
TUG1 can promote proliferation and migration of pancreatic cancer
via EMT pathway. Eur Rev Med Pharmacol Sci. 21:2377–2384.
2017.PubMed/NCBI
|
|
101
|
Li Z, Zhao L and Wang Q: Overexpression of
long non-coding RNA HOTTIP increases chemoresistance of
osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J
Transl Res. 8:2385–2393. 2016.PubMed/NCBI
|
|
102
|
Lang HL, Hu GW, Zhang B, Kuang W, Chen Y,
Wu L and Xu GH: Glioma cells enhance angiogenesis and inhibit
endothelial cell apoptosis through the release of exosomes that
contain long non-coding RNA CCAT2. Oncol Rep. 38:785–798. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Liu T, Zhang X, Gao S, Jing F, Yang Y, Du
L, Zheng G, Li P, Li C and Wang C: Exosomal long noncoding RNA
CRNDE-h as a novel serum-based biomarker for diagnosis and
prognosis of colorectal cancer. Oncotarget. 7:85551–85563. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Isin M, Uysaler E, Ozgur E, Koseoglu H,
Sanli O, Yucel OB, Gezer U and Dalay N: Exosomal lncRNA-p21 levels
may help to distinguish prostate cancer from benign disease. Front
Genet. 6:1682015.PubMed/NCBI
|
|
105
|
Wang Y, Liu XJ and Yao XD: Function of
PCA3 in prostate tissue and clinical research progress on
developing a PCA3 score. Chin J Cancer Res. 26:493–500.
2014.PubMed/NCBI
|
|
106
|
Godinho M, Meijer D, Setyono-Han B,
Dorssers LC and van Agthoven T: Characterization of BCAR4, a novel
oncogene causing endocrine resistance in human breast cancer cells.
J Cell Physiol. 226:1741–1749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Jang SY, Kim G, Park SY, Lee YR, Kwon SH,
Kim HS, Yoon JS, Lee JS, Kweon YO, Ha HT, et al: Clinical
significance of lncRNA-ATB expression in human hepatocellular
carcinoma. Oncotarget. 8:78588–78597. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia
R, Liu YW, Liu XH, Zhang EB, Lu KH and Shu YQ: Long noncoding RNA
ANRIL promotes non-small cell lung cancer cell proliferation and
inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer
Ther. 14:268–277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Pang Q, Ge J, Shao Y, Sun W, Song H, Xia
T, Xiao B and Guo J: Increased expression of long intergenic
non-coding RNA LINC00152 in gastric cancer and its clinical
significance. Tumour Biol. 35:5441–5447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Xu N, Chen F, Wang F, Lu X, Wang X, Lv M
and Lu C: Clinical significance of high expression of circulating
serum lncRNA RP11-445H22.4 in breast cancer patients: A Chinese
population-based study. Tumour Biol. 36:7659–7665. 2015. View Article : Google Scholar : PubMed/NCBI
|