Introduction
Colorectal cancer (CRC) is one of the most malignant
cancer types worldwide. In developed countries, ~100 million people
suffer from CRC each year, and the mortality rate has increased in
recent years (1,2). Metastasis is the major cause of
mortality in patients with CRC and increases the risk of tumor
recurrence (3). Currently,
chemotherapy or radiotherapy following surgical resection are the
main treatment options (4). The
most commonly used first line chemotherapeutic drugs are
5-fluorouracil, oxaliplatin and irinotecan (5). Although progress in diagnosis and
treatment of cancer has been made in the past decade, the overall
survival rate for patients with CRC remains unfavorable and a
relatively high proportion of patients become chemoresistant
(6). Therefore, early screening
and novel therapeutic approaches are essential for improved
prognosis of patients with colorectal cancer.
Herbal medicines have long been accepted as useful
adjuvant therapeutic agents for a number of diseases. The use of
berberine as an alternative medicine is widespread in the developed
world (7). Berberine is a natural
isoquinoline alkaloid derived from Berberis species,
including Coptis chinensis, which has been used to treat
intestinal infections, particularly bacterial diarrhea, for
thousands of years in China (8).
Additionally, it has been reported to have antitumor effects in
various types of cancer including melanoma, glioma, lung and breast
cancer and hepatocellular carcinoma (9–12).
Berberine may exhibit an anticancer effect in CRC (13,14);
however, the underlying functional mechanism has not been fully
elucidated.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs >200 nucleotides in length and serve an
important role in regulating gene expression (15). They can regulate target gene
expression level in the cytoplasm by serving as microRNA
(miRNA/miR) sponges (16), and
silence or activate target genes through modification at the
promoter region (17,18). lncRNAs may directly interact with
signaling pathways to serve as prognostic predictors or therapeutic
targets for different types of cancer (19,20).
Furthermore, previous reports indicated that lncRNAs may
participate in chemoresistance (21,22).
However, their roles in the function of berberine are poorly
understood. The identification of lncRNAs involved in the function
of berberine may enhance human cancer treatment and aid to overcome
chemoresistance.
In the current study, the regulatory function of
lncRNA cancer susceptibility 2 (CASC2) in the tumor-suppressive
role of berberine in CRC was investigated. The results revealed
that berberine suppressed cell viability and promoted apoptosis of
human CRC cancer cell lines in a concentration-dependent manner.
Furthermore, berberine treatment increased the endogenous level of
lncRNA CASC2, and this upregulation of lncRNA CASC2 promoted
berberine-induced cell cytotoxicity by interacting with enhancer of
zeste 2 polycomb repressive complex 2 subunit (EZH2) and silencing
the expression of BCL2.
Materials and methods
Ethics statement and tissue
samples
The study included 60 patients with CRC [male/female
ratio, 36/24; age range, 42–73 (median age, 56)] who underwent
partial or total surgical resection at the West China Second
University Hospital (Chengdu, China) between June 2014 and July
2016. Paired cancer tissues and normal control tissues (>2 cm
distal from the cancer area) were collected by surgical resection.
Tissue specimens were stored in liquid nitrogen during
transportation. The current study was approved by the West China
Second University Hospital. All participants signed informed
consent prior to using the tissues for scientific research.
Cell culture
Five human CRC cell lines (HT-29, HCT116, SW480,
SW620 and LoVo) and one human fetal normal colonic cell line (FHC)
were purchased from the American Tissue Culture Collection. Human
CRC cells were cultured in DMEM (BioWhittaker; Lonza Group, AG)
containing 10% FBS (Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin and streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
in a humidified incubator at 37°C and 5% CO2. Normal
colon FHC cells were grown in DMEM/F12 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS, 10 ng/ml cholera
toxin, 5 µg/ml transferrin, 5 µg/ml insulin, 100 ng/ml
hydrocortisone and additional 10 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, at 37°C in 5%
CO2 and 95% air. Berberine was purchased from
Sigma-Aldrich (Merck KGaA; cat. no. B3251) and used at the
concentration of 50 µM (diluted with PBS and added into culture
medium).
Expression profile analysis of
lncRNAs
Total RNA was extracted from HT29 cells treated with
berberine (50 µM for 48 h) or normal culture medium using the
RNeasy Plus Mini kit (Qiagen, Inc.) according to the manufacturer's
protocol. lncRNAs were sequenced using the high-throughput,
high-sensitivity HiSeq 2500 sequencing platform (Illumina, Inc.).
cDNA was sonicate-fragmented (10-cycles of 30 sec on and 30 sec off
at 4°C; Bioruptor®; Diagenode, Inc.) to an average size
of 250 bp to construct a cDNA library. Data processing and
statistical analysis for the RNA-sequencing data were performed (R
software version 3.5.2, Lucent Technologies Inc.) and a heat map
was subsequently generated by using ggplot2 R package (http://cran.r-project.org/web/packages/ggplot2/).
Vector construction and cell
transfection
The synthetic silencing oligonucleotides used for
small interfering RNAs (siRNAs), si-CASC2 and si-EZH2, were
synthesized by Sangon Biotech Co., Ltd. Negative control siRNA
(cat. no. 12935-110; Invitrogen; Thermo Fisher Scientific, Inc.)
was used. The sequences of small-interfering RNAs are presented in
Table I. The CASC2 overexpression
vector (p-CASC2) was generated by cloning the specific sequences of
CASC2 into a pcDNA 3.1 vector (synthesized by Sangon Biotech Co.,
Ltd.). The vectors were labeled with green fluorescence protein
(GFP) to confirm the transfection. For overexpression experiments,
CRC cells in 6-well plate were transfected with the plasmids (2,000
ng/well) using TransFast™ Transfection Reagent (Promega
Corporation) according to the manufacturer's protocol. For
knockdown experiments, a total of 5×105 HT29 or HCT116
cells were seeded into each well of a 6-well plate and transfected
with the oligoribonucleotides (100 nM) upon reaching 70–80%
confluence. The change in the expression levels of the target genes
was determined by the reverse-transcription quantitative PCR
(RT-qPCR) at 24 h after transfection. The transfected cells were
subsequently used for functional assays.
 | Table I.Primer and siRNA sequences. |
Table I.
Primer and siRNA sequences.
| A, RT-qPCR
primers |
|---|
|
|---|
| Name | Sequence
(5′-3′) |
|---|
| CASC2
(Forward) |
GCACATTGGACGGTGTTTCC |
| CASC2
(Reverse) |
CCCAGTCCTTCACAGGTCAC |
| EZH2 (Forward) |
GGCTCCTCTAACCATGTTTACAACT |
| EZH2 (Reverse) | AGCGGTTTTGACACTCTG
AACTAC |
| BCL2 (Forward) | TCT
TCCAGGAACCTCTGTGATG |
| BCL2 (Reverse) |
CAATGCCGCCATCGCTTACACC |
| GAPDH
(Forward) |
GCACCGTCAAGGCTGAGAAC |
| GAPDH
(Reverse) |
ATGGTGGTGAAGACGCCAGT |
|
| B, RIP-qPCR
primers |
|
| Name | Sequence
(5′-3′) |
|
| CASC2
(Forward) |
GCACATTGGACGGTGTTTCC |
| CASC2
(Reverse) |
CCCAGTCCTTCACAGGTCAC |
| lncRNA control
(Forward) |
GGGTGTTTACGTAGACCAGAA |
| lncRNA control
(Reverse) |
CTTCCAAAAGCCTTCTGCCTTA |
| β-actin
(Forward) |
CTCGCTTCGGCAGCACATATAC |
| β-actin
(Reverse) |
AACGCTTCACGAATTTGCGTGT |
|
| C, ChIP-qPCR
primers |
|
| Name | Sequence
(5′-3′) |
|
| BCL2-A
(Forward) |
GAGGAGCCATCCGCACATCA |
| BCL2-A
(Reverse) |
AGCTTAGACTGTAAGCTGGT |
| BCL2-B
(Forward) |
AGTCCCACAACAGCATAGGG |
| BCL2-B
(Reverse) |
TCCCTAGGTCAGGACCACCT |
| BCL2-C
(Forward) |
CTCCAGCTTGGGTGAAAGAG |
| BCL2-C
(Reverse) |
GGGCTTTTACACTTGGCTAG |
| GAPDH-D
(Forward) |
AGGGAAGCTGACAGGGATGGCG |
| GAPDH-D
(Reverse) |
ATCGAAGATGGACGAGTGGGTA |
| GAPDH-E
(Forward) |
CCCCGCTACTCCTCCTCCTAAG |
| GAPDH-E
(Reverse) |
TCCACGACCAGTTGTCCATTCC |
|
| D,
siRNA |
|
| Name | Sequence
(5′-3′) |
|
| si-CASC2-1 |
UGAAAAGAGCCGUGAGCUA |
| si-CASC2-2 |
AAATAAAGATGGTGGAATG |
| si-CASC2-3 |
CUGCAAGGCCGCAUGAUGA |
| si-EZH2 |
AUCAGCUCGUCUGAACCUCUU |
| si-NC (GFP) |
GGCUACGUCCAGGAGCGCACC |
RNA extraction and RT-qPCR
Total RNA was extracted from cells or tissue samples
by using the RNeasy Plus Mini kit (Qiagen, Inc.) according to the
manufacturer's protocol. The 20 µl RT reactions were performed
using a PrimeScript® RT reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China) and incubated for 30 min at
37°C and 5 sec at 85°C. Subsequently, 2 µl of diluted RT product
was mixed with 23 µl reaction buffer (Takara Inc.) to a final
volume of 25 µl. The primer sequences are presented in Table I. All reactions labeled with
SYBRGreen (Takara Inc.) were carried out using an Eppendorf
Mastercycler EP Gradient S (Eppendorf) under the following
conditions: 95°C for 30 sec, followed by 45 cycles of 95°C for 5
sec and 60°C for 30 sec. The expression levels of detected RNAs
were normalized to GAPDH using the comparative 2−ΔΔCq
method (23).
Cell viability assay
The cell viability following treatment with
berberine was assayed using the MTT assay (Dojindo Molecular
Technologies, Inc.). In brief, 3×104 cells were seeded
into a 96-well plate and treated with 0–100 µM berberine (cat. no.
B3251; Sigma-Aldrich; Merck KGaA) or PBS control for 12–72 h. The
cells were treated with the 10 µl MTT reagent and incubated for an
additional 2 h at room temperature. Purple formazan was then
dissolved by adding 150 µl DMSO (Sigma-Aldrich; Merck KGaA) to each
well. The optical density at a wavelength of 450 nm was
subsequently measured with a spectrophotometer (Thermo Fisher
Scientific, Inc.).
Apoptosis assay
Cells in 6-well plate (1–5×106/ml) were
collected following berberine treatment (50 µM) or transient
transfection for 48 h, and washed with PBS and 0.025% trypsin-EDTA
to obtain single-cell suspensions. Cells were fixed in ice-cold 70%
ethanol (60 min at 4°C) and stained with an Annexin V FITC and
propidium iodide solution kit (Sigma-Aldrich; Merck KGaA) for 20
min at room temperature. Apoptosis was detected using a flow
cytometer (BD FACSCalibur™; BD Biosciences) and analysed using
Modfit LT version 5.0 software (Verity Software House Inc.).
Cell migration and invasion assay
Cell migration was evaluated by performing a
wound-healing assay. The cells were seeded at a density of
5×105 cell/well into six-well plates. After 12 h of
treatment with 50 µM berberine and/or transfection with si-CASC2,
the layer of cells was scratched using a sterile 20 µl pipette tip
to form wounds. The non-adherent cells were washed away with
culture medium, and the cells were cultured for 48 h followed by
measurement of the gap area. Cell invasive ability was evaluated
using a Transwell invasion assay using Boyden chambers (8 µm pore
size; BD Biosciences). The membranes were coated with Matrigel.
Cells in serum-free media were placed into the upper chamber. Media
containing 10% FBS was added to the lower chamber. Following 24 h
of incubation at 37°C, the cells that had invaded through the
membrane were fixed with 100% methanol for 20 min at room
temperature and stained with 0.1% crystal violet for 10 min at room
temperature and 3 random fields were imaged using an inverted
microscope at 10× magnification (Leica Microsystems, Inc., Buffalo
Grove, IL, USA).
RNA immunoprecipitation (RIP)
HT29 and HCT116 cells were rinsed with cold PBS and
fixed using 1% formaldehyde for 10 min. Following 10 min
centrifugation at 1,5000 × g and 4°C, cell pellets were collected
and re-suspended in NP-40 lysis buffer (Beyotime Institute of
Biotechnology). The RIP assay was performed using the Magna RIP™
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore)
according to the manufacturer's protocol. Total RNA was
immunoprecipitated overnight at 4°C with an antibody against EZH2
(1:50, cat. no. ab186006; Abcam) or negative control IgG (1:50,
cat. no. 12-370, EMD Millipore). The magnetic bead bound complexes
were immobilized with a magnet and unbound materials were washed
off. RNA was extracted and analyzed by RT-qPCR.
Chromatin immunoprecipitation
(ChIP)
An EZ-Magna ChIP kit (EMD Millipore) was used
according to the manufacturer's protocol. Briefly, HT29 cells
(1×106/ml) were treated with 4% paraformaldehyde and
incubated for 10 min at 4°C to generate DNA-protein cross-links.
Cell lysates were then sonicated for with a 10 sec on and 10 sec
off mode for 12 cycles to generate chromatin fragments of 200–300
bp and immunoprecipitated overnight at 4°C with anti-EZH2 antibody
(1:50, cat. no. ab186006; Abcam), anti-H3K27me3 antibody (1:50,
cat. no. ab6002; Abcam) or the negative control IgG (1:50, cat. no.
12-370; EMD Millipore). Two sites in the GAPDH promoter region were
used as a control for the identification of interactions between
GAPDH and BCL2. Precipitated DNA was recovered and analyzed by
RT-qPCR.
Western blot and antibodies
RIPA assay buffer (Sigma Aldrich; Merck KGaA) was
used to extract proteins from the cells. Protein concentration was
measured using a bicinchoninic acid assay (Sigma Aldrich; Merck
KGaA). The same quantity of protein (25 µg) was transferred onto
PVDF membranes following the separation process by performing a
routine SDS-PAGE with 10% gel. The membrane was blocked with 5% (5
g/100 ml) nonfat dry milk in TBS plus Tween buffer for 2 h at room
temperature. The membrane was incubated with primary antibodies
against EZH2 (1:1,000; cat. no. ab186006; Abcam), BCL2 (1:1,000;
cat. no. ab32124; Abcam) and β-actin (1:1,000; cat. no. ab8227;
Abcam) overnight at 4°C. Following primary incubation, membranes
were incubated with horseradish peroxidase-conjugated secondary
antibodies (1:5,000; cat. no. 7074; Cell Signaling Technology,
Inc.) for 1 h at room temperature. Protein bands were detected
using ECL reagent (Amersham; GE Healthcare). Gray analysis by image
analysis was performed using the software Gel-Pro Analyzer (version
4.0; United States Biochemical) after scanning.
Statistical analysis
Data are presented as the median and interquartile
range. The Mann-Whitney U test was used for the comparison of
datasets containing two independent groups, and the Wilcoxon
signed-rank test was used for comparison of paired samples. The
Kruskal-Wallis test followed by the Bonferroni post hoc analysis
was used for evaluating the differences among multiple groups. The
Spearman's test was used to analyze the correlation between CASC2
and BCL2 mRNA expression. Statistical analysis was performed using
GraphPad Prism (version 6; GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Berberine suppresses cell viability
and promotes apoptosis in CRC cells
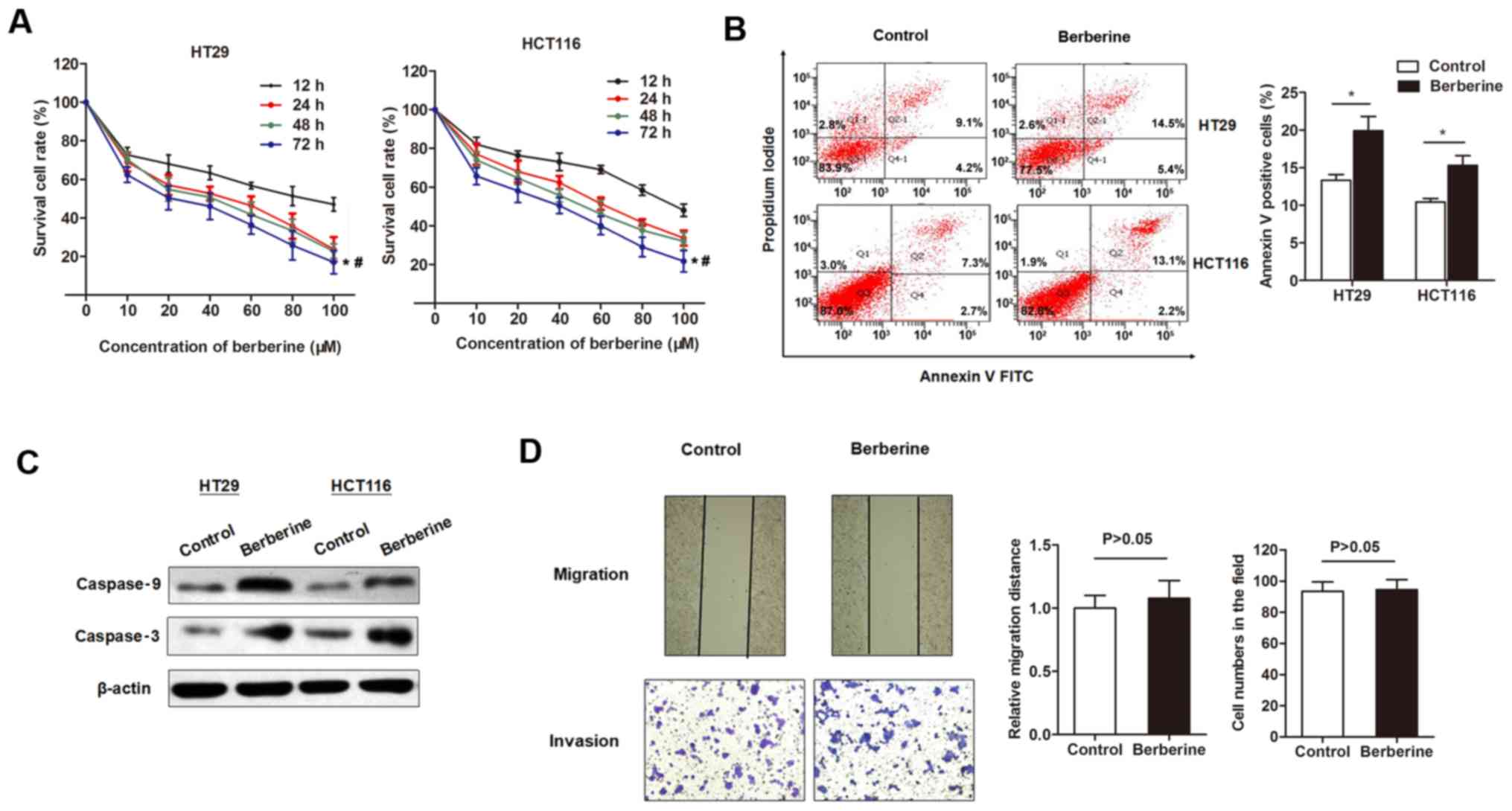
The inhibitory effects of berberine on the cell
viability of CRC cell lines was investigated. The MTT assay
indicated that berberine decreased the in vitro cell
viability of two CRC cell lines in a dose-dependent manner, with a
half maximal inhibitory concentration of 43.77 µM for HT29 and
56.44 µM for HCT116 at 48 h post-treatment (Fig. 1A). Flow cytometry analysis was
subsequently used to detect cell apoptosis. The percentage of
apoptotic cells was significantly increased following treatment
with 50 µM berberine for 48 h compared with controls (Fig. 1B). To investigate whether the
increased cell apoptosis was due to activation of apoptotic
signaling pathways, the expression levels of several apoptotic
proteins were detected. Western blotting revealed that the
expression levels of cleaved caspase-3 and −9 were upregulated by
the treatment of berberine at 50 µM for 48 h (Fig. 1C). The effect of berberine on
migration and invasion capacity of HT29 cells were assessed by
performing scratch and Transwell assays, respectively. The results
revealed that berberine treatment did not have a significant effect
on cell migration and invasion compared with controls (Fig. 1D).
lncRNA expression profile of CRC cell
lines
On the basis of the aforementioned observations, the
pathways underlying berberine-induced apoptosis were further
investigated. lncRNAs are important regulators for the
proliferation and apoptosis of cancer cells as well as chemotherapy
resistance (24). Therefore, the
current study aimed to identify specific lncRNAs altered by
berberine treatment. High-throughput lncRNA sequencing was
performed using 50 µM berberine-treated HT29 cells and cells
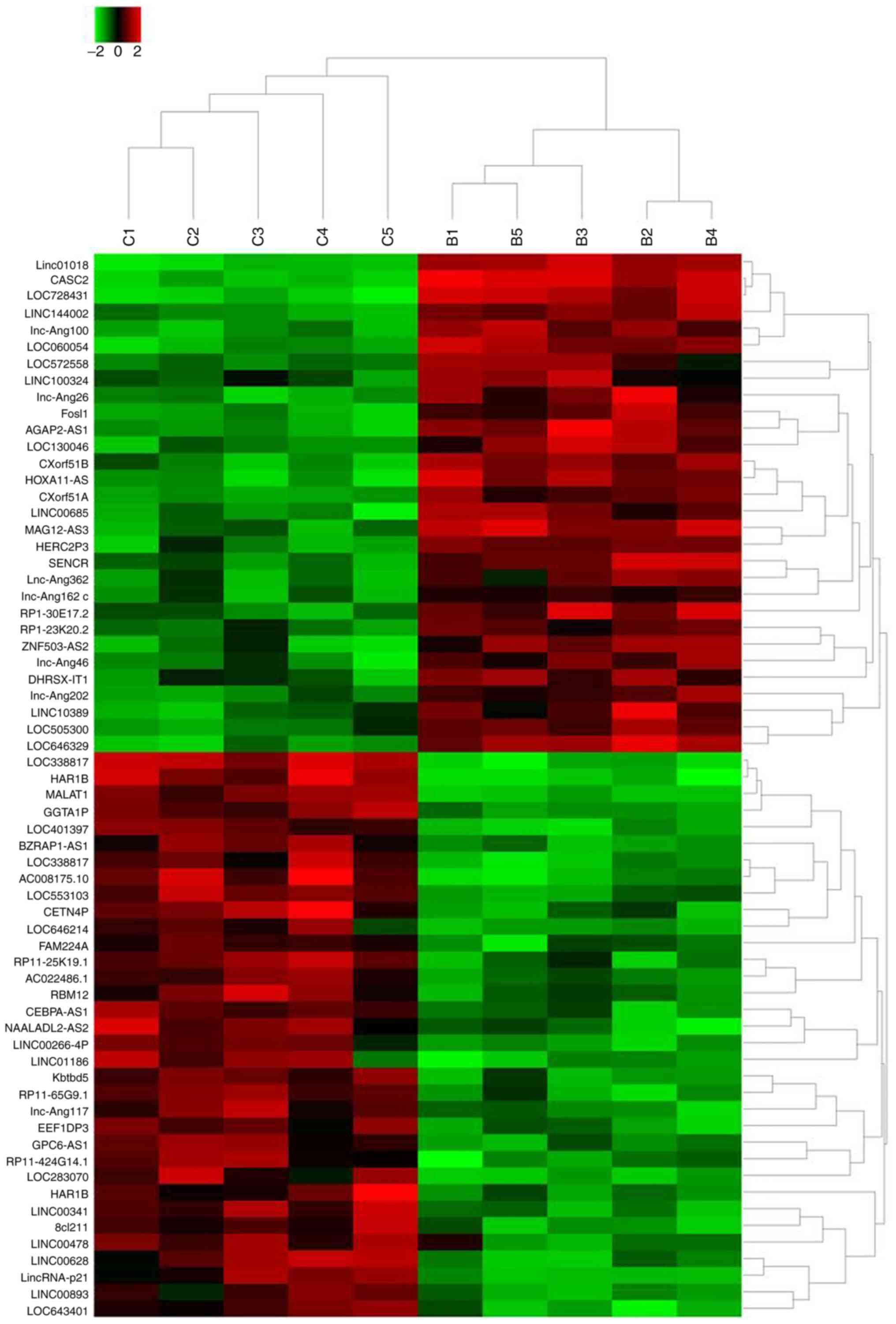
cultured with normal condition. The results revealed that a total
of 64 lncRNAs exhibited >2-fold change in expression between
berberine-treated cells and non-treated cells (Fig. 2). Specifically, the expression of
30 lncRNAs was increased following the treatment of berberine
compared with control treatment. LINC01018 had 49.7953-fold higher
expression, ranking as the most differentially expressed, followed
by lncRNA CASC2 and LOC728431. In addition, a total of 34 lncRNAs
exhibited decreased expression levels following berberine
treatment. LOC338817 showed the lowest expression level
(55.4954-fold lower compared with controls), followed by HAR1B and
lncRNA MALAT1 (Table II).
 | Table II.Candidate lncRNAs selected on a basis
of the sequencing analysis. |
Table II.
Candidate lncRNAs selected on a basis
of the sequencing analysis.
| lncRNA | Location | Regulation (B vs.
C) | Fold change | P-value |
|---|
| LINC01018 | Chr5p15.31 | Up | 49.7953 | 0.00030542 |
| CASC2 | Chr10p26.11 | Up | 35.5740 | 0.00079371 |
| LOC728431 | Chr1p34.3 | Up | 24.1439 | 0.00135860 |
| LOC338817 | Chr12p13.2 | Down | 55.4954 | 0.00012671 |
| HAR1B | Chr20q13.33 | Down | 44.3649 | 0.00039834 |
| MALAT1 | Chr11p13.1 | Down | 31.7852 | 0.00068024 |
Expression level of lncRNA CASC2 in
CRC cell lines is increased following berberine treatment
The aforementioned dysregulated lncRNA(s) were
validated using qPCR. Compared with the RNA sequencing data, only
lncRNA CASC2 showed a statistically significant increased
expression level in HT29 cells following berberine treatment
compared with controls. Expression of the other five lncRNAs showed
little difference between the two group of cells when analyzed by
RT-qPCR (Fig. 3A-F). Therefore,
the functional role of CASC2 was investigated further. Previous
reports indicated that lncRNA CASC2 may be involved in cell
apoptosis, proliferation and cell cycle regulation in cancer
(25,26). Therefore, berberine may decrease
CRC cell viability and promote apoptosis through the upregulation
of lncRNA CASC2. To investigate this hypothesis, the expression
level of lncRNA CASC2 in CRC cell lines was determined. The results
revealed that the expression level of lncRNA CASC2 was decreased in
the five CRC cell lines used in the present study compared with the
normal colon epithelial cell line FHC (Fig. 3G). In addition, CASC2 was
downregulated in primary CRC tissues when compared with non-tumor
tissues (Fig. 3H). Furthermore,
lncRNA CASC2 was upregulated in HT29 cells following treatment with
berberine for 48 h in a dose-dependent manner (Fig. 3I).
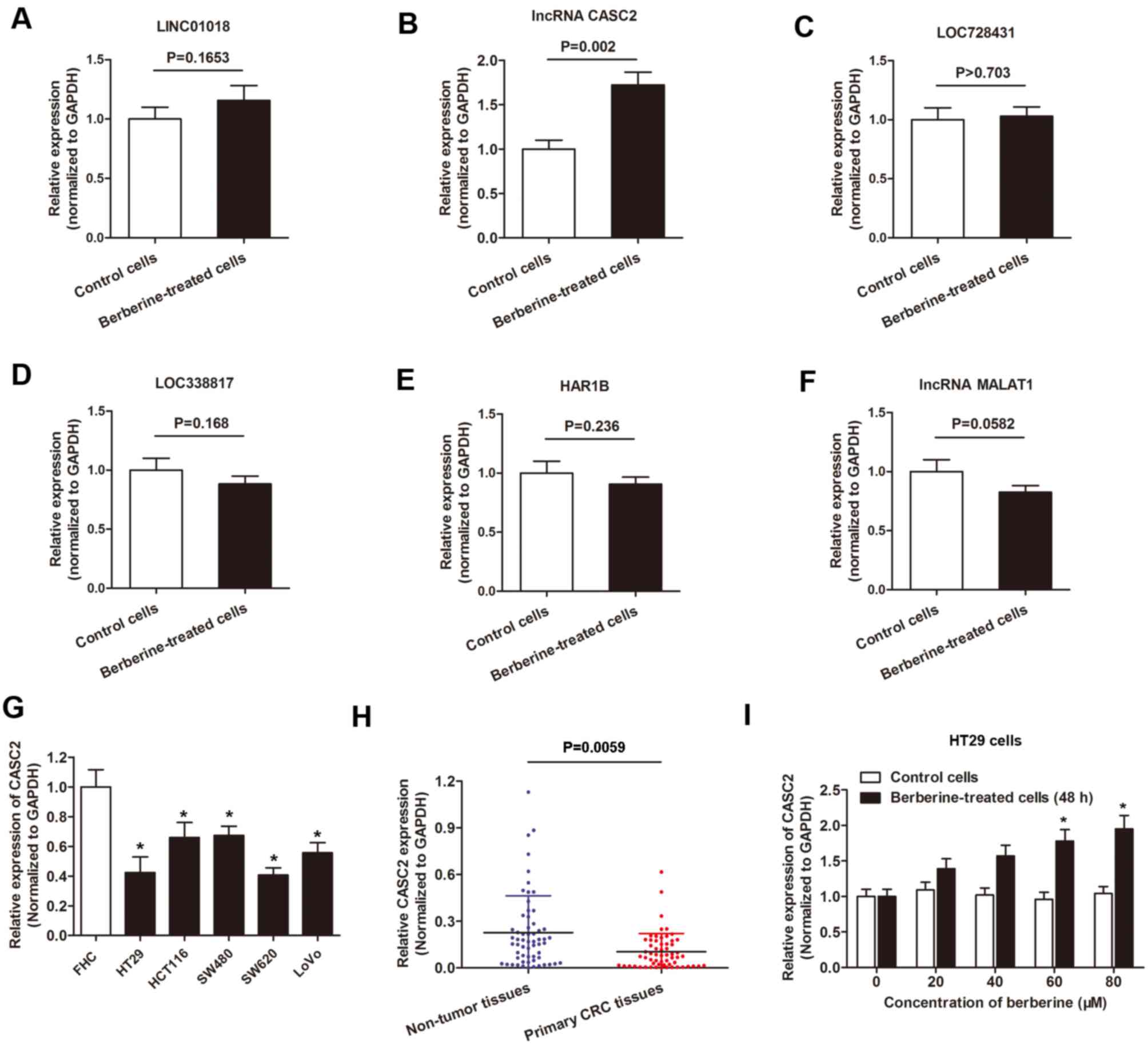 | Figure 3.lncRNA CASC2 was upregulated by
berberine treatment in CRC. A-F. RT-qPCR analysis of the six
indicated lncRNAs, (A) LINC01018, (B) CASC2, (C) LOC728431, (D)
LOC33817, (E) HAR1B, (F) MALAT1, in HT29 cells cultured with
berberine-containing medium or berberine-free medium. (G) lncRNA
CASC2 expression was measured using RT-qPCR in the indicated cell
lines. *P<0.05 vs. FHC cells. (H) lncRNA CASC2 level was
detected via RT-qPCR in paired CRC tissues and adjacent non-tumor
tissues obtained from patients with CRC. The expression of lncRNA
CASC2 was significantly downregulated in primary CRC tissues when
compared with noncancerous tissues. (I) lncRNA CASC2 level was
increased in HT29 cells following treatment with berberine at
increasing concentrations for 48 h. *P<0.05 vs. control cells.
lncRNA, long non-coding RNA; CASC2, cancer susceptibility 2; CRC,
colorectal cancer; RT-qPCR, reverse-transcription quantitative PC;
LINC01018, long intergenic non-protein coding RNA 1018; LOC728431,
long intergenic non-protein coding RNA 1137; MALAT1, metastasis
associated lung adenocarcinoma transcript 1. |
Berberine suppresses cell viability
and promotes apoptosis by inducing expression of lncRNA CASC2
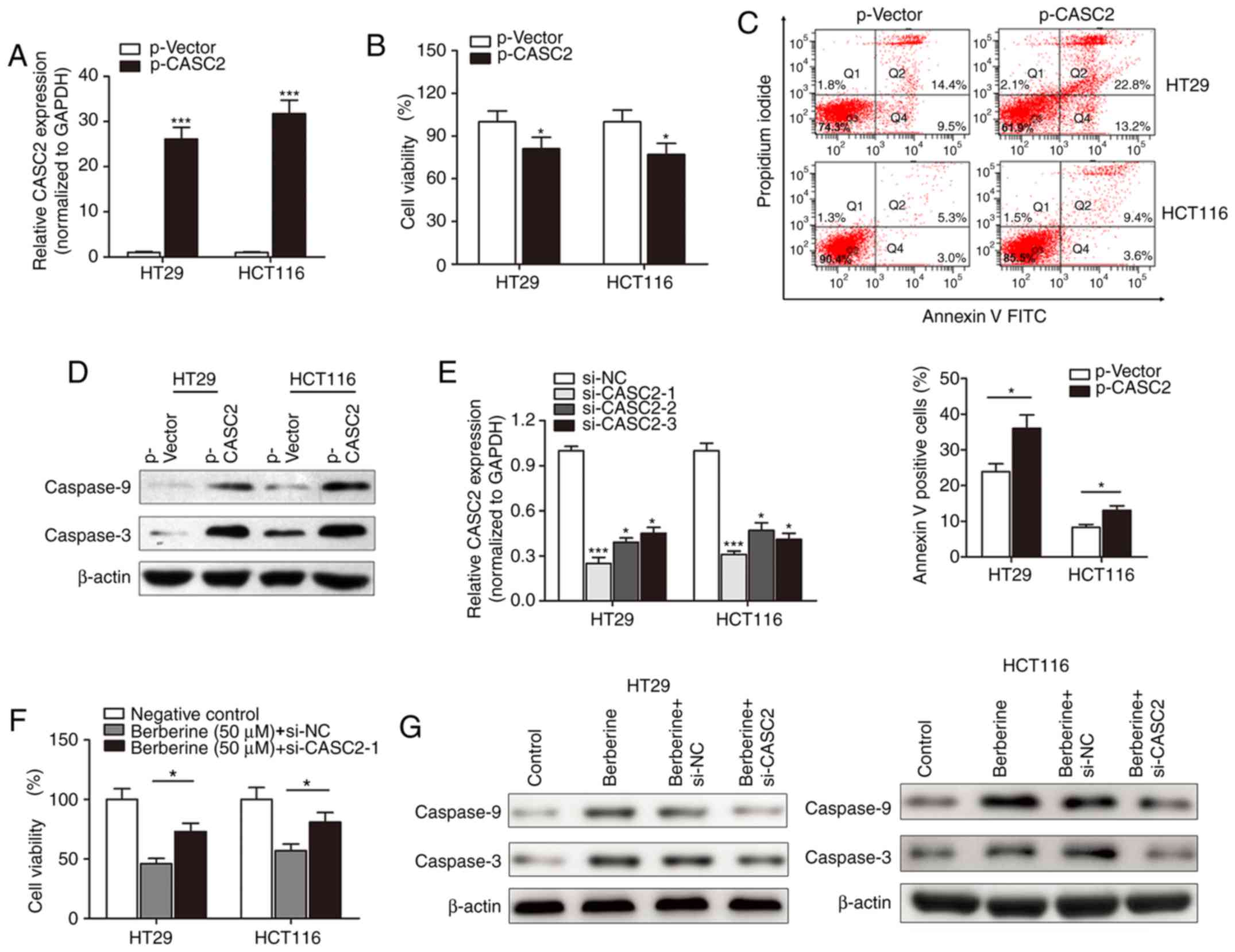
Following the investigation of the expression of
lncRNA CASC2 in berberine-treated CRC cells, the functional role of
CASC2 in berberine-induced cell apoptosis and cytotoxicity was
determined. lncRNA CASC2 was significantly upregulated in HT29 and
HCT116 cells following transfection with p-CASC2 compared with the
control vector (Fig. 4A). The MTT
assay suggested that lncRNA CASC2 suppressed cell viability in HT29
and HCT116 cells (Fig. 4B). Flow
cytometry revealed that overexpression of CASC2 significantly
increased the number of HT29 and HCT116 apoptotic cells compared
with cells transfected with control vectors (Fig. 4C). Western blotting indicated that
p-CASC2 resulted in increased expression of the pro-apoptotic
proteins, cleaved caspase-3 and −9 compared with the control vector
(Fig. 4D). Subsequently, si-CASC
was used to determine whether lncRNA CASC2 was associated with the
function of berberine. si-CASC2-1 demonstrated the best silencing
effect when compared with the other two inhibitors, and was thus
used for subsequent experiments (Fig.
4E). si-CASC2-1 reduced the berberine-induced inhibition of
cell viability in HT29 and HCT116 cell lines (Fig. 4F). Similarly, si-CASC2-1 attenuated
the berberine-induced increase in expression of cleaved caspase-3
and −9 (Fig. 4G).
BCL2 is required for the functional
effect induced by berberine and lncRNA CASC2
As the berberine/CASC2 signaling pathway affected
apoptosis in CRC cell lines, the expression of apoptosis-targeted
genes was investigated. Significant differences in several
apoptosis-related targets regulated by lncRNA CASC2, including
BCL2, which is a well-known antiapoptotic protein (27,28),
were identified (Table III).
Thus, lncRNA CASC2 may promote apoptosis via targeting the
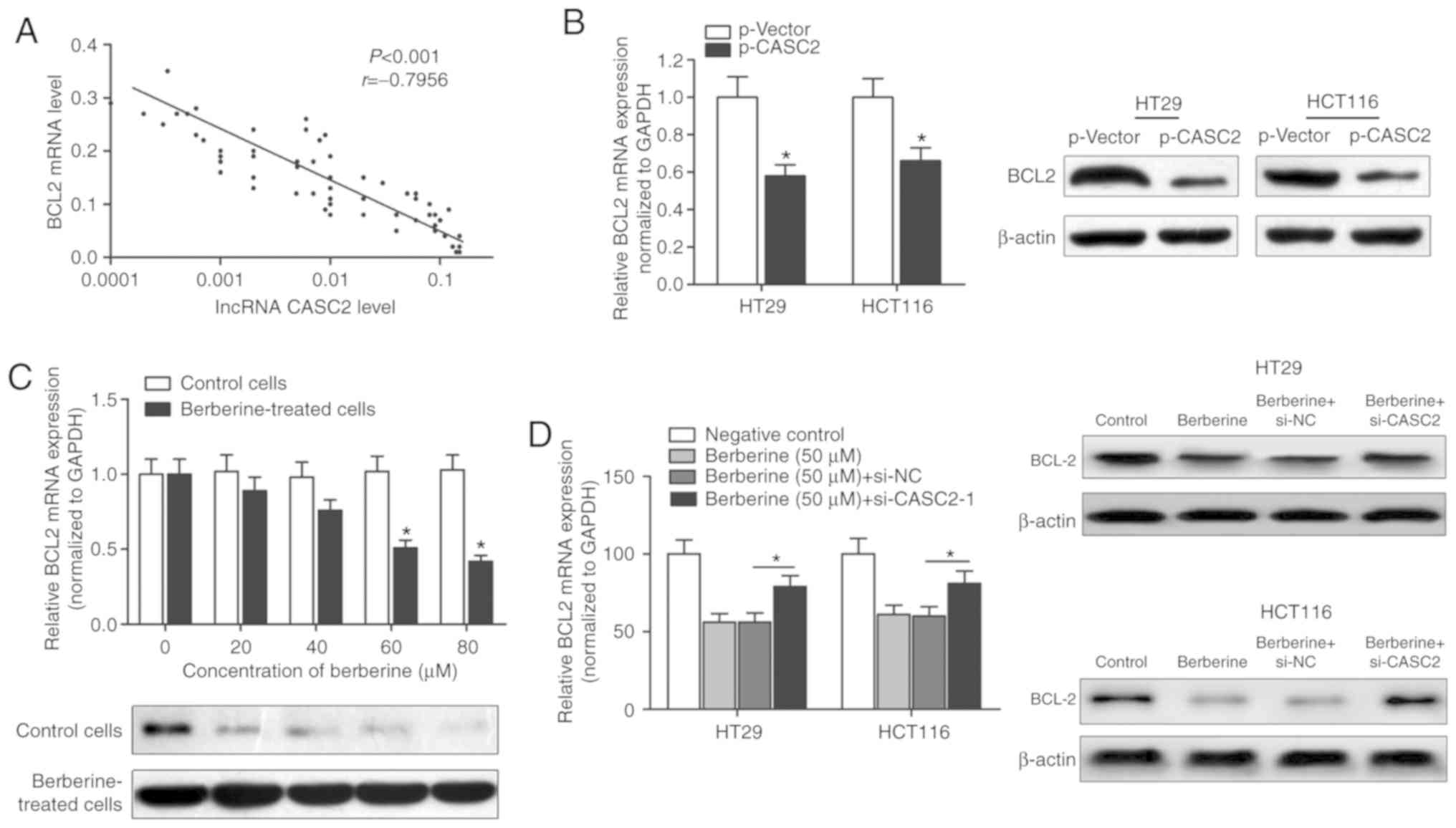
antiapoptotic gene, BCL2. The expression level of BCL2 in CRC
tissue samples was determined using RT-qPCR. A significant negative
correlation between the expression of BCL2 and lncRNA CASC2 was
identified (Fig. 5A). Moreover,
overexpression of CASC2 decreased the expression of BCL2 at the
transcript and protein levels, as demonstrated by RT-qPCR and
western blot analysis (Fig. 5B).
BCL2 mRNA and protein levels were decreased in HT29 cells treated
with berberine in a dose-dependent manner (Fig. 5C). However, transfection with
si-CASC-1 reversed the berberine-induced decrease of BCL2 mRNA and
protein levels (Fig. 5D),
suggesting that berberine exerted its proapoptotic effect via a
CASC2-BCL2 signaling mechanism.
 | Table III.Long non-coding RNA CASC2 regulated
target genes associated with apoptosis. |
Table III.
Long non-coding RNA CASC2 regulated
target genes associated with apoptosis.
| Gene symbol | Gene name | Location | Fold change
(p-CASC2/negative control, all P<0.005) |
|---|
| BCL2 | BCL2 apoptosis
regulator | Chr18q21.33 | 0.008 |
| BCL2A1 | BCL2 related
protein A1 | Chr15q25.1 | 0.06 |
| PARP2 | Poly (ADP-ribose)
polymerase 2 | Chr14q11.2 | 0.19 |
| BIRC3 | Baculoviral IAP
repeat containing 3 | Chr11q22.2 | 0.52 |
| BAX | BCL2-associated X,
apoptosis regulator | Chr19q13.33 | 25.39 |
| MCL1 | MCL1 apoptosis
regulator, BCL2 family member | Chr1q21.2 | 19.31 |
| BAK1 | BCL2
antagonist/killer 1 | Chr6p21.31 | 10.48 |
| CASP9 | Caspase 9 | Chr1p36.21 | 9.56 |
| CASP3 | Caspase 3 | Chr4p35.1 | 7.38 |
EZH2 associates with lncRNA CASC2,
which subsequently suppresses BCL2 expression
A RIP assay was performed to identify whether there
was a direct interaction between lncRNA CASC2 and EZH2. CASC2 was
pulled down by EZH2 while no enrichment was identified when using
an IgG antibody as a control (Fig.
6A). Moreover, no enrichment was identified when using the EZH2
antibody to pull down β-actin or lncRNA control, suggesting that
the association between EZH2 and CASC2 was specific (Fig. 6B). Furthermore, the role of EZH2 in
the regulation of BCL2 expression was evaluated. si-EZH2 was used
to silence EZH2 in HT29 and HCT116 cells (Fig. 6C). Knockdown of EZH2 in HT29 and
HCT116 cells upregulated the expression level of BCL2 (Fig. 6D and E). A ChIP assay was performed
to detect the effect of lncRNA CASC2 on histone modification in the
BCL2 gene promoter in HT29 cells. A set of detection sites were
used to design specific primers for the amplification of BCL2
promoter sequences pulled down by anti-EZH2 or anti-H3K27-me3
antibodies (Fig. 6F).
Overexpression of CASC2 significantly increased the enrichment of
BCL2 (detection site A, B and C) when using the anti-EZH2 and
anti-H3K27me3 antibodies, while no significant change in the
enrichment level was identified for GAPDH (detection site D and E;
Fig. 6G and H). In addition,
lncRNA CASC2 had little effect on the enrichment of BCL2 when using
the anti-IgG antibody (Fig. 6I).
To conclude, the current study revealed that lncRNA CASC2
downregulated BCL2 expression level in CRC by binding with the
promoter region of BCL2 in an EZH2-dependent manner.
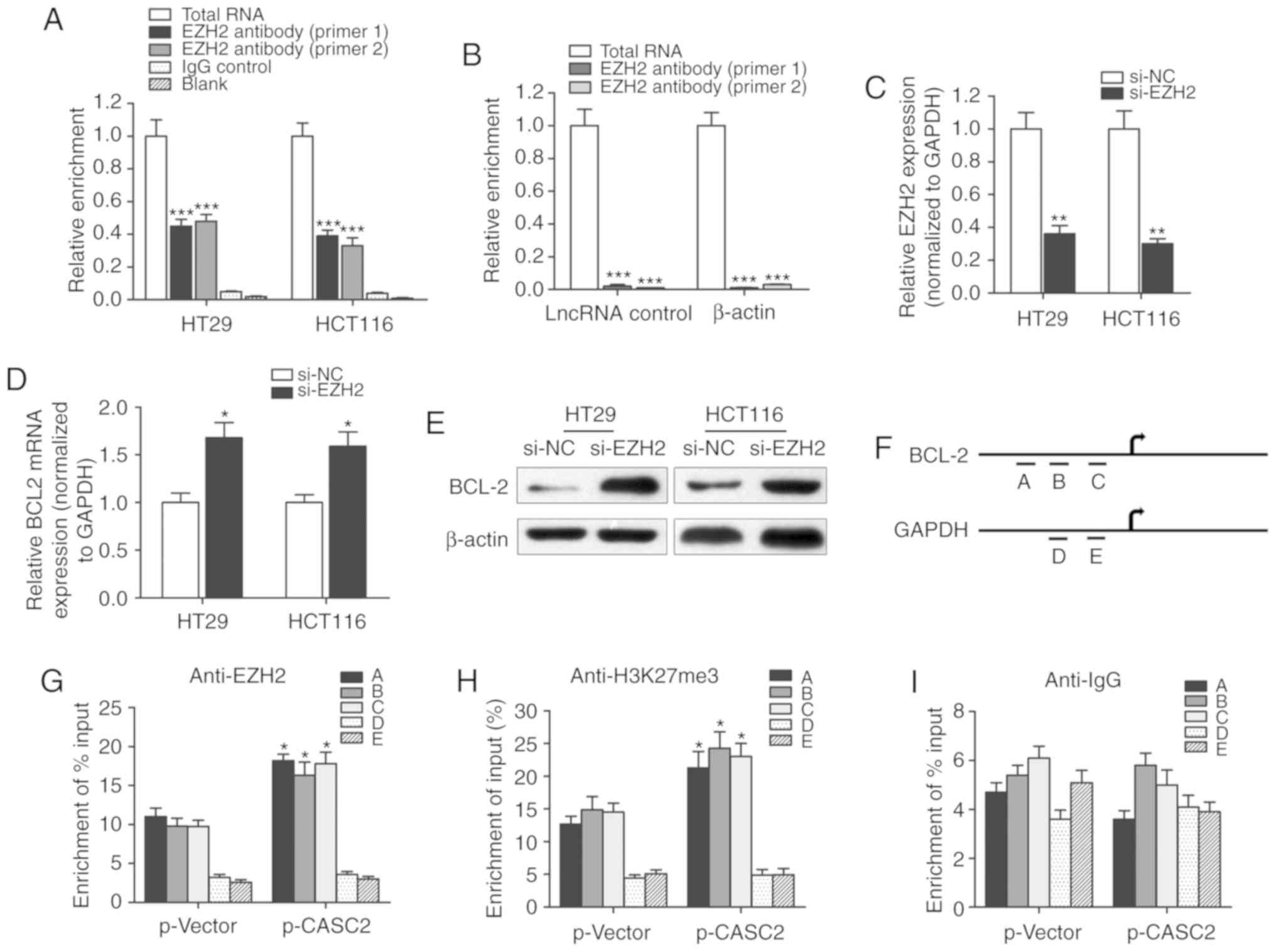 | Figure 6.lncRNA CASC2 decreased the expression
level of BCL2 by binding with EZH2. (A) RIP experiments were
performed to verify the direct interaction between EZH2 and lncRNA
CASC2 by using an anit-EZH2 antibody. Primers used for amplifying
CASC2 were generated to verify the enrichment of lncRNA CASC2
pulled down by the anti-EZH2 antibody. ***P<0.001 vs. control
IgG group. (B) lncRNA control or β-actin was used as a negative
control for CASC2. A RIP assay was performed using EZH2 antibody.
***P<0.001 vs. total RNA group. (C) The silencing effect of
siRNA against EZH2 was determined via quantitative PCR. **P<0.01
vs. si-NC group. BCL2 (D) mRNA and (E) protein levels were measured
in cells with EZH2 knockdown. *P<0.05 vs. si-NC group. (F)
Chromatin immunoprecipitation analysis of HT29 cells overexpressing
CASC2 was performed with generated primers to detect the
amplification at the BCL2 promoter (detection sites A, B and C) and
GAPDH promoter (detection sites D and E) regions. Measurement of
BCL2 sequences with (G) EZH2, (H) H3K27m3 and (I) IgG is presented
as relative to total input. *P<0.05 compared with the p-Vector
group, respectively. lncRNA, long non-coding RNA; CASC2, CASC2,
cancer susceptibility 2; EZH2, enhancer of zeste 2 polycomb
repressive complex 2 subunit; RIP, RNA immunoprecipitation; siRNA,
small interfering RNA; NC, negative control; H3K27m3, H3 lysine 27
trimethylation. |
Discussion
Patients with advanced colon cancer that develop
resistance to chemotherapy currently have limited therapeutic
options in the clinic; therefore, many patients turn to alternative
treatments (29). In recent years,
the interest in herbal remedies has grown rapidly in the
industrialized world, s these drugs are increasingly considered to
be effective and safe alternatives to synthetic drugs. The present
study focused on berberine, one of the few well-established plant
products supported by clinical trials (30,31),
which is the standardized extract from Coptis chinensis. The
results obtained in the current study indicated that berberine
inhibited the viability and promoted apoptosis of CRC cell lines.
High throughput RNA sequencing identified lncRNA CASC2 as a
potential functional target of berberine, and that CASC2 is
involved in the chemopreventive functions of berberine. The
antiapoptotic protein BCL2 was investigated and it was revealed
that lncRNA CASC2 exerted its effect by suppressing BCL2 expression
via EZH2.
Alkaloids are used in traditional medicine for the
treatment of many diseases. These compounds are synthesized in
plants as secondary metabolites and have several effects on
cellular metabolism (32). The
medicinal alkaloid isolated from Coptis chinensis,
berberine, has attracted attention in the scientific community
(33). For example, Piyanuch et
al (34) demonstrated that
berberine upregulated the expression levels of growth
differentiation factor 15 and activating transcription factor 3 in
colorectal cancer. Liu et al (35) suggested that berberine may exert
antitumor effects in CRC by regulating miR-429. Chuang et al
(36) indicated an anticancer role
of berberine via the suppression of cell viability of
hepatocellular cancer cells and the upregulation of the protein
expression of multiple tumor suppressor genes. The role of
berberine in CRC was further explored by Liu et al (14), which revealed that berberine
inhibited the invasion and metastasis of CRC cells via the
prostaglandin-endoperoxide synthase 2/prostaglandin E2 mediated
Janus kinase 2/STAT3 signaling pathway. However, the mechanism
underling the cytotoxic effect of berberine and whether one or a
cluster of lncRNAs are involved in this process remains unknown.
The present study suggested that berberine suppresses the viability
of CRC cell lines and promotes cell apoptosis in a dose-dependent
manner. Moreover, RNA sequencing indicated that a number of lncRNAs
may be important regulators of the berberine-dependent pathway.
During the past years, miRNAs and lncRNAs have been
investigated as potential diagnostic predictors or therapeutic
targets for a number of different diseases (37). lncRNAs may have novel regulatory
roles in cancer and may serve as potential prognostic and
therapeutic targets in clinical practice (38). However, the specific functional
association between lncRNAs and berberine in cancer is not well
known. Therefore, the current study sought to identify and validate
specific lncRNAs, which may be important for the antitumor effects
exhibited by berberine. By combining RNA sequencing array with
RT-qPCR validation, lncRNA CASC2 was identified as an lncRNA that
was regulated by berberine treatment. Furthermore, the gain- and
loss-of-function assays revealed that berberine exerted its
anticancer effect by upregulating lncRNA CASC2 expression.
lncRNA CASC2, a novel human lncRNA mapping to 10q26
in humans, has been originally characterized as a downregulated
gene acting as a tumor suppressor gene in endometrial cancer
(39), and recently in other
cancers (25,40,41).
Exogenous expression of CASC2 significantly inhibited the growth of
undifferentiated endometrial cancer cells (39). Fan et al (42) revealed that CASC2 promoted the
apoptosis of hepatocellular carcinoma cells by targeting miR-24. Ba
et al (43) reported that
the decreased expression of lncRNA CASC2 facilitated osteosarcoma
growth and invasion by regulating miR-181a. Huang et al
(44) demonstrated that lncRNA
CASC2 inhibited CRC cell proliferation and tumor growth by sponging
miR-18a and silencing the STAT3 gene. The aforementioned studies
demonstrated a decreased expression of CASC2 in cancer tissues
compared with normal tissues. The results obtained in the current
study revealed that CASC2 was downregulated in CRC cells compared
with normal epithelial cells, consistent with the aforementioned
studies. Furthermore, the current study investigated the regulatory
mechanism that may account for the role of CASC2 following
treatment with berberine. The antiapoptotic gene, BCL2, was
identified as a functional target of CASC2. Gain- and
loss-of-function experiments revealed that berberine exerted its
proapoptotic effect via a CASC2-BCL2 signaling mechanism.
Antiapoptosis is established as an important factor
in the development of drug resistance, and disruptions of the
apoptotic pathway have been demonstrated to suppress cell
cytotoxicity and promote drug resistance (45). BCL2 was identified as the most
dysregulated protein following silencing of lncRNA CASC2 in the
current study. BCL-2 family proteins, which include BCL-2, BCL-XL,
BCL-W, MCL1 apoptosis regulator BCL2 family member and BCL2 related
protein A1, share structural homology in the BCL2 homology 1, 2, 3
and 4 domains. BCL2 is an important cell death regulator, and is
involved in the control of the release of cytochrome c from
mitochondria in the intrinsic apoptotic pathway (46–48).
Thus, the current study investigated whether lncRNA CASC2 promoted
apoptosis via antiapoptotic gene, BCL2. Currently, the specific
mechanism by which lncRNA CASC2 regulates BCL2 remains unclear.
Consequently, the current study focused on EZH2, which is
frequently associated with other lncRNAs and affects cancer
progression by altering H3K27 trimethylation and silencing
transcription. lncRNAs may exert their function by recruiting
RNA-binding portions, including EZH2, thus leading to gene
methylation and chromatin modifications. EZH2 is often
overexpressed in different types of human cancer and may promote
cell proliferation, invasion and tumor angiogenesis. The RIP
experiment in the current study revealed that lncRNA CASC2 was
physically associated with EZH2 in CRC cells. It is known that EZH2
enhances methylation of H3K27, leading to gene silencing involved
in cancer progression and cell apoptosis (49). This accords with the ChIP results
of the present study, which indicated that overexpression of CASC2
increased binding of EZH2 at the promoter region of the BCL2 gene,
causing elevated H3K27me3 formation, and potentially inhibiting the
antiapoptotic effect of BCL2.
The present study had limitations. The cytotoxic
effect of berberine requires further validation in in vivo
models. Experimental conditions for the treatment of CRC tumors
with berberine in mice, including the concentration, duration of
treatment and administration route of berberine are to be
determined. The diagnostic and prognostic functions of CASC2
expression in patients with CRC receiving berberine treatment
remain unclear. Future experiments focused on the clinical
importance of CASC2 in berberine treatment are required.
In conclusion, the current study revealed that
berberine decreases cell viability and promote apoptosis of CRC
cell lines in vitro. Moreover, mechanistic research
suggested that berberine exerted its anticancer effect in CRC by
increasing the expression level of lncRNA CASC2 and suppressing
BCL2 in an EZH2-dependent manner. Thus, berberine may be a
potential alternative treatment drug for patients with CRC, and
lncRNA CASC2 may serve as an important therapeutic target to
improve the anticancer effect of berberine.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science
Foundation of the Natural Science Foundation of China (grant no.
81301399) and the Sichuan Science and Technology Department Key
Research and Development Project (grant no. 2017SZ0122).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WD and LM acquired the data, and created a draft of
the manuscript; YC and YL collected clinical samples and performed
the experimental assays; PC, HX and XW analyzed and interpreted the
data, and performed statistical analysis; WD, LM, YC and HX
reviewed the manuscript, and produced the figures and tables. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Clinical
Research Ethics Committee of the West China Second University
Hospital. All participants signed informed consent prior to using
the tissues for scientific research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Engholm G, Kejs AM, Brewster DH, Gaard M,
Holmberg L, Hartley R, Iddenden R, Møller H, Sankila R, Thomson CS
and Storm HH: Colorectal cancer survival in the Nordic countries
and the United Kingdom: Excess mortality risk analysis of 5 year
relative period survival in the period 1999 to 2000. Int J Cancer.
121:1115–1122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jansen MC, Bueno-de-Mesquita HB, Buzina R,
Fidanza F, Menotti A, Blackburn H, Nissinen AM, Kok FJ and Kromhout
D: Dietary fiber and plant foods in relation to colorectal cancer
mortality: The Seven Countries Study. Int J Cancer. 81:174–179.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tomida C, Aibara K, Yamagishi N, Yano C,
Nagano H, Abe T, Ohno A, Hirasaka K, Nikawa T and Teshima-Kondo S:
The malignant progression effects of regorafenib in human colon
cancer cells. J Med Invest. 62:195–198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goldberg RM, Sargent DJ, Morton RF, Fuchs
CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC and Alberts
SR: A randomized controlled trial of fluorouracil plus leucovorin,
irinotecan, and oxaliplatin combinations in patients with
previously untreated metastatic colorectal cancer. J Clin Oncol.
22:23–30. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hiraki M, Nishimura J, Takahashi H, Wu X,
Takahashi Y, Miyo M, Nishida N, Uemura M, Hata T, Takemasa I, et
al: Concurrent targeting of KRAS and AKT by miR-4689 Is a novel
treatment against mutant KRAS colorectal cancer. Mol Ther Nucleic
Acids. 4:e2312015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wasserman I, Lee LH, Ogino S, Marco MR, Wu
C, Chen X, Datta J, Sadot E, Szeglin B, Guillem JG, et al: smad4
loss in colorectal cancer patients correlates with recurrence, loss
of immune infiltrate, and chemoresistance. Clin Cancer Res.
25:1948–1956. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li D, Zhang Y, Liu K, Zhao Y, Xu B, Xu L,
Tan L, Tian Y, Li C, Zhang W, et al: Berberine inhibits
colitis-associated tumorigenesis via suppressing inflammatory
responses and the consequent EGFR signaling-involved tumor cell
growth. Lab Invest. 97:1343–1353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li G, Zhao M, Qiu F, Sun Y and Zhao L:
Pharmacokinetic interactions and tolerability of berberine chloride
with simvastatin and fenofibrate: An open-label, randomized,
parallel study in healthy Chinese subjects. Drug Des Devel Ther.
13:129–139. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Choi MS, Yuk DY, Oh JH, Jung HY, Han SB,
Moon DC and Hong JT: Berberine inhibits human neuroblastoma cell
growth through induction of p53-dependent apoptosis. Anticancer
Res. 28:3777–3784. 2008.PubMed/NCBI
|
|
10
|
Wang N, Feng Y, Zhu M, Tsang CM, Man K,
Tong Y and Tsao SW: Berberine induces autophagic cell death and
mitochondrial apoptosis in liver cancer cells: The cellular
mechanism. J Cell Biochem. 111:1426–1436. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Y, Wang K, Gu C, Yu G, Zhao D, Mai
W, Zhong Y, Liu S, Nie Y and Yang H: Berberine, a natural plant
alkaloid, synergistically sensitizes human liver cancer cells to
sorafenib. Oncol Rep. 40:1525–1532. 2018.PubMed/NCBI
|
|
12
|
Li J, Liu F, Jiang S, Liu J, Chen X, Zhang
S and Zhao H: Berberine hydrochloride inhibits cell proliferation
and promotes apoptosis of non-small cell lung cancer via the
suppression of the MMP2 and Bcl-2/Bax signaling pathways. Oncol
Lett. 15:7409–7414. 2018.PubMed/NCBI
|
|
13
|
Wu K, Yang Q, Mu Y, Zhou L, Liu Y, Zhou Q
and He B: Berberine inhibits the proliferation of colon cancer
cells by inactivating Wnt/β-catenin signaling. Int J Oncol.
41:292–298. 2012.PubMed/NCBI
|
|
14
|
Liu X, Ji Q, Ye N, Sui H, Zhou L, Zhu H,
Fan Z, Cai J and Li Q: Berberine inhibits invasion and metastasis
of colorectal cancer cells via COX-2/PGE2 mediated JAK2/STAT3
signaling pathway. PLoS One. 10:e01234782015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tuck AC and Tollervey D: A
transcriptome-wide atlas of RNP composition reveals diverse classes
of mRNAs and lncRNAs. Cell. 154:996–1009. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gong C and Maquat LE: lncRNAs
transactivate STAU1-mediated mRNA decay by duplexing with 3′UTRs
via Alu elements. Nature. 470:284–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Engreitz JM, Pandya-Jones A, McDonel P,
Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander
ES, et al: The Xist lncRNA exploits three-dimensional genome
architecture to spread across the X chromosome. Science.
341:12379732013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gayen S, Maclary E, Buttigieg E, Hinten M
and Kalantry S: A primary role for the Tsix lncRNA in maintaining
random X-chromosome inactivation. Cell Rep. 11:1251–1265. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bai WL, Zhao SJ, Wang ZY, Zhu YB, Dang YL,
Cong YY, Xue HL, Wang W, Deng L, Guo D, et al: lncRNAs in secondary
hair follicle of cashmere goat: Identification, expression, and
their regulatory network in Wnt signaling pathway. Anim Biotechnol.
29:199–211. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mullin NK, Mallipeddi NV, Hamburg-Shields
E, Ibarra B, Khalil AM and Atit RP: Wnt/β-catenin signaling pathway
regulates specific lncRNAs that impact dermal fibroblasts and skin
fibrosis. Front Genet. 8:1832017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bester AC, Lee JD, Chavez A, Lee YR,
Nachmani D, Vora S, Victor J, Sauvageau M, Monteleone E, Rinn JL,
et al: An integrated genome-wide CRISPRa approach to functionalize
lncRNAs in drug resistance. Cell. 173:649–664 e20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu B, Xu M, Shi H, Gao X and Liang P:
Genome-wide identification of lncRNAs associated with
chlorantraniliprole resistance in diamondback moth Plutella
xylostella (L.). BMC Genomics. 18:3802017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang L, Geng Z, Meng X, Meng F and Wang
L: Screening for key lncRNAs in the progression of gallbladder
cancer using bioinformatics analyses. Mol Med Rep. 17:6449–6455.
2018.PubMed/NCBI
|
|
25
|
Li P, Xue WJ, Feng Y and Mao QS: Long
non-coding RNA CASC2 suppresses the proliferation of gastric cancer
cells by regulating the MAPK signaling pathway. Am J Transl Res.
8:3522–3529. 2016.PubMed/NCBI
|
|
26
|
Yu Y, Liang S, Zhou Y, Li S, Li Y and Liao
W: HNF1A/CASC2 regulates pancreatic cancer cell proliferation
through PTEN/Akt signaling. J Cell Biochem. 120:2816–2827. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Durai R, Yang SY, Sales KM, Seifalian AM,
Goldspink G and Winslet MC: Insulin-like growth factor binding
protein-4 gene therapy increases apoptosis by altering Bcl-2 and
Bax proteins and decreases angiogenesis in colorectal cancer. Int J
Oncol. 30:883–888. 2007.PubMed/NCBI
|
|
28
|
Jiang M and Milner J: Bcl-2 constitutively
suppresses p53- dependent apoptosis in colorectal cancer cells.
Genes Dev. 17:832–837. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pretner E, Amri H, Li W, Brown R, Lin CS,
Makariou E, Defeudis FV, Drieu K and Papadopoulos V: Cancer-related
overexpression of the peripheral-type benzodiazepine receptor and
cytostatic anticancer effects of Ginkgo biloba extract (EGb 761).
Anticancer Res. 26:9–22. 2006.PubMed/NCBI
|
|
30
|
Ming J, Xu S, Liu C, Liu X, Jia A and Ji
Q: Effectiveness and safety of bifidobacteria and berberine in
people with hyperglycemia: Study protocol for a randomized
controlled trial. Trials. 19:722018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Sun J, Zhang YJ, Chai QY, Zhang
K, Ma HL, Wu XK and Liu JP: The effect of berberine on insulin
resistance in women with polycystic ovary syndrome: Detailed
statistical analysis plan (SAP) for a multicenter randomized
controlled trial. Trials. 17:5122016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wink M: Quinolizidine alkaloids:
Biochemistry, metabolism, and function in plants and cell
suspension cultures. Planta Med. 53:509–514. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Q, Piao XL, Piao XS, Lu T, Wang D
and Kim SW: Preventive effect of Coptis chinensis and berberine on
intestinal injury in rats challenged with lipopolysaccharides. Food
Chem Toxicol. 49:61–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Piyanuch R, Sukhthankar M, Wandee G and
Baek SJ: Berberine, a natural isoquinoline alkaloid, induces NAG-1
and ATF3 expression in human colorectal cancer cells. Cancer Lett.
258:230–240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu H, Huang C, Wu L and Wen B: Effect of
evodiamine and berberine on miR-429 as an oncogene in human
colorectal cancer. Onco Targets Ther. 9:4121–4127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chuang TY, Wu HL, Min J, Diamond M, Azziz
R and Chen YH: Berberine regulates the protein expression of
multiple tumorigenesis-related genes in hepatocellular carcinoma
cell lines. Cancer Cell Int. 17:592017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tukiainen T, Villani AC, Yen A, Rivas MA,
Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A,
et al: Landscape of X chromosome inactivation across human tissues.
Nature. 550:244–248. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sarfi M, Abbastabar M and Khalili E: Long
noncoding RNAs biomarker-based cancer assessment. J Cell Physiol.
Mar 5–2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Baldinu P, Cossu A, Manca A, Satta MP,
Sini MC, Rozzo C, Dessole S, Cherchi P, Gianfrancesco F, Pintus A,
et al: Identification of a novel candidate gene, CASC2, in a region
of common allelic loss at chromosome 10q26 in human endometrial
cancer. Hum Mutat. 23:318–326. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gao Z, Wang H, Li H, Li M, Wang J, Zhang
W, Liang X, Su D and Tang J: Long non-coding RNA CASC2 inhibits
breast cancer cell growth and metastasis through the regulation of
the miR-96-5p/SYVN1 pathway. Int J Oncol. 53:2081–2090.
2018.PubMed/NCBI
|
|
41
|
Xue Z, Zhu X and Teng Y: Long non-coding
RNA CASC2 inhibits progression and predicts favorable prognosis in
epithelial ovarian cancer. Mol Med Rep. 18:5173–5181.
2018.PubMed/NCBI
|
|
42
|
Fan JC, Zeng F, Le YG and Xin L: lncRNA
CASC2 inhibited the viability and induced the apoptosis of
hepatocellular carcinoma cells through regulating miR-24-3p. J Cell
Biochem. 119:6391–6397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ba Z, Gu L, Hao S, Wang X, Cheng Z and Nie
G: Downregulation of lncRNA CASC2 facilitates osteosarcoma growth
and invasion through miR-181a. Cell Prolif. 51:2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang G, Wu X, Li S, Xu X, Zhu H and Chen
X: The long noncoding RNA CASC2 functions as a competing endogenous
RNA by sponging miR-18a in colorectal cancer. Sci Rep. 6:265242016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Johnson MI and Hamdy FC: Apoptosis
regulating genes in prostate cancer (review). Oncol Rep. 5:553–557.
1998.PubMed/NCBI
|
|
46
|
Waters JS, Webb A, Cunningham D, Clarke
PA, Raynaud F, di Stefano F and Cotter FE: Phase I clinical and
pharmacokinetic study of bcl-2 antisense oligonucleotide therapy in
patients with non-Hodgkin's lymphoma. J Clin Oncol. 18:1812–1823.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lessene G, Czabotar PE and Colman PM:
BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov.
7:989–1000. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen Y, Zhou R, Yi Z, Li Y, Fu Y, Zhang Y,
Li P, Li X and Pan Y: Porphyromonas gingivalis induced inflammatory
responses and promoted apoptosis in lung epithelial cells infected
with H1N1 via the Bcl-2/Bax/caspase-3 signaling pathway. Mol Med
Rep. 18:97–104. 2018.PubMed/NCBI
|
|
49
|
Yoo KH and Hennighausen L: EZH2
methyltransferase and H3K27 methylation in breast cancer. Int J
Biol Sci. 8:59–65. 2012. View Article : Google Scholar : PubMed/NCBI
|