Introduction
Respiratory infections caused by various viruses,
such as influenza, respiratory syncytial virus, parainfluenza,
rhinovirus and human coronavirus (including severe acute
respiratory syndrome coronavirus 2), pose significant public health
challenges (1). Currently,
specific antiviral treatments for a number of these viruses are
lacking, often leaving symptomatic therapy as the primary approach
for managing viral respiratory infections. Furthermore, the ongoing
threat of potential pandemics underscores the need to explore novel
approaches to combat pulmonary viral infections. Because epithelial
cells in the respiratory tract are vulnerable to various viral
pathogens, a detailed understanding of the immune system in the
airway epithelium is essential for developing new therapeutic
strategies.
The innate immune system is a frontline defense
against invading microbes, orchestrating subsequent adaptive immune
responses (2). Recognition of
pathogen-associated molecular patterns by pattern-recognition
receptors triggers the initiation of innate immune responses. Among
these receptors, Toll-like receptor 3 (TLR3) serves as a key
receptor for virus-derived double-stranded RNA (dsRNA) (3), and is prominently expressed in airway
epithelial cells. Activation of TLR3 by dsRNA leads to the
activation of NF-κB and interferon regulatory factor 3, resulting
in the production of cytokines and chemokines that are pivotal in
orchestrating immune and inflammatory responses (4–6).
Central to this antiviral defense mechanism is the induction of
type I interferons (IFNs) by TLR3 signaling, with IFN-β emerging as
a principal type I IFN in bronchial epithelial cells (7). Following IFN-β production, hundreds
of IFN-stimulated genes (ISGs) are upregulated, forming a complex
network that modulates both innate and adaptive immune responses
(8). For example, IFN-β induces
the expression of C-X-C motif chemokine ligand 10 (CXCL10), a
chemokine that promotes lymphocyte chemotaxis (9), and melanoma
differentiation-associated gene 5 (MDA5), a dsRNA receptor and
signaling molecule (10).
Furthermore, ISG56 and ISG60 have also been speculated to
contribute to antiviral immunity alongside other ISGs (11).
Despite extensive studies (12,13)
on the molecular mechanisms underlying inflammatory responses and
innate antiviral immunity in airway epithelial cells, the specific
roles of ISGs remain incompletely understood. ISG56, also known as
IFN-induced protein with tetratricopeptide repeats 1 (IFIT1),
encodes a protein with a helix-turn-helix four-repeat motif that
mediates various antiviral responses (11). A previous study has shown that
ISG56 mediates CXCL10 expression in BEAS-2B bronchial epithelial
cells during TLR3 signaling (14).
ISG60 is another IFIT protein, also known as retinoic
acid-inducible gene G (15) and
IFIT3 (16), which induces IFN-β
signaling and suppresses adenovirus early gene expression in
alveolar adenocarcinoma cell-derived 293FT cells, A549 cells and
immortalized normal human diploid fibroblasts (17). These studies suggest that ISG60 may
have a crucial role in host defense against adenovirus infection;
however, the mechanisms by which ISG60 exerts its antiviral
function via TLR3 signaling in airway epithelial cells remain
unknown. The present study aimed to investigate the function and
expression of ISG60 in BEAS-2B cells primed with a TLR3 ligand,
polyinosinic-polycytidylic acid (poly IC).
Materials and methods
Cell culture
Noncancerous bronchial epithelial cell-derived
BEAS-2B cells (American Type Culture Collection) were cultured at
37°C and 5% CO2 in Dulbecco's modified Eagle's medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (Biowest), 100 U/ml penicillin, 100 µg/ml streptomycin B and
250 ng/ml Amphotericin B (Nacalai Tesque, Inc.) as previously
described (14). Poly IC (cat. no.
P9582; MilliporeSigma) was added to the cell culture medium for
treatment at 37°C. To examine the concentration-dependent effect of
poly IC, the cells were treated with 3, 5, 10, 30 and 50 µg/ml poly
IC for 16 h (for mRNA expression analysis) or 24 h (for protein
expression analysis). In time course experiments, the cells were
treated with 30 µg/ml poly IC for 2, 4, 8, 16 and 24 h. For RNA
interference, cells at 50–70% density cultured for 24 h in
antibiotic-free medium were transfected at 37°C with either
negative control small interfering RNA (siRNA) (cat. no. 1027281;
Qiagen GmbH), or siRNAs against IFN-β (custom synthesized by
Invitrogen; Thermo Fisher Scientific, Inc.) (18), ISG56 (cat. no. SI02660777; Qiagen
GmbH) or ISG60 (cat. no. SI04197788 as ISG60 si-1; cat. no.
SI03152737 as ISG60 si-2; Qiagen GmbH) at a concentration of 10
pmol/ml using Lipofectamine® RNAiMAX (Invitrogen; Thermo
Fisher Scientific, Inc.). After 4 h of transfection, the medium was
changed, and the cells were subjected to subsequent experiments 48
h after transfection. The siRNA sequences for IFN-β, ISG56 and
ISG60 were as follows. IFN-β siRNA, sense
5′-CCAUGAGCUACAACUUGCUUGGAUU-3′, antisense
5′-AAUCCAAGCAAGUUGUAGUCUAUGG-3′; ISG56 siRNA, sense
5′-GGAUCAGAUUGAAUUCCUATT-3′, antisense 5′-UAGGAAUUCAAUCUGAUCCAA-3′;
ISG60 siRNA-1, sense 5′-AGAUGAUUGAAGCACUAAATT-3′, antisense
UUUAGUGCUUCAAUCAUCUCT-3′; ISG60 siRNA-2, sense
5′-GUCAUGGACUAUUCGAAUATT-3′ and antisense
5′-UAUUCGAAUAGUCCAUAGCAT-3′. The cells were also treated with
recombinant human [r(h)]IFN-β (ProSpec-Tany TechnoGene Ltd.) at
37°C for 16 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA was extracted from the cultured cells using an
Illustra RNAspin kit (GE Healthcare). To denature RNA, the RNA
solution was heated at 70°C for 5 min and was then cooled
immediately on ice. Subsequently, RT was performed at 37°C for 1 h
using oligo(dT)18 primer (FASMAC), M-MLV reverse
transcriptase (Thermo Fisher Scientific, Inc.) and dNTP mix (Thermo
Fisher Scientific, Inc.), followed by inactivation of the enzyme at
70°C for 10 min. The cDNA for 18S, CXCL10, IFN-β, ISG56, ISG60 and
MDA5 were amplified using SsoAdvanced Universal SYBR Green Supermix
(Bio-Rad Laboratories, Inc.) with a qPCR system (Bio-Rad
Laboratories, Inc.). The thermocycling conditions were as follows:
Pre-denaturation at 95°C for 30 sec; followed by 40 cycles at 95°C
for 10 sec, 60°C for 10 sec and 72°C for 20 sec; and final
extension at 72°C for 5 min. 18S ribosomal RNA was utilized as an
internal control. The 2−ΔΔCq method (19) was used for quantification. CXCL10
and IFN-β mRNA expression levels are presented in arbitrary units,
as these genes were not detectable in the cells not stimulated with
poly IC. The mRNA expression levels of the other genes are
expressed as a fold increase compared with unstimulated cells. The
following primers (custom composites purchased from FASMAC) were
used: 18S (NR_146146.1), forward (F) 5′-ACTCAACACGGGAAACCTCA-3′,
reverse (R) 5′-AACCAGACAAATCGCTCCAC-3′; CXCL10 (NM_001565.4), F
5′-TTCAAGGAGTACCTCTCTCTAG-3′, R 5′-CTGGATTCAGACATCTCTTCTC-3′; IFN-β
(NM_002176.4), F 5′-CCTGTGGCAATTGAATGGGAGGC-3′, R
5′-CCAGGCACAGTGACTGTACTCCTT-3′; ISG56 (NM_001270930.2), F
5′-TAGCCAACATGTCCTCACAGAC-3′, R 5′-TCTTCTACCACTGGTTTCATGC-3′; ISG60
(NM_001031683.4), F: 5′-GAACATGCTGACCAAGCAGA-3′, R
5′-CAGTTGTGTCCACCCTTCCT-3′; MDA5 (XM_047445407.1), F
5′-GTTGAAAAGGCTGGCTGAAAAC-3′, R 5′-TCGATAACTCCTGAACCACTG-3′.
Western blotting
After cultivation, the cells were washed twice using
phosphate-buffered saline, scraped from the dish and Laemmli sample
buffer (Cell Signaling Technologies, Inc.) was used for protein
extraction. The BCA protein assay kit (FUJIFILM Wako Pure Chemical
Corporation) was used for protein quantification. Proteins (20
mg/lane) in the lysates were resolved using SDS-PAGE on 5–20% gels
(ATTO Corporation) and were transferred to polyvinylidene
difluoride membranes (Merck KGaA). The membranes were blocked with
Tris-buffered saline containing 5% nonfat dry milk at 25°C for 2 h,
and were then incubated with the following primary antibodies:
Rabbit anti-ISG56 (1:3,000; cat. no. GTX118713; GeneTex, Inc.),
rabbit anti-ISG60 (1:5,000; cat. no. GTX112442; GeneTex, Inc.),
rabbit anti-signal transducer and activator transcription 1 (STAT1)
(1:10,000; cat. no. sc-592; Santa Cruz Biotechnology, Inc.), mouse
anti-phosphorylated (P)-STAT1 (1:5,000; cat. no. sc-136229; Santa
Cruz Biotechnology, Inc.) and rabbit anti-actin IgG (1:5,000; cat.
no. A5060; MilliporeSigma) at 4°C overnight. After washing, the
membranes were incubated with horseradish peroxidase (HRP)-labeled
secondary anti-mouse (1:10,000; cat. no. A28177; Thermo Fisher
Scientific, Inc.) or anti-rabbit (1:10,000; cat. no. 458; Medical
& Biological Laboratories Co., Ltd.) IgG at 25°C for 1 h. The
Luminata Crescendo Western HRP Substrate (Merck KGaA) was utilized
for detection.
Enzyme-linked immunosorbent assay
(ELISA)
After incubation, the conditioned medium was
recovered and centrifuged at 10,000 × g at 4°C for 10 min. IFN-β
and CXCL10 protein concentrations in the collected supernatant were
assessed using commercial sandwich ELISA kits from PBL Assay
Science (cat. no. 41435-1) and R&D Systems, Inc. (cat. no.
DIP100), respectively. The assays were performed according to the
manufacturers' protocols.
Statistical analysis
The results of RT-qPCR and ELISA are shown using box
plots, which are presented as the median (center of the box), 25th
percentile (bottom of the box), 75th percentile (top of the box)
and range (minimum and maximum), or as the mean ± standard
deviation. The Mann-Whitney U-test, or one-way ANOVA and Dunnett's
post hoc test were utilized for statistical analyses using EZR
software version 1.65 (20)
(Saitama Medical Center, Jichi Medical University, Saitama, Japan).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of ISG60 in BEAS-2B cells
treated with poly IC
In unstimulated BEAS-2B cells, only small amounts of
ISG60 protein and mRNA were expressed. The protein and mRNA
expression levels of ISG60 increased in cells supplemented with
poly IC. At ≤10 µg/ml, poly IC did not affect the protein and mRNA
expression levels of ISG60, whereas ISG60 expression was markedly
upregulated in response to 30–50 µg/ml poly IC (Fig. 1A and B). Therefore, the cells were
supplemented with 30 µg/ml poly IC in subsequent experiments. The
mRNA expression levels of ISG60 were markedly increased at 4–16 h
and declined at 24 h in cells treated with poly IC (Fig. 1C). Furthermore, IFN-β mRNA was not
detected in unstimulated cells. Increased expression levels of
ISG60 protein were observed in cells treated with poly IC at 8 h,
which progressively increased until 24 h (Fig. 1D). Transient expression of IFN-β
mRNA was detected 2 h after poly IC treatment (Fig. 1E). IFN-β protein levels were
transiently detected in the conditioned medium from cells treated
with poly IC; they peaked at 4 h and quickly decreased thereafter
(Fig. 1F).
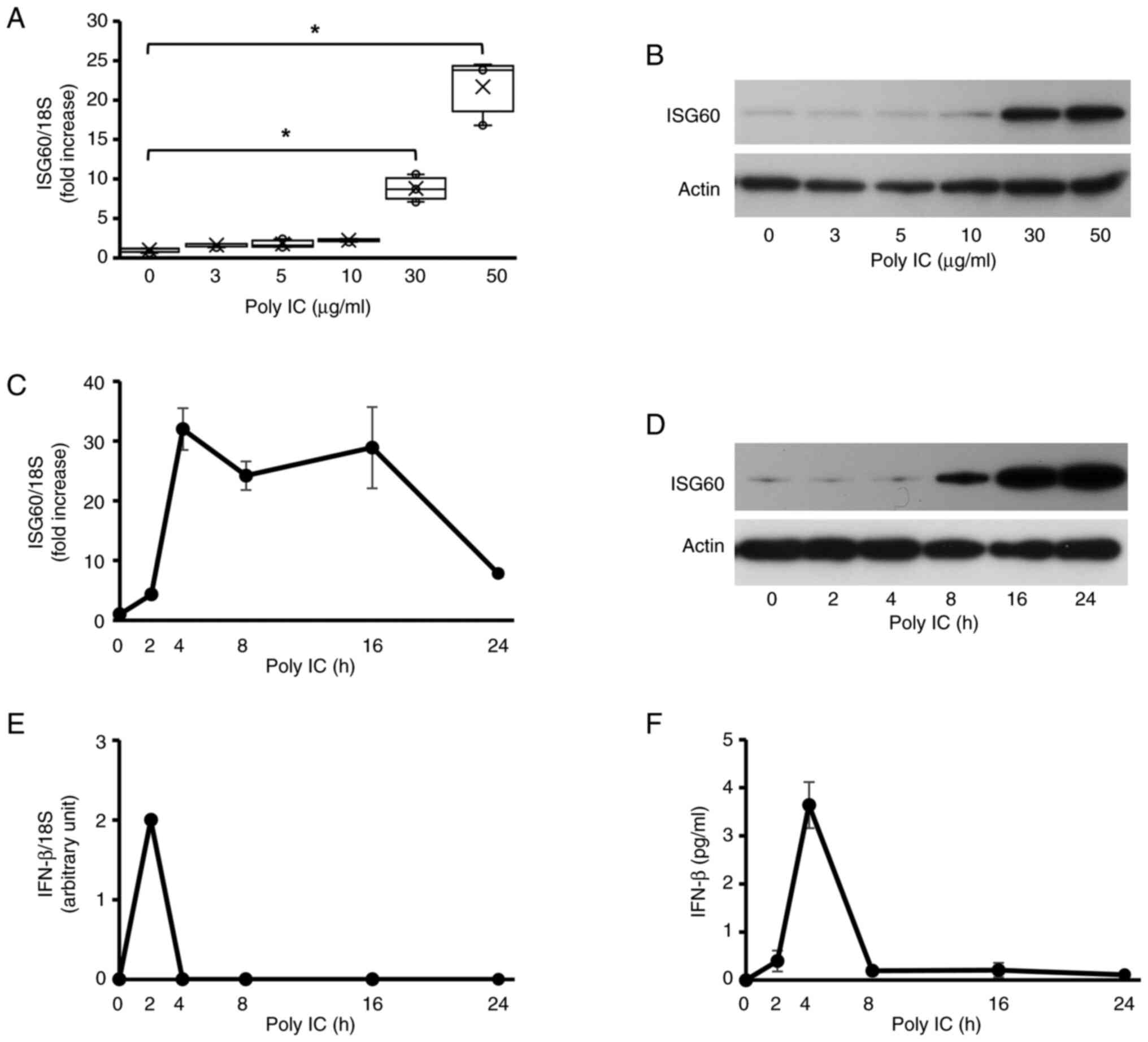 | Figure 1.Poly IC induces the expression of
ISG60 in BEAS-2B bronchial epithelial cells. (A and B) BEAS-2B
cells were cultured and stimulated with different concentrations of
the Toll-like receptor 3 agonist poly IC. (A) After 16 h, RNA was
extracted from the cells, and RT-qPCR analysis of ISG60 mRNA was
performed. Data are presented as box plots showing the median
(center of the box), 25th percentile (bottom of the box), 75th
percentile (top of the box) and range (minimum and maximum), and
were analyzed using one-way ANOVA and Dunnett's test, n=3.
*P<0.05. (B) Cellular proteins were extracted after 24 h, and
western blotting was performed for ISG60 and actin. (C-F) Cells
were stimulated with 30 µg/ml poly IC for up to 24 h. (C) ISG60
mRNA expression levels were detected using RT-qPCR analysis. (D)
ISG60 protein expression levels in cell lysates were detected using
western blotting. (E) IFN-b mRNA expression was examined using
RT-qPCR analysis. (F) IFN-β protein concentration in the
conditioned medium was measured using enzyme-linked immunosorbent
assay. (C, E and F) Data are presented as the mean ± SD (n=3).
IFN-β, interferon β; ISG60, IFN-stimulated gene 60; poly IC,
polyinosinic-polycytidylic acid; RT-qPCR, reverse
transcription-quantitative PCR. |
IFN-β serves a role in poly IC-induced
expression of ISG60 in BEAS-2B cells
As shown in Fig. 1E and
F, IFN-β mRNA and protein was not detected in cells without
poly IC stimulation, thus suggesting that no IFN-β mRNA and protein
are expressed in unstimulated BEAS-2B cells. In the medium
collected from cells transfected with control siRNA or IFN-β siRNA
without poly IC-stimulation, IFN-β protein was not detected
(Fig. 2A), and IFN-β mRNA
expression was also not detected in those cells (Fig. 2B). Transfection of BEAS-2B cells
with siRNA against IFN-β resulted in almost complete knockdown of
IFN-β protein and mRNA expression in poly IC-stimulated cells,
suggesting that transfection of IFN-β siRNA was successful
(Fig. 2A and B). In addition,
knockdown of IFN-β almost completely suppressed ISG60 mRNA
(Fig. 2C) and protein (Fig. 2D) expression upon poly IC
treatment. By contrast, treatment of BEAS-2B cells with 1 ng/ml
r(h)IFN-β increased the expression levels of ISG60 mRNA (Fig. 2E) and protein (Fig. 2F).
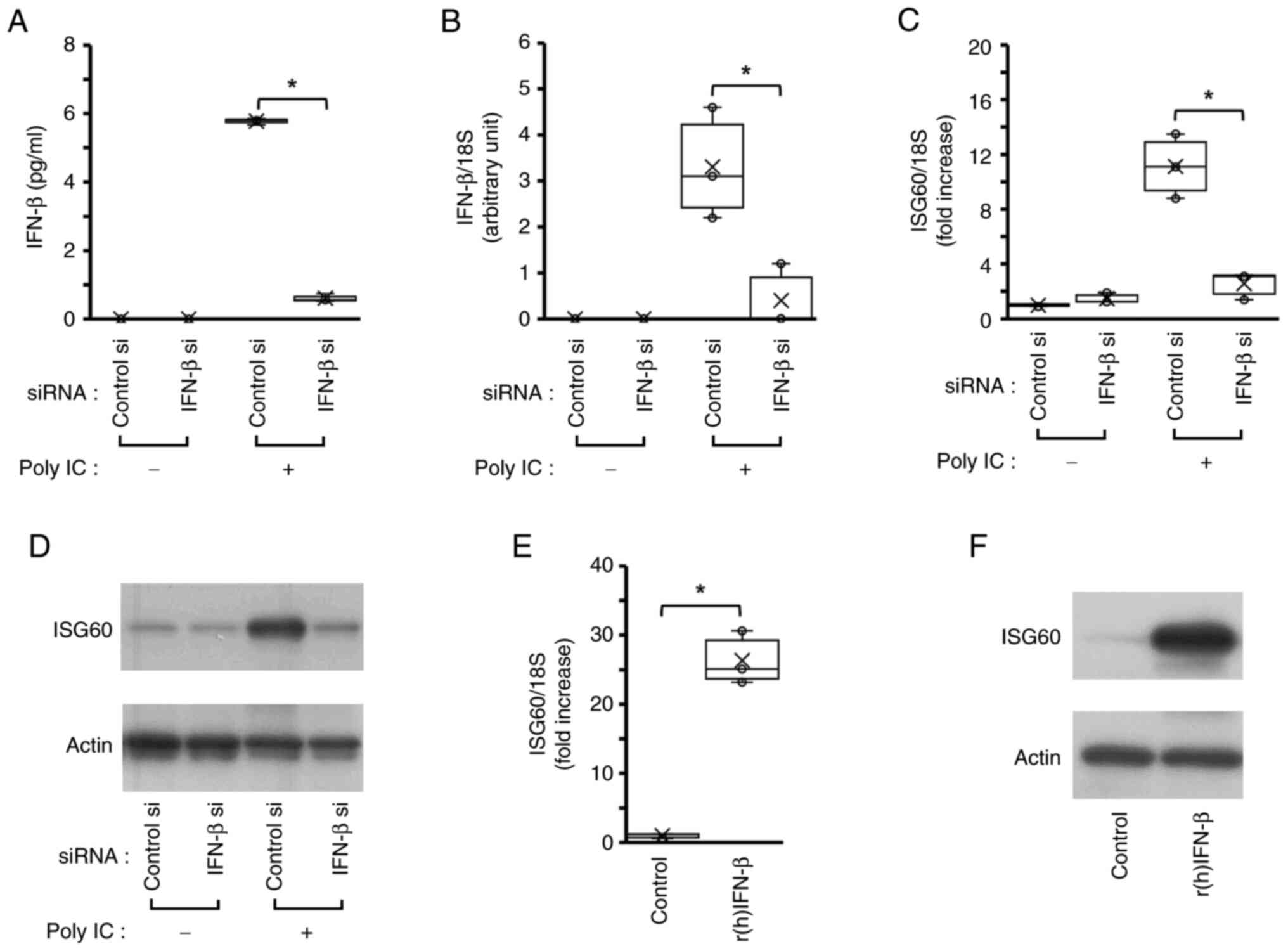 | Figure 2.IFN-β is involved in poly IC-induced
ISG60 expression. The cells were transfected with a specific siRNA
against IFN-β and were incubated for 24 h. Then, the cells were
treated with 30 µg/ml poly IC. (A) After 4 h incubation, the medium
was collected and IFN-β concentration was measured using an
enzyme-linked immunosorbent assay. (B) After 2 h incubation, RNA
was extracted and IFN-β mRNA expression was examined using RT-qPCR.
(C) After 16 h incubation, RNA was extracted, and ISG60 mRNA
expression was examined. (D) ISG60 protein expression was analyzed
using western blotting after 24 h. The cells were treated with
r(h)IFN-β for 16 h and were subjected to (E) RT-qPCR and (F)
western blotting. *P<0.05 using Mann-Whitney U-test; n=3. IFN-β,
interferon β; ISG, IFN-stimulated gene; poly IC,
polyinosinic-polycytidylic acid; r(h), recombinant human; RT-qPCR,
reverse transcription-quantitative PCR; si, small interfering. |
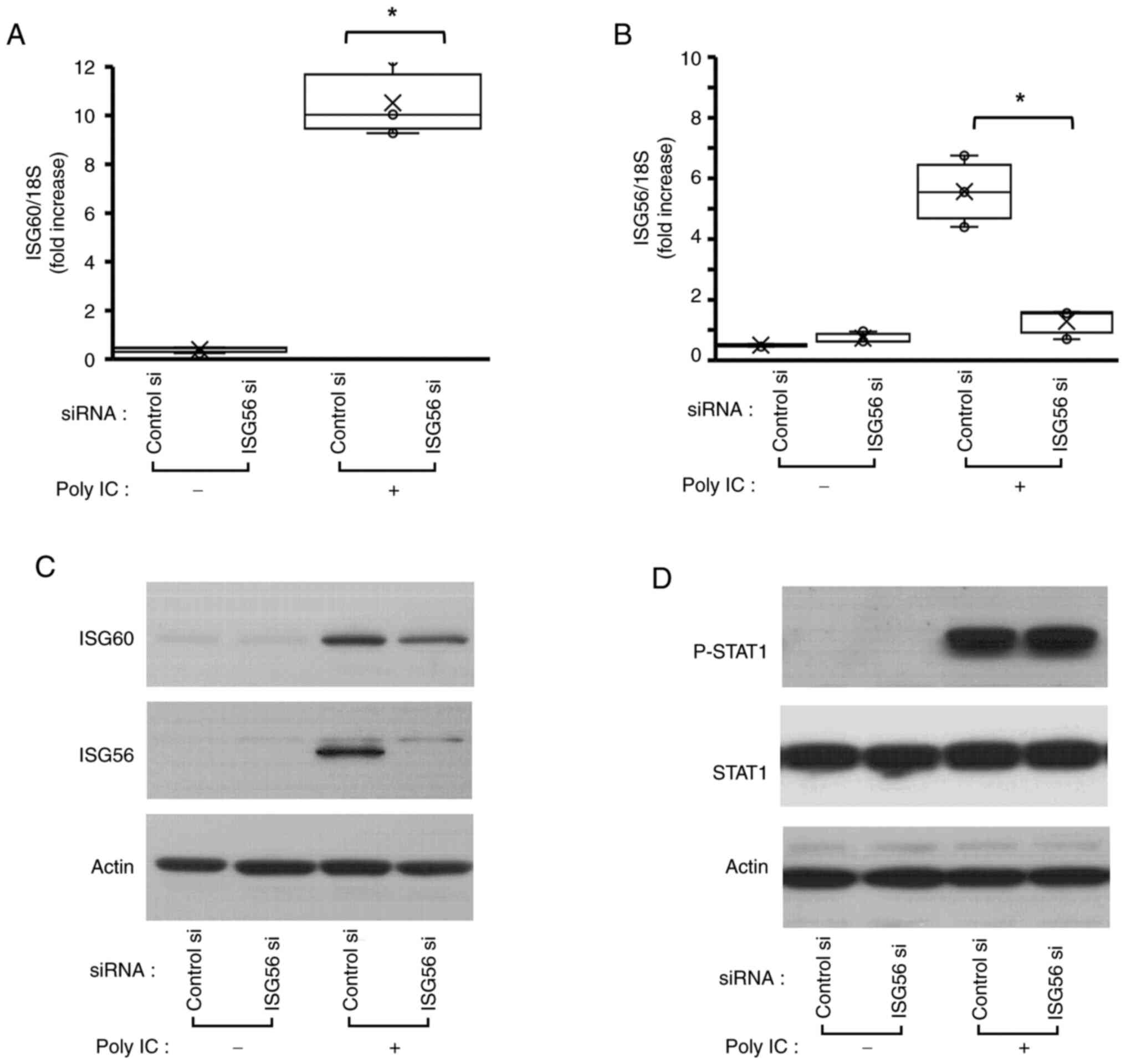
ISG56 is associated with ISG60
expression
The present study assessed the function of ISG56 on
ISG60 expression. ISG56 mRNA and protein knockdown was confirmed
using RT-qPCR (Fig. 3B) and
western blotting in poly IC-treated cells, respectively (Fig. 3C). Notably, there was no
significant difference between groups in unstimulated cells,
because ISG56 protein expression was not detectable in unstimulated
cells (Fig. 3C). ISG56 knockdown
markedly reduced poly IC-induced expression of ISG60 mRNA (Fig. 3A) and slightly reduced poly
IC-induced ISG60 protein (Fig.
3C). However, P-STAT1 expression was not affected by ISG56
knockdown (Fig. 3D).
ISG60 is implicated in poly
IC-mediated expression of ISG56 and CXCL10, but does not affect
IFN-β expression and STAT1 phosphorylation
Transfection of the cells with ISG60 si-1 and ISG60
si-2, followed by western blotting, demonstrated that these two
siRNAs markedly decreased poly IC-induced ISG60 expression,
suggesting that ISG60 expression was successfully knocked down
(Fig. 4A). Furthermore, ISG60
knockdown decreased poly IC-induced protein and mRNA expression
levels of ISG56 (Fig. 4A and D),
and protein concentration and mRNA expression levels of CXCL10
(Fig. 4C and D), whereas no
notable change was observed in P-STAT1 or STAT1 protein expression
(Fig. 4B). Furthermore, knockdown
of ISG60 using ISG60 si-1 did not affect the mRNA expression levels
of IFN-β (Fig. 4D). In order to
examine whether ISG60 selectively inhibits poly IC-induced
expression of ISG56 and CXCL10, or inhibits the expression of all
ISGs, the present study next examined the effect of ISG60 knockdown
on the poly IC-induced expression of another ISG, MDA5. MDA5 was
chosen because it is one of the key ISGs in antiviral innate immune
reactions (21). The results
revealed that poly IC-induced MDA5 mRNA expression was not affected
by transfection with ISG60 si-1 (Fig.
4D). These results suggested that ISG56 and CXCL10 were
selectively regulated by ISG60. Transfection of cells with ISG60
si-2 slightly decreased the mRNA expression levels of IFN-β and
MDA5, and this may be due to weak non-specific effects.
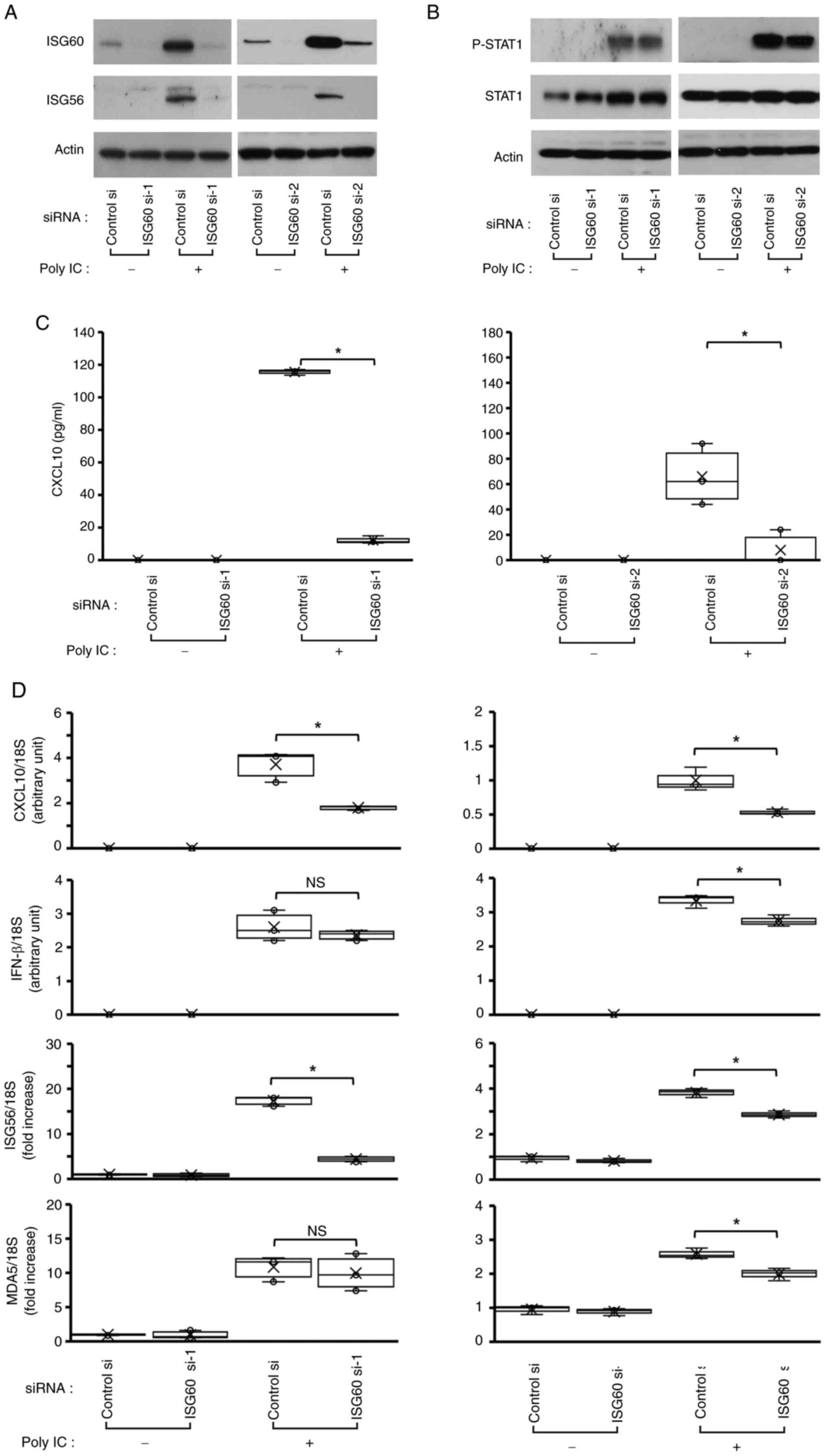 | Figure 4.ISG60 is partially involved in poly
IC-induced expression of CXCL10 and ISG56. The cells transfected
with two different ISG60 siRNAs were treated with poly IC. (A)
After incubating for 24 h, the cells were lysed and subjected to
western blotting for ISG56, ISG60 and actin. (B) After incubating
for 6 h, the cells were lysed and subjected to western blotting for
P-STAT1, STAT1 and actin, (C) After 24 h incubation, the
cell-conditioned medium was collected, and the concentration of
CXCL10 protein was measured using an enzyme-linked immunosorbent
assay. (D) After incubating for 2 h (for IFN-β mRNA analysis) or 16
h (for CXCL10, ISG56 and MDA5 mRNA analysis), RNA was extracted
from the cells, and reverse transcription-quantitative PCR was
performed. *P<0.05 using Mann-Whitney U-test; n=3. CXCL10, C-X-C
motif chemokine ligand 10; IFN-β, interferon β; ISG, IFN-stimulated
gene; MDA, melanoma differentiation-associated gene 5; NS, not
significant; P-, phosphorylated; poly IC,
polyinosinic-polycytidylic acid; si, small interfering; STAT1,
signal transducer and activator transcription 1. |
Discussion
Previous studies have shown that TLR3 is expressed
in various cell types, including dendritic cells (22), fibroblasts (23), intestinal epithelial cells
(24) and airway epithelial cells
(25). TLR3 serves a physiological
function in antiviral innate immunity and is involved in various
inflammatory conditions (26); for
example, TLR3-deficient mice have been reported to be susceptible
to encephalomyocarditis virus compared with wild-type mice, whereas
they were resistant to influenza virus (27). TLR3 activates the signal
transduction cascade after recognizing virus-derived dsRNA to
facilitate the transcription of type I IFNs. IFN-β is a type I IFN
mainly expressed in bronchial epithelial cells, which induces the
expression of ISGs that function in antiviral host defense
reactions.
The present study confirmed that the expression of
ISG60 in BEAS-2B cells was time- and concentration-dependent when
stimulated by poly IC. This finding is consistent with those of
previous studies showing poly IC-induced ISG60 expression in U373MG
astrocytoma cells (28) and
hCMEC/D3 human brain microvascular endothelial cells (29). These findings suggested that ISG60
may have a role in congenital antiviral immune responses in various
tissues, including bronchial epithelial cells. Because ISG60 is a
member of the ISG family, the present study investigated whether
de novo-produced IFN-β contributes to poly IC-induced ISG60
expression using RNA interference against IFN-b. As a result, IFN-β
knockdown largely suppressed poly IC-induced ISG60 expression, and
the induction of ISG60 protein and mRNA expression was observed in
BEAS-2B cells treated with r(h)IFN-β. These results indicated that
poly IC-induced ISG60 expression may be dependent on newly
synthesized IFN-β, and ISG60 was implicated in TLR3/IFN-β-mediated
antiviral reactions in BEAS-2B cells.
CXCL10, a CXC chemokine, is a robust chemoattractant
of T lymphocytes and natural killer cells, which serves various
roles in the pathogenesis of inflammatory diseases and infections
(30). The expression of CXCL10
must be tightly regulated to induce chemotaxis of appropriate
numbers of lymphocytes to the site of infection (31). Insufficient CXCL10 expression can
exacerbate viral infections, while excessive CXCL10 can lead to
excessive lymphocyte infiltration, resulting in tissue injury
(32). Moreover, CXCL10 protein
levels in serum and the severity of acute respiratory virus
infection have been shown to be correlated (33). Therefore, ‘fine-tuning’ CXCL10
expression is crucial for appropriate immune responses in the
airway. In the current study, ISG60 knockdown using two different
siRNAs partially decreased poly IC-induced CXCL10 mRNA expression
and protein levels. These findings indicated that ISG60 may not be
the primary regulator of CXCL10 expression, but may be implicated
in the ‘fine-tuning’ of TLR3-mediated CXCL10 expression in BEAS-2B
cells. This finding is consistent with that of a previous study in
U373 astrocytoma cells (28).
Phosphorylation of STAT1 is an important step in the
signaling pathway induced by IFN-β. In the present study, poly
IC-induced IFN-β expression and subsequent STAT1 phosphorylation
were not affected by ISG60 knockdown in BEAS-2B cells. This finding
suggested that ISG60 may be involved in a reaction downstream of
the IFN-β/STAT1 axis. However, this finding is in contrast to the
results of a previous study in U373 cells, in which ISG60
positively regulated STAT1 phosphorylation (28). Additionally, in hCMEC/D3 brain
endothelial cells, ISG60 negatively regulated TLR3-induced
signaling (29). These differences
indicated that ISG60 may function differently in various cell
types.
ISG60, also known as IFIT3 (34), is a tetratricopeptide repeat
protein that participates in various biological processes,
including protein-protein and protein-RNA interactions within
multi-molecule complexes. These proteins regulate chaperones,
transcription, splicing, cell cycle control, protein transport, and
phosphate turnover (35). In
addition, ISG56/IFIT1 is known to be linked to innate immunity
(11). In a previous study, we
reported that ISG56 may be implicated in poly IC-induced CXCL10
expression in BEAS-2B cells (14).
ISG56 is considered a partner molecule of ISG60 (11); therefore, this relationship was
further assessed in the present study. ISG60 knockdown decreased
poly IC-induced ISG56 expression, whereas poly IC-induced
expression of MDA5, another innate immunity-related ISG, remained
unchanged upon ISG60 knockdown. These findings suggested that ISG60
may selectively regulate ISG56 expression. By contrast, ISG56
knockdown decreased ISG60 expression, whereas the P-STAT1
expression did not change. These findings indicated that ISG60 and
ISG56 are mutually regulated downstream of P-STAT1 in poly
IC-treated BEAS-2B cells.
Because IFIT proteins, including ISG60 and ISG56,
are known to have broad-spectrum antiviral functions (36), the results of the present study
suggested that ISG60 and ISG56 may serve crucial roles in antiviral
innate immune reactions in airway epithelial cells. Gene expression
is regulated by protein complexes containing transcriptional
factors, transcriptional regulators, coactivators and RNA
polymerase. In addition, other components, including non-coding
RNAs, are involved in the precise control of the gene expression.
It has been reported that IFIT proteins regulate antiviral immune
reactions by mediating a variety of protein-protein and protein-RNA
interactions (11). Therefore, it
was hypothesized that ISG60, as a component of a multiple protein
complex, may regulate the expression of ISG56 and CXCL10 via
protein-protein and/or protein-RNA interactions. However, the
present study has some limitations. First, the study did not
clarify the specific molecular mechanisms by which ISG60 regulates
the poly IC-induced expression of CXCL10, or by which ISG60 and
ISG56 regulate each other. Future studies should assess the
detailed pathways and interactions involved. Second, the present
study did not examine an experimental model with ISG60
upregulation. Third, this study only used an in vitro cell
culture system. Future studies using lentiviral transfection to
upregulate ISG60 or animal models may enhance the relevance and
applicability of the present findings.
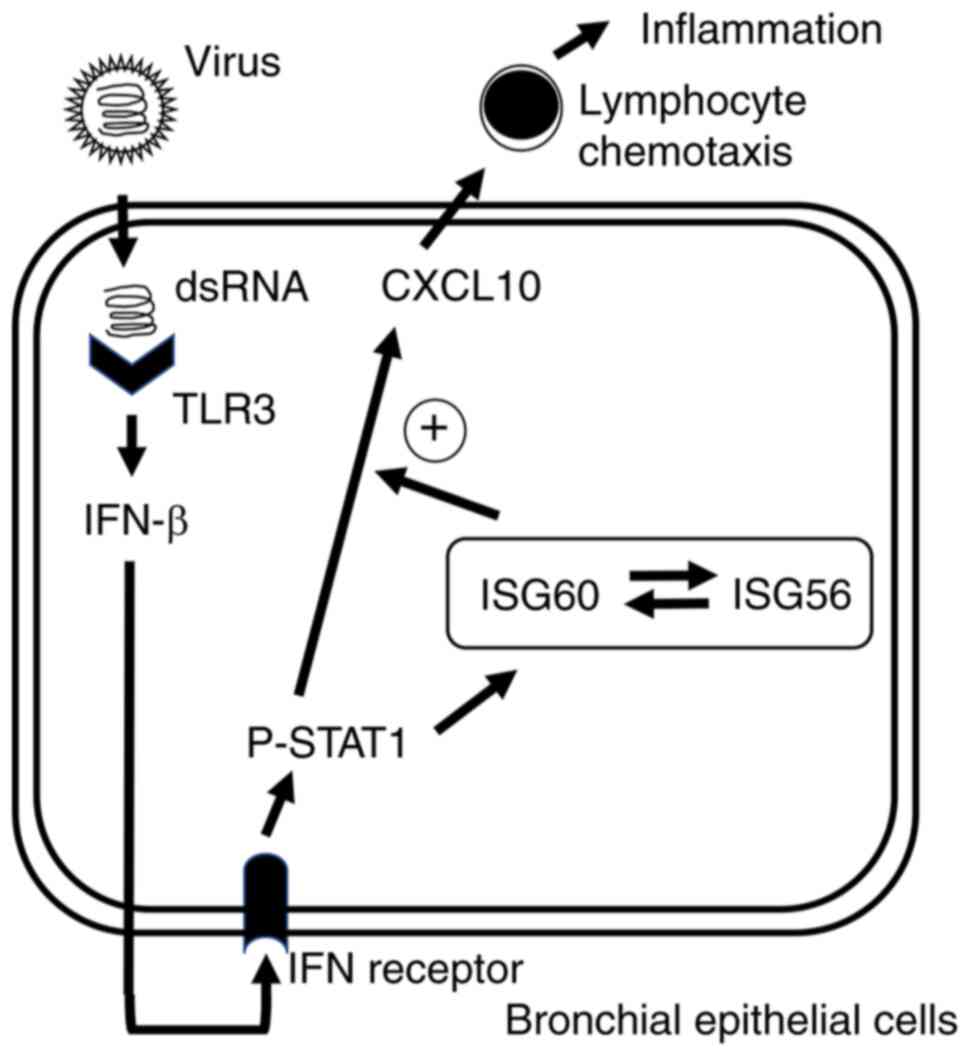
In conclusion, the present study demonstrated that
ISG60 was upregulated by TLR3 signaling and may have a role in the
‘fine-tuning’ of CXCL10 expression in BEAS-2B cells. The present
study reported that the TLR3/IFN-β/ISG60/CXCL10 axis (Fig. 5) may be a novel mechanism to
enhance antiviral innate immune responses in bronchial epithelial
cells. This axis may be important in antiviral immunity in the
respiratory tract and appropriate activation of this axis could
contribute to the host defense against viral infection. However,
excess or dysregulated activation of this axis may lead to excess
accumulation of lymphocytes; since excess accumulation of
lymphocytes induces the excess activation of cytokine networks
(37), dysregulated activation of
this axis may exacerbate inflammation and airway injury induced by
viral infection. Therefore, this axis may be involved in both
physiological antiviral innate immune reaction and pathological
inflammation induced by viral infection in the airways. The present
findings suggested that ISG60 may serve as a potential target for
developing new therapeutic strategies against viral respiratory
infections.
Acknowledgements
The authors would like to thank Ms. Michiko Nakata
(Department of Neuropathology, Hirosaki University Graduate School
of Medicine) for technical support.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YT and TI contributed to all experiments and
prepared the manuscript. YK performed the ELISA. MT performed the
western blotting. TS, MD and MS performed the RNA extraction and
RT-qPCR analyses. SK and KS performed the cell culturing. TI and ST
designed the study. All authors have read and approved the final
version of the manuscript. YT and TI confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CXCL10
|
C-X-C motif chemokine ligand 10
|
|
dsRNA
|
double-stranded RNA
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
IFN
|
interferon
|
|
IFIT1
|
IFN-induced protein with
tetratricopeptide repeats 1
|
|
ISG
|
IFN-stimulated gene
|
|
MDA5
|
melanoma differentiation-associated
gene 5
|
|
P-STAT1
|
phosphorylated-signal transducer and
activating transcription-1
|
|
poly IC
|
polyinosinic-polycytidylic acid
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
r(h)IFN-β
|
recombinant human IFN-β
|
|
siRNA
|
small interfering RNA
|
|
TLR3
|
Toll-like receptor 3
|
References
|
1
|
Bakaletz LO: Viral-bacterial co-infections
in the respiratory tract. Curr Opin Microbiol. 35:30–35. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Medzhitov R and Janeway C Jr: Innate
immunity: Impact on the adaptive immune response. Curr Opin
Immunol. 9:4–9. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okahira S, Nishikawa F, Nishikawa S,
Akazawa T, Seya T and Matsumoto M: Interferon-beta induction
through toll-like receptor 3 depends on double-stranded RNA
structure. DNA Cell Biol. 24:614–623. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guillot L, Le Goffic R, Bloch S, Escriou
N, Akira S, Chignard M and Si-Tahar M: Involvement of toll-like
receptor 3 in the immune response of lung epithelial cells to
double-stranded RNA and influenza A virus. J Biol Chem.
280:5571–5580. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu M and Levine SJ: Toll-like receptor,
RIG-I-like receptors and the NLRP3 inflammasome: Key modulators of
innate immune responses to double-stranded RNA viruses. Cytokine
Growth Factor Rev. 22:63–72. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kawai T and Akira S: The role of
pattern-recognition receptors in innate immunity: Update on
Toll-like receptors. Nat Immunol. 11:373–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hsu ACY, Parsons K, Barr I, Lowther S,
Middleton D, Hansbro PM and Wark PAB: Critical role of constitutive
type I interferon response in bronchial epithelial cell to
influenza infection. PLoS One. 7:e329472012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Majde JA: Viral double-stranded RNA,
cytokines, and the flu. J Interferon Cytokine Res. 20:259–272.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hamamdzic D, Phillips-Dorsett T,
Altman-Hamamdzic S, London SD and London L: Reovirus triggers cell
type-specific proinflammatory responses dependent on the autocrine
action of IFN-beta. Am J Physiol Lung Cell Mol Physiol.
280:L18–L29. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Imaizumi T, Aizawa-Yashiro T, Tsuruga K,
Tanaka H, Matsumiya T, Yoshida H, Tatsuta T, Xing F, Hayakari R and
Satoh K: Melanoma differentiation-associated gene 5 regulates the
expression of a chemokine CXCL10 in human mesangial cells:
Implications for chronic inflammatory renal diseases. Tohoku J Exp
Med. 228:17–61. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fensterl V and Sen GC: The ISG56/IFIT1
gene family. J Interferon Cytokine Res. 31:71–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vareille M, Kieninger E, Edwards MR and
Regamey N: The airway epithelium: Soldier in the fight against
respiratory viruses. Clin Microbiol Rev. 24:210–219. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sanders CJ, Doherty PC and Thomas PG:
Respiratory epithelial cells in innate immunity to influenza virus
infection. Cell Tissue Res. 343:13–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shiratori T, Imaizumi T, Hirono K,
Kawaguchi S, Matsumiya T, Seya K and Tasaka S: ISG56 is involved in
CXCL10 expression induced by TLR3 signaling in BEAS-2B bronchial
epithelial cells. Exp Lung Res. 46:195–202. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu M, Tong JH, Mao M, Kan LX, Liu MM, Sun
YW, Fu G, Jing YK, Yu L, Lepaslier D, et al: Cloning of a gene
(RIG-G) associated with retinoic acid-induced differentiation of
acute promyelocytic leukemia cells and representing a new member of
a family of interferon-stimulated genes. Proc Natl Acad Sci USA.
94:7406–7411. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou X, Michal JJ, Zhang L, Ding B, Lunney
JK, Liu B and Jiang Z: Interferon induced IFIT family genes in host
antiviral defense. Int J Biol Sci. 9:200–208. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chikhalya A, Dittmann M, Zheng Y, Sohn SC,
Rice CM and Hearing P: Human IFIT3 protein induced interferon
signaling and inhibits adenovirus immediate early gene expression.
mBio. 12:e02829212021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Imaizumi T, Tanaka H, Matsumiya T, Yoshida
H, Tanji K, Tsuruga K, Oki E, Aizawa-Yashiro T, Ito E and Satoh K:
Retinoic acid-inducible gene-I is induced by double-stranded RNA
and regulates the expression of CC chemokine ligand (CCL) 5 in
human mesangial cells. Nephrol Dial Transplant. 25:3534–3539. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schoggins JW, Wilson SJ, Panis M, Murphy
MY, Jones CT, Bieniasz P and Rice CM: A diverse range of gene
products are effectors of the type I interferon antiviral response.
Nature. 472:481–485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Muzio M, Bosisio D, Polentarutti N,
D'amico G, Stoppacciaro A, Mancinelli R, van't Veer C, Penton-Rol
G, Ruco LP, Allavena P and Mantovani A: Differential expression and
regulation of toll-like receptors (TLR) in human leukocytes:
Selective expression of TLR3 in dendritic cells. J Immunol.
164:5998–6004. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matsumoto M, Kikkawa S, Kohase M, Miyake K
and Seya T: Establishment of a monoclonal antibody against human
Toll-like receptor 3 that blocks double-stranded RNA-mediated
signaling. Biochem Biophys Res Commun. 293:1364–1369. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cario E and Podolsky DK: Differential
alteration in intestinal epithelial cell expression of toll-like
receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect
Immun. 68:7010–7017. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser
BJ, Valverde RA, Zlotnick A, Mukhopadhyay S, Ranjith-Kumar CT and
Kao CC: Viral double-strand RNA-binding proteins can enhance innate
immune signaling by toll-like Receptor 3. PLoS One. 6:e258372011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsumoto M and Seya T: TLR3: Interferon
induction by double-stranded RNA including poly(I:C). Adv Drug
Deliv Rev. 60:805–812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang SY, Herman M, Ciancanelli MJ, Pérez
de Diego R, Sancho-Shimizu V, Abel L and Casanova JL: TLR3 immunity
to infection inmice and humans. Curr Opin Immunol. 25:19–33. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Imaizumi T, Yoshida H, Hayakari R, Xing F,
Wang L, Matsumiya T, Tanji K, Kawaguchi S, Murakami M and Tanaka H:
Interferon-stimulated gene (ISG) 60, as well as ISG56 and ISG54,
positively regulates TLR3/IFN-β/STAT1 axis in U373MG human
astrocytoma cells. Neurosci Res. 105:35–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Imaizumi T, Sassa N, Kawaguchi S,
Matsumiya T, Yoshida H, Seya K, Shiratori T, Hirono K and Tanaka H:
Interferon-stimulated gene 60 (ISG60) constitutes a negative
feedback loop in the downstream of TLR3 signaling in hCMEC/D3
cells. J Neuroimmunol. 324:16–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu M, Guo S, Hibbert JM, Jain V, Singh N,
Wilson NO and Stiles JK: CXCL10/IP-10 in infectious diseases
pathogenesis and potential therapeutic implications. Cytokine
Growth Factor Rev. 22:121–130. 2011.PubMed/NCBI
|
|
31
|
Dwinell MB, Lugering N, Eckmann L and
Kangnoff MF: Regulated production of interferon-inducible T-cell
chemoattractants by human intestinal epithelial cells.
Gastroenterology. 120:49–59. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Frangogiannis NG, Mendoza LH, Smith CW,
Michael LH and Entman ML: Induction of the synthesis of the C-X-C
chemokine interferon-gamma-inducible protein-10 in experimental
canine endotoxemia. Cell Tissue Res. 302:365–376. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hayney MS, Henriquez KM, Barnet JH, Ewers
T, Champion HM, Flannery S and Barrett B: Serum IFN-γ-induced
protein 10 (IP-10) as a biomarker for severity of acute respiratory
infection in healthy adults. J Clin Virol. 90:32–37. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schmeisser H, Mejido J, Balinsky CA,
Morrow AN, Clark CR, Zhao T and Zoon KC: Identification of alpha
interferon-induced genes associated with antiviral activity in
Daudi cells and characterization of IFIT3 as a novel antiviral
gene. J Virol. 84:10671–10680. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blatch GL and Lässle M: The
tetratricopeptide repeat: A structural motif mediating
protein-protein interactions. Bioessays. 21:932–939. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Diamond MS and Farzan M: The
broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat
Rev Immunol. 13:46–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang Y, Liu J, Zhang D, Xu Z, Ji J and Wen
C: Cytokine storm in COVID-19: The current evidence and treatment
strategies. Front Immunol. 11:17082020. View Article : Google Scholar : PubMed/NCBI
|