Introduction
Glioma is the most common type of primary malignant
brain tumor (1–3). In the United States, the annual
morbidity rate of glioma is 0.5%. There are ~18,000 new glioma
cases and 13,000 mortaloties annually. The prognosis of
glioblastoma remains particularly poor and its 5-year survival rate
is <3%, which is only second to pancreatic and lung cancer
(4). Although anticancer regimens and
surgery are used to treat glioma, tumor cells often migrate into
brain parenchyma and thus patients succumb to the disease extremely
quickly as invasive cells are able to escape radiation exposure and
resection (5–7). Currently, no effective treatments for
glioma have been identified as the potential mechanism of the
disease and prognostic factors of glioma remain unclear (8). Therefore, the development of novel
diagnostic methods to effectively manage gliomas is urgently
required.
C-terminal-binding proteins (CTBPs) were originally
identified based on their ability to bind the C-terminus of type
2/5 adenovirus 243R E1A proteins as cellular binding partners
(9). CTBPs are involved in multiple
biological processes via a number of transcription factors. In the
nucleus, CTBPs act as transcriptional corepressors, while in the
cytoplasm they regulate the endocytic membranes and fission of
Golgi (10–12). In vertebrates, CTBP has two isoforms,
CTBP1 and CTBP2, located at human chromosomes 4 and 10,
respectively and multiple mechanisms have indicated that these
proteins are also involved in the Wnt signaling pathways and cell
cycle regulation (13,14) by acting as direct corepressors of
transcription factor (TCF)-3 and TCF-4 (15). These functions have been demonstrated
in a number of human cells, including colorectal cancer cell lines
(15). Although CTBP1 and CTBP2 share
similarities in gene expression and function, there remains a
difference between the two. A recent study revealed that dorsal
root ganglia express CTBP1, whereas migrating neural crest cells
express CTBP2 (16). Notably, the
majority of CTBP1 mutant mice are fertile and viable, however,
CTBP2 mutations lead to embryonic lethality and delayed neural
development (17). CTBP2 proteins
also exhibit a critical function in tumorigenesis (18). The overexpression of CTBP2 reduces
phosphatase and tensin homolog levels in tumor cells (19).
At present, literature regarding the expression of
CTBP2 proteins in cancer is limited; CTBP2 expression in brain
tumors has not been reported previously. In the present study,
CTBP2 protein expression in gliomas was investigated. This study
evaluated whether CTBP2 is expressed in glioma and whether it is
involved in the proliferation and differentiation of the disease,
as well as its association with glioma grade. The aims of the
present study were to investigate the association between CTBP2
expression and clinicopathological features in glioma patients and
to identify potential therapeutic strategies for the disease.
Patients and methods
Patients
A total of 60 glioma patients that underwent
subtotal resection at The Second Affiliated Hospital of Nantong
University (Nantong, China) between February 2005 and February 2011
were included in the present study. The patient cohort included 31
male and 29 female patients with a median age of 42 years (range,
18–70 years). No patients had received preoperative chemotherapy
and after the surgical incision had healed at post-operative day
15–20, all patients were treated with tridimensional radiotherapy
with 2 Gy/day, 5 days/week, continuous radiotherapy for 5–7 weeks
and a total dose of 60–66 Gy. A week after radiation treatment, the
patients were administered oral chemotherapy consisting of
lomustine at a dose of 0.005–0.006 mg/m2/day, 5 days/week, with a
treatment interval of 23 days and a treatment cycle of 28 days. The
treatment lasted for 3–5 cycles according to the patients'
tolerance. Patients that succumbed due to other diseases or causes
were excluded from the study. The functional impairment of all
patients was assessed using Karnofsky performance status (KPS)
scores (20): 30 patients exhibited a
KPS score of <80 and 30 patients exhibited a KPS score of
>80. Histological grading and classification were conducted in
all gliomas according to the World Health Organization (WHO) 2007
central nervous system tumor classification system (21). A total of 25 patients exhibited low
grade (I–II) malignant glioma and 35 patients exhibited high grade
(III–IV) malignant glioma. A total of 8 brain tissues that were
obtained from patients undergoing intracranial decompression due to
brain trauma served as the control group. Postoperative pathology
confirmed that all control tissues exhibited no glial cell
proliferation.
Tissue preparation
A total of 60 glioma tissue samples were obtained
from glioma patients that underwent subtotal resection and 8
control brain tissues were obtained from brain injury patients that
underwent intracranial decompression. Necrotic tissue was removed
from specimens immediately following isolation. Each patient
specimen was divided into two sections: One section was immediately
frozen at −80°C for subsequent use and the other section was fixed
with 10% neutral formalin, paraffin-embedded and cut into 5-mm
sections. Consent for the collection of specimens was obtained from
all patients and the study was approved by the Ethics Committee of
The Second Affiliated Hospital of Nantong University.
Reagents
Mouse anti-human monoclonal CTBP2 antibody (catalog
no. ab128871) was obtained from Abcam (Cambridge, MA, USA),
biotin-labeled anti-mouse immunoglobulin G (IgG; catalog no.
BA1001) was obtained from Boster Biological Engineering Co., Ltd.,
(Wuhan, China). β-actin monoclonal antibody (catalog no. AF0003)
was purchased from Beyotime Institute of Biotechnology (Shanghai,
China). Polymerized peroxidase-labeled goat anti-mouse IgG (catalog
no. A0216; Beyotime Institute of Biotechnology) was used for
immunohistochemical staining. Donkey anti-mouse IgG biotinylated
antibody (catalog no. BAF018; R&D Systems, Shanghai, China) and
western immunoblot kit (sodium dodecyl sulfate-polyacrylamide gel
electrophoresis gel preparation kit; catalog no. P0012A; Beyotime
Institute of Biotechnology) and RIPA lysis solution (catalog no.
P0013C; Beyotime Institute of Biotechnology) were used for western
blotting.
Western blot analysis
Total protein was extracted from the frozen tumor
specimens using a Nuclear and Cytoplasmic Protein Extraction kit
and stored at −70°C after the concentration was determined by
ultraviolet spectrophotometry. Separating gel and stacking gel, as
well as the electrophoresis buffer, were configured according to
the instructions of the western immunoblot kit (22), followed by sample loading and
electrophoresis. After electrophoresis, proteins were transferred
to nitrocellulose membranes, blocked with skimmed milk powder and
incubated with mouse anti-human monoclonal CTBP2 antibody
(dilution, 1:1,000) at 4°C overnight. The membrane was washed 3
times, for 10 min each, followed by incubation with HRP-labeled
donkey anti-mouse IgG at 4°C for ~4 h. Next, the membrane was
washed and DAB (Beyotime Institute of Biotechnology) was added as
the chromogen. After washing the membrane, enhanced
chemiluminescence reagent (Beyotime Institute of Biotechnology) was
added for development. β-actin served as an internal control
(dilution, 1:1,000). All the western band densities were analyzed
using ImageQuant LAS 4000 (GE Healthcare Life Science, Shanghai,
China) and Quantity One v4.62 software (Bio-Rad, Hercules, CA,
USA).
Immunohistochemical staining
Formalin-fixed paraffin embedded 5-mm sections were
dewaxed conventionally and treated with 3%
H2O2 to inactivate the endogenous
peroxidases, followed by blocking with fetal bovine serum (Gibco;
Thermo Fisher Scientific, Waltham, MA, USA). Following incubation
with mouse anti-human monoclonal CTBP2 antibody (dilution, 1:100)
overnight at room temperature the sections were incubated with
polymerized peroxidase-labeled goat anti-mouse IgG (dilution,
1:100). DAB was then added as the chromogen and sections were
dehydrated for transparent mounting. For the evaluation of CTBP2
expression in different grades of tumors, 10 randomly selected
visual fields per section were examined at ×400 magnifications on a
light microscope (Leica, Wetzlar, Germany).
Cells that exhibited yellow, brown or reddish brown
cytoplasmic staining were regarded as positively-stained cells.
Immunohistochemistry scores were calculated based on the staining
intensity and the percentage of positive cells. Staining intensity
was defined as follows: 0, negative staining; 1, weak staining; 2,
moderate staining; and 3, strong staining. The number of positive
cells was defined as follows: 0, 0% positive cells; 1, <25%
positive cells; 2, 25–50% positive cells; 3, 51–80% positive cells;
and 4, >80% positive cells. Five fields of vision were randomly
selected under high magnification (×400) and 200 cells were counted
in each field of vision. The number of positive cells were counted
to calculate the mean. Based on the sum of the staining intensity
and number of positive cell scores, immunohistochemistry staining
scores were as follows: 0–2, negative (−); 3, weak (+); 4–5,
moderate (++); and 6–7, strong (+++).
Statistical analysis
All data analysis was performed using SPSS 19.0
statistical software (SPSS, Inc., Chicago, IL, USA) and P<0.05
was considered to indicate a statistically significant difference.
Data are expressed as the mean ± standard deviation. The
correlation between CTBP2 protein expression and
clinicopathological parameters was analyzed using the χ2
test and Spearman's rank correlation. One way analysis of variance
was used to compare differences between groups. Kaplan-Meier
survival curves were generated and survival rates were assessed
using the log-rank test.
Results
CTBP2 protein expression is higher in
glioma tissues than normal brain tissues
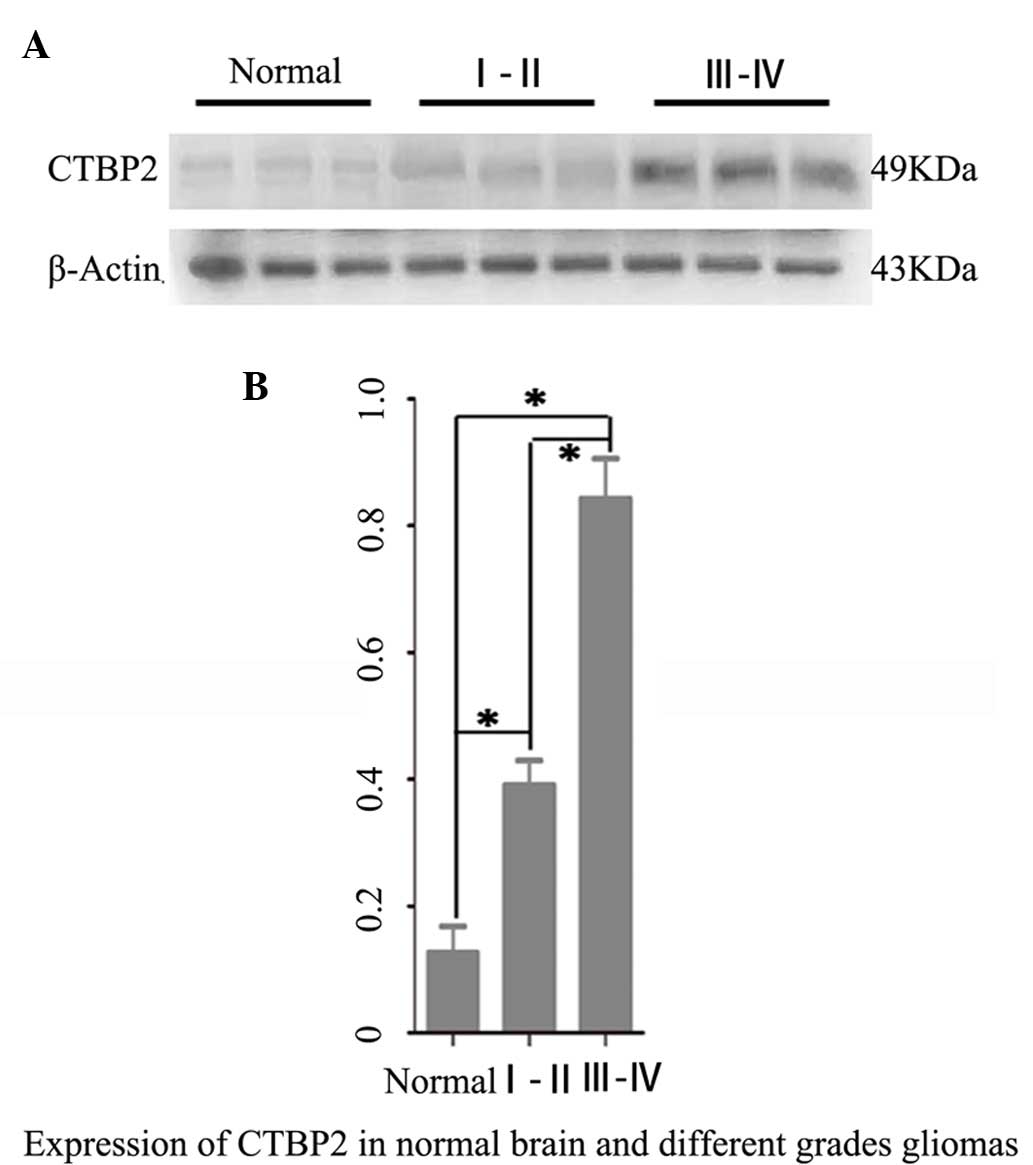
Western blot analysis revealed that CTBP2 protein
expression levels in both the high grade (WHO III–IV) glioma
(0.843±0.354) and low grade (WHO I–II) glioma groups (0.402±0.132)
were significantly higher than that in brain tissues of the normal
brain tissue control group (0.134±0.073) (P=0.005; P=0.008).
Furthermore, the CTBP2 protein expression level in the high grade
glioma group was significantly higher than that in the low grade
glioma group (P=0.042) (Fig. 1).
CTBP2 positive expression rate is
higher in glioma tissues than normal brain tissues
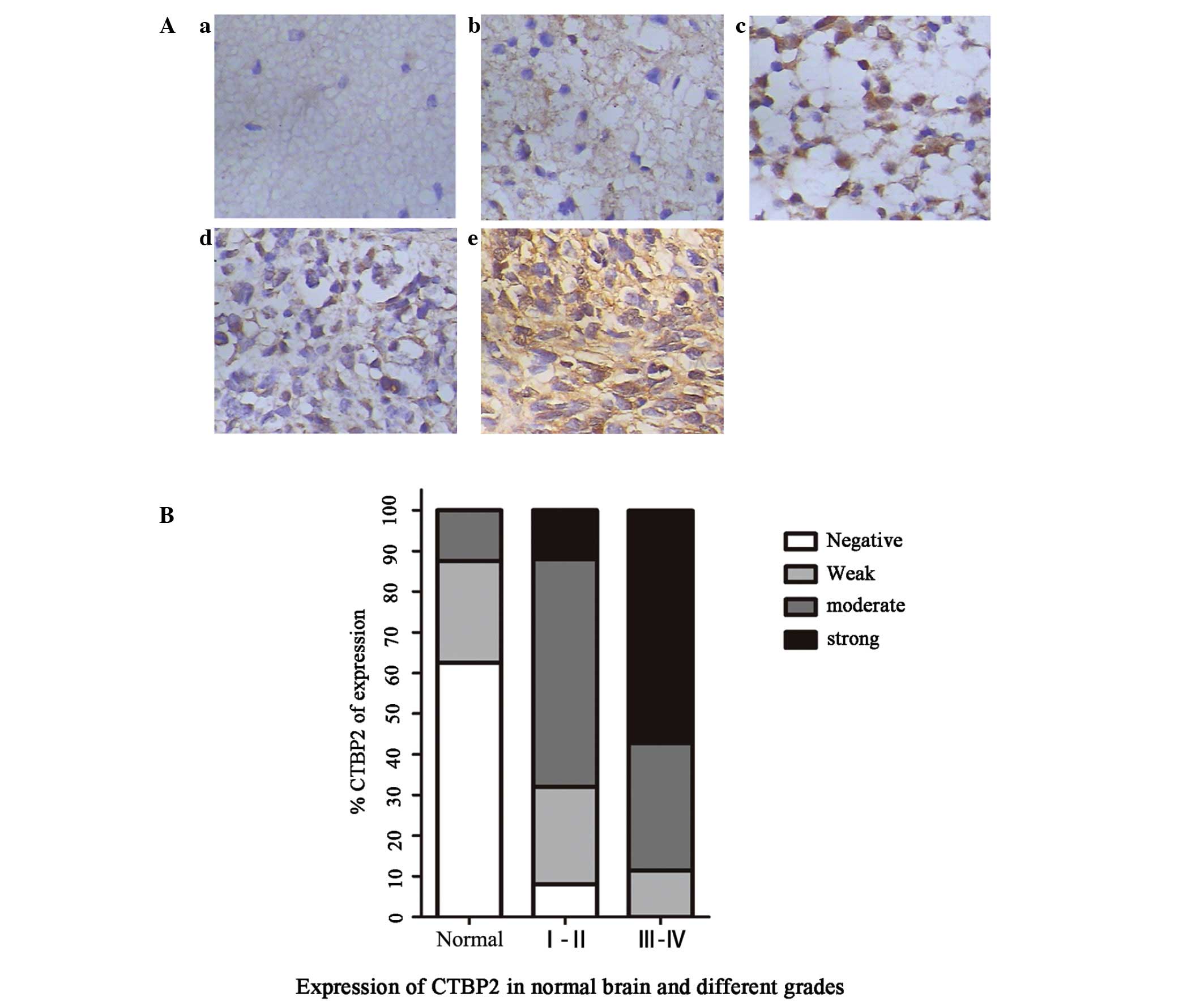
Immunohistochemical staining revealed that CTBP2
protein was predominantly localized in the nucleus of glioma cells
and was also marginally expressed in the cytoplasm (Fig. 2). The positive expression rate of
CTBP2 in all glioma patients was 96.67%, while the positive
expression rate of CTBP2 in the control group was 37.5%. The strong
positive expression rate of CTBP2 in the control, low grade glioma
and high grade glioma groups was 0.00, 12.00 and 57.14%,
respectively. The negative expression rate of CTBP2 was 62.5% in
the control group and 0.00% in the high grade glioma group
(P<0.05). These results demonstrated that the positive
expression of CTBP2 in the high grade (WHO III–IV) glioma group was
significantly higher than that in the low grade (WHO I–II) glioma
and control groups (P=0.003) (Table
I).
 | Table I.CTBP2 expression scores in NB tissue
and WHO grade I–II and III–IV gliomas. |
Table I.
CTBP2 expression scores in NB tissue
and WHO grade I–II and III–IV gliomas.
|
|
| CTBP2 expression
score, n |
|---|
|
|
|
|
|---|
| Group | n | – | + | ++ | +++ |
|---|
| NB | 8 | 5 | 2 | 1 | 0 |
| Grade I–II | 25 | 2 | 6 | 14 | 3 |
| Grade III–IV | 35 | 0 | 4 | 11 | 20 |
High CTBP2 expression is associated
with high glioma grade and poor prognosis of glioma patients
All glioma patients were divided into high CTBP2
expression (number of positive cells, >50%) and low CTBP2
expression (number of positive cells, <50%) groups according to
the expression of CTBP2 identified by immunohistochemistry. No
significant associations between CTBP2 expression and gender, age,
tumor size, location or KPS scores were identified. However, CTBP2
expression was significantly correlated with pathological WHO
glioma grade (P=0.001) (Table II).
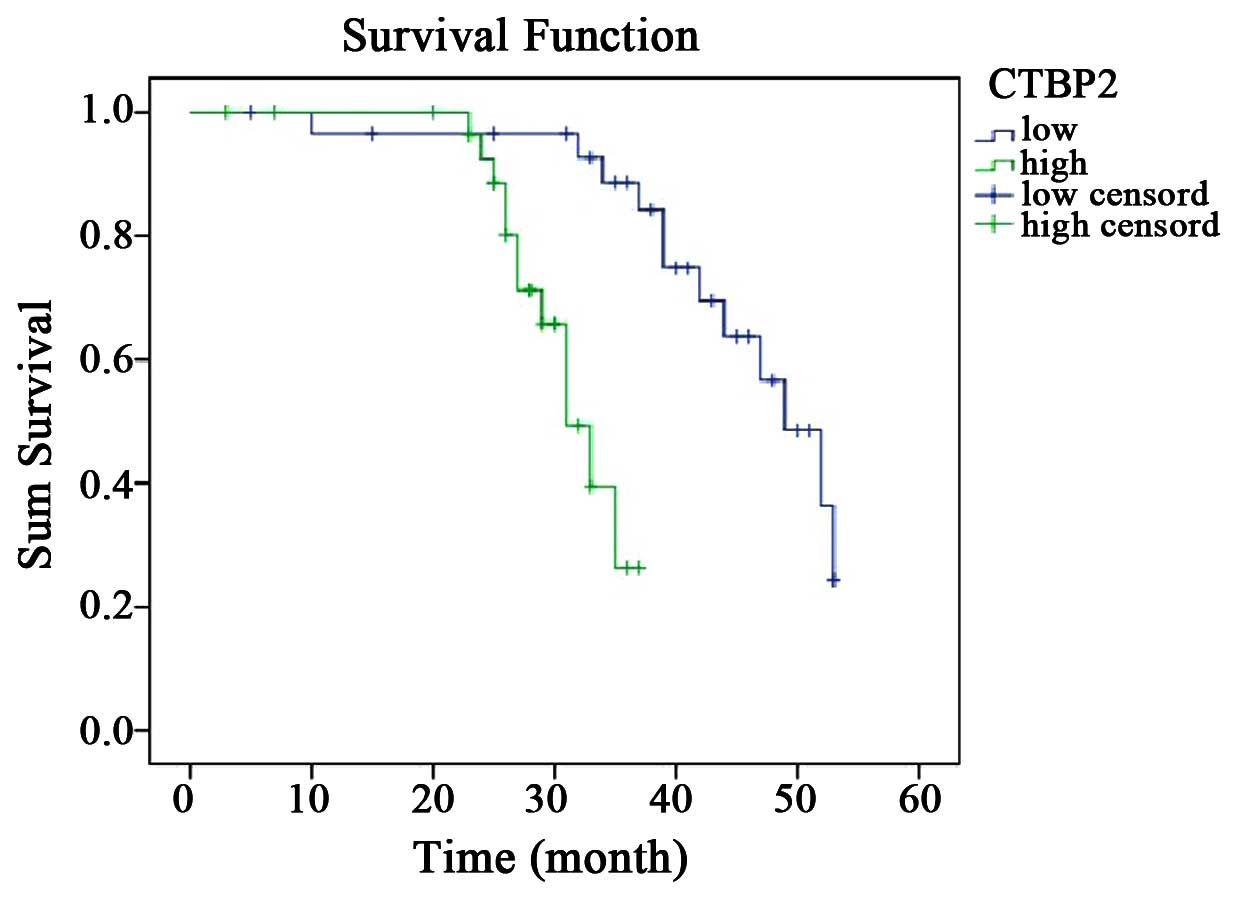
Notably, survival analysis revealed that the low CTBP2 expression
group exhibited a significantly longer survival time and better
prognosis than the high CTBP2 expression group (P=0.001) (Fig. 3).
 | Table II.Association between CTBP2 expression
and clinicopathological characteristics of 60 glioma patients. |
Table II.
Association between CTBP2 expression
and clinicopathological characteristics of 60 glioma patients.
|
|
| CTBP2 expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameter | n | Low, n | High, n | P-value |
|---|
| Gender |
|
|
| 0.438 |
| Male | 31 | 14 | 17 |
|
|
Female | 29 | 16 | 13 |
|
| Age, years |
|
|
| 0.436 |
|
<50 | 33 | 18 | 15 |
|
|
>50 | 27 | 12 | 15 |
|
| KPS score |
|
|
| 0.302 |
|
<80 | 30 | 13 | 17 |
|
|
>80 | 30 | 17 | 13 |
|
| Tumor location |
|
|
| 0.501 |
|
Frontal | 27 | 12 | 15 |
|
|
Temporal | 23 | 11 | 12 |
|
|
Other | 10 | 7 | 3 |
|
| Tumor diameter,
cm |
|
|
| 0.602 |
|
<4 | 26 | 12 | 14 |
|
|
>4 | 34 | 18 | 16 |
|
| WHO grade |
|
|
| 0.001 |
|
I–II | 25 | 19 | 6 |
|
|
III–IV | 35 | 11 | 24 |
|
Discussion
Despite recent advances in diagnostic modalities and
therapeutic strategies, such as radiation therapy and
chemotherapeutic regimens, glioma remains one of the most lethal
types of human cancer (23,24). The median survival time of glioma
patients is <2 years and the 5-year survival rate is among the
lowest for all cancers, at <3% (25). In recent years, studies have been
conducted to identify a potential therapeutic target for human
malignant gliomas. An increased understanding of the biology and
molecular mechanisms underlying glioma development and progression
is required for advances in the treatment of glioma.
CTBPs function in the nucleus as transcriptional
co-repressors via their interaction with adenovirus E1A (26). Extensive molecular and cellular
analyses have identified CTBPs as regulators of pathways that are
critical for tumor initiation, progression, cell migration, cell
apoptosis and response to therapy (15,27).
However, data regarding the expression and regulation of CTBPs in
human cancers and the involvement of CTBPs in glioma is limited. By
contrast to CTBP1, which is expressed from embryo to adult, CTBP2
is predominantly expressed during embryogenesis (16). Additionally, previous studies have
demonstrated that the CTBP2 protein is involved in pathways
associated with tumorigenesis, including transforming growth
factor-β and Wnt signaling pathways and cell cycle regulation
(10,11). Thus, these findings are consistent
with those of the present study which revealed that CTBP2 is highly
expressed in malignant gliomas and increases with ascending tumor
WHO grade. However, CTBP2 expression in astrocytic tumors has not
yet been fully characterized and to date its function in glioma
cells has not been reported.
The results of the present study revealed that CTBP2
expression was markedly increased in malignant gliomas and directly
correlated with WHO glioma grade. Immunostaining revealed that
CTBP2 was predominantly localized in the nuclei. By evaluating the
association between CTBP2 expression and clinicopathological
variables, it was demonstrated that CTBP2 expression correlated
with WHO glioma grade and patient survival. The increased CTBP2
expression in gliomas was significantly associated with higher WHO
grade and poorer disease-specific survival of patients (P=0.001).
However, further studies that investigate CTBP2 expression and its
potential prognostic value are required in the future.
Although CTBP2 has been reported to be highly
expressed in a number of cancers (10,28), its
expression in glioma tissues with varying degrees of malignancy has
not been examined. A previous study demonstrated that CTBP2, as a
coregulator of nuclear transcription, directly interacts with
various targets to promote astrocytical activation and
proliferation (29). We postulate
that CTBP2 may be re-expressed in glioma, and that the pattern of
expression may correlate with the degree of malignant potential.
This hypothesis was confirmed by the current study, which revealed
that increased CTBP2 expression correlates with a higher WHO glioma
grade and poor patient survival. However, the detailed mechanism
underlying transcriptional regulation of CTBP2 in glioma remains
unclear. CTBP2 functions as a corepressor for a substantial number
of transcriptional repressors and consequently regulates diverse
cellular processes. p53, which is a target of CTBP2, suppresses the
sensitivity of breast cancer cells to mechanistically diverse
cancer chemotherapeutic agents, (30). Furthermore, CTBP2 is involved in the
repression of p53-inducible pro-apoptotic genes, including Bax and
Noxa, which are involved in the induction of cellular apoptosis
(31,32). These findings support the results that
CTBP2 was highly expressed in glioblastomas compared with low grade
gliomas and normal brain tissues. Recent studies have demonstrated
that CTBP2, as a negative transcriptional regulator of p16INK4A,
may modulate cell proliferation and contribute to the progression
of esophageal squamous cell carcinoma (33). These findings indicate that CTBP2
provides prognostic information and thus, targeting CTBP2 may
present a novel approach for glioma treatment. However, the
biological functions of CTBP2 and its involvement in tumorigenesis
require further investigation. In addition, the mechanism by which
CTBP2 and p53 or p16 coordinately modulate the growth of glioma
also requires investigation in the future.
In conclusion, the results of the present study
revealed that the expression of CTBP2 was increased in high grade
glioblastomas compared with low grade glioblastomas and normal
brain tissue and that high CTBP2 expression was associated with a
poorer disease-specific survival. Gliomas exhibit poor prognosis
due to their invasive potential and resistance to current
therapeutic modalities. The results of this study indicate that
CTBP2 may present a potential diagnostic marker and a novel
therapeutic target in the treatment of glioma.
Acknowledgements
The authors would like to thank the Department of
Neurosurgery, Affiliated Hospital of Medical College Qingdao
University (Qingdao, China) for providing technical assistance and
equipment support.
Glossary
Abbreviations
Abbreviations:
|
CTBPs
|
C.terminal-binding proteins
|
|
KPS
|
Karnofsky performance status
|
References
|
1
|
Claes A, Idema AJ and Wesseling P: Diffuse
glioma growth: A guerilla war. Acta Neuropathol. 114:443–458. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wen PY, Fine HA, Black PM, Shrieve DC,
Alexander E III and Loeffler JS: High-grade astrocytomas. Neurol
Clin. 13:875–900. 1995.PubMed/NCBI
|
|
3
|
Kleihues P, Soylemezoglu F, Schäuble B,
Scheithauer BW and Burger PC: Histopathology, classification, and
grading of gliomas. Glia. 15:211–221. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ostrom QT, Gittleman H, Farah P, et al:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol.
15(Suppl 2): ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bao S, Wu Q, Li Z, Sathornsumetee S, Wang
H, McLendon RE, Hjelmeland AB and Rich JN: Targeting cancer stem
cells through L1CAM suppresses glioma growth. Cancer Res.
68:6043–6048. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan X, Curtin J, Xiong Y, Liu G, Hogiu S
Waschsmann, Farkas DL, Black KL and Yu JS: Isolation of cancer stem
cells from adult glioblastoma multiforme. Oncogene. 23:9392–9400.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holland EC: Glioblastoma multiforme: The
terminator. Proc Natl Acad Sci USA. 97:6242–6244. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boyd JM, Subramanian T, Schaeper U, La
Regina M, Bayley S and Chinnadurai G: A region in the C-terminus of
adenovirus 2/5 E1a protein is required for association with a
cellular phosphoprotein and important for the negative modulation
of T24-ras mediated transformation, tumorigenesis and metastasis.
EMBO J. 12:469–478. 1993.PubMed/NCBI
|
|
10
|
Bergman LM and Blaydes JP: C-terminal
binding proteins: Emerging roles in cell survival and
tumorigenesis. Apoptosis. 11:879–888. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chinnadurai G: The transcriptional
corepressor CtBP: A foe of multiple tumor suppressors. Cancer Res.
69:731–734. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Corda D, Colanzi A and Luini A: The
multiple activities of CtBP/BARS proteins: The Golgi view. Trends
Cell Biol. 16:167–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meloni AR, Smith EJ and Nevins JR: A
mechanism for Rb/p130-mediated transcription repression involving
recruitment of the CtBP corepressor. Proc Natl Acad Sci USA.
96:9574–9579. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Izutsu K, Kurokawa M, Imai Y, Maki K,
Mitani K and Hirai H: The corepressor CtBP interacts with Evi-1 to
repress transforming growth factor beta signaling. Blood.
97:2815–2822. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hamada F and Bienz M: The APC tumor
suppressor binds to C-terminal binding protein to divert nuclear
beta-catenin from TCF. Dev Cell. 7:677–685. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Van Hateren N, Shenton T and Borycki AG:
Expression of avian C-terminal binding proteins (Ctbp1 and Ctbp2)
during embryonic development. Dev Dyn. 235:490–495. 2006.
View Article : Google Scholar
|
|
17
|
Hildebrand JD and Soriano P: Overlapping
and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2
during mouse development. Mol Cell Biol. 22:5296–5307. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chinnadurai G: CtBP, an unconventional
transcriptional corepressor in development and oncogenesis. Mol
Cell. 9:213–224. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paliwal S, Kovi RC, Nath B, Chen YW, Lewis
BC and Grossman SR: The alternative reading frame tumor suppressor
antagonizes hypoxia-induced cancer cell migration via interaction
with the COOH-terminal binding protein corepressor. Cancer Res.
67:9322–9329. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mor V, Laliberte L, Morris JN and Wiemann
M: The Karnofsky Performance Status Scale. An examination of its
reliability and validity in a research setting. Cancer.
53:2002–2007. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fuller GN and Scheithauer BW: The 2007
Revised World Health Organization (WHO) Classification of Tumours
of the Central Nervous System: Newly codified entities. Brain
Pathol. 17:304–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu R, Liu X, Zheng Y, Gu J, Xiong S,
Jiang P, Jiang X, Huang E, Yang Y, Ge D and Chu Y: MicroRNA-7
sensitizes non-small cell lung cancer cells to paclitaxel. Oncol
Lett. 8:2193–2200. 2014.PubMed/NCBI
|
|
23
|
Soffietti R, Bertero L, Pinessi L and Rudà
R: Pharmacologic therapies for malignant glioma: A guide for
clinicians. CNS Drugs. 28:1127–1137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu CX, Lin GS, Lin ZX, Zhang JD, Chen L,
Liu SY, Tang WL, Qiu XX and Zhou CF: Peritumoral edema on magnetic
resonance imaging predicts a poor clinical outcome in malignant
glioma. Oncology letters. 10:2769–2776. 2015.PubMed/NCBI
|
|
25
|
Chen J, Li Y, Yu TS, McKay RM, Burns DK,
Kernie SG and Parada LF: A restricted cell population propagates
glioblastoma growth after chemotherapy. Nature. 488:522–526. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kovi RC, Paliwal S, Pande S and Grossman
SR: An ARF/CtBP2 complex regulates BH3-only gene expression and
p53-independent apoptosis. Cell Death Differ. 17:513–521. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mirnezami AH, Campbell SJ, Darley M,
Primrose JN, Johnson PW and Blaydes JP: Hdm2 recruits a
hypoxia-sensitive corepressor to negatively regulate p53-dependent
transcription. Curr Biol. 13:1234–1239. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chan CB, Liu X, Jang SW, Hsu SI, Williams
I, Kang S, Chen J and Ye K: NGF inhibits human leukemia
proliferation by downregulating cyclin A1 expression through
promoting acinus/CtBP2 association. Oncogene. 28:3825–3836. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hübler D, Rankovic M, Richter K, Lazarevic
V, Altrock WD, Fischer KD, Gundelfinger ED and Fejtova A:
Differential spatial expression and subcellular localization of
CtBP family members in rodent brain. PLoS One. 7:e397102012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Birts CN, Harding R, Soosaipillai G,
Halder T, Azim-Araghi A, Darley M, Cutress RI, Bateman AC and
Blaydes JP: Expression of CtBP family protein isoforms in breast
cancer and their role in chemoresistance. Biol Cell. 103:1–19.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paliwal S, Pande S, Kovi RC, Sharpless NE,
Bardeesy N and Grossman SR: Targeting of C-terminal binding protein
(CtBP) by ARF results in p53-independent apoptosis. Mol Cell Biol.
26:2360–2372. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Muniz VP, Barnes JM, Paliwal S, Zhang X,
Tang X, Chen S, Zamba KD, Cullen JJ, Meyerholz DK, Meyers S, et al:
The ARF tumor suppressor inhibits tumor cell colonization
independent of p53 in a novel mouse model of pancreatic ductal
adenocarcinoma metastasis. Mol Cancer Res. 9:867–877. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guan C, Shi H, Wang H, Zhang J, Ni W, Chen
B, Hou S, Yang X, Shen A and Ni R: CtBP2 contributes to malignant
development of human esophageal squamous cell carcinoma by
regulation of p16INK4A. J Cell Biochem. 114:1343–1354. 2013.
View Article : Google Scholar : PubMed/NCBI
|