Introduction
Despite the improvements in diagnostic and screening
techniques in recent years, and the increased availability in
vaccines, cervical cancer remains the second most common type of
cancer affecting the female reproductive system (1), and the fourth leading cause of
cancer-associated mortalities in women worldwide (2). Epidemiological studies have indicated
that human papillomavirus is an important risk factor, and possibly
the most required etiological agent, in the development of cervical
cancer (3–5). However, this agent alone is insufficient
to trigger cervical cancer development (6). Thus, a number of previous studies have
speculated that other genetic events may affect this malignant
transformation (7–10). However, the genetic basis underlying
cervical tumorigenesis and progression is largely unknown.
The mammalian transcription factor forkhead box M1
(FOXM1) belongs to the extensive family of forkhead transcription
factors, which harbor 100 amino acids and an evolutionarily
conserved DNA binding domain called forkhead or winged-helix domain
(11–13). FOXM1 is a dynamic cancer-associated
biomarker involved in the regulation of various biological
processes, including cell cycle progression, differentiation,
metastasis, invasion and angiogenesis (14–18). In a
previous study, the present authors demonstrated that FOXM1 was
overexpressed in cervical cancer tissues, and its nuclear
expression was observed to be correlated with the pathological
grade of the tumor (19).
Furthermore, FOXM1 may be involved in the regulation of cancer
invasion and metastasis by regulating the expression and activity
of matrix metalloproteinase (MMP)-2, MMP-9 and vascular endothelial
growth factor (VEGF) (17). Chan
et al (20) and He et
al (21) also indicated that
FOXM1 is important in the tumorigenesis and development of cervical
cancer. However, the molecular mechanism underlying the modulation
of FOXM1 expression remains unclear. Teh et al (22) demonstrated that the overexpression of
glioma-associated oncogene 1 (GLI1) significantly elevated the
messenger RNA (mRNA) levels and transcriptional activity of FOXM1
in numerous human tumor cell lines. Since then, various studies
have attempted to identify an association between FOXM1 and
hedgehog (Hh) signaling in human tumors (23–25).
The Hh signaling pathway, which was first reported
by Nüsslein-Volhard and Wieschaus in 1980 (26), regulates growth and patterning during
organogenesis, and its malfunction leads to multiple human
disorders, including birth defects and cancer (27–29). In
humans, this signaling pathway involves three ligands, namely Sonic
Hh (SHh), Indian Hh (IHh) and Desert Hh (DHh). Among these three
ligands, SHh is the most widely expressed in mammalian tissues,
while IHh is specific for bone development and DHh expression is
restricted to gonads (29,30). The receptor for Hh ligands is a
protein with 12 transmembrane domains termed patched 1 (PTCH1),
which is located in the cell membrane (31). Another transmembrane protein called
smoothened (SMO) is also involved in the Hh signaling pathway,
while GLIs are zinc-finger proteins that function as translational
modulators of this pathway (32).
In the absence of Hh ligands, PTCH1 inhibits the
activity of SMO, while the binding of Hh ligands to PTCH1 relieves
the inhibition of SMO caused by PTCH1, and SMO subsequently
activates the translational modulators GLI1, GLI2 and GLI3
(33). This cascade of events
ultimately results in the regulation of the corresponding target
genes (33). Certain components of
the Hh signaling pathway such as PTCH1 and GLI1 are direct
transcriptional targets, thus establishing a feedback loop that
regulates the level of activity of this pathway (34).
Various studies on the role of the Hh signaling
pathway in cervical cancer have been previously conducted, but the
precise molecular mechanism underlying the processes of metastasis
and invasion in cervical cancer remains unclear. In addition, the
association between the Hh signaling pathway and FOXM1 in cervical
cancer is largely unknown. Therefore, the expression of Hh
signaling molecules (including SHh, PTCH1, SMO and GLI1) and FOXM1
was analyzed in the present study in a tissue microarray of
cervical cancer using immunochemistry. In addition, the association
between these molecules and the clinicopathological parameters of
patients with cervical cancer (including pathological grade,
clinical stage and lymph node metastasis), as well as the
association between the Hh pathway and FOXM1 expression were also
evaluated in the present study.
Materials and methods
Patients and tissue microarray
All patients underwent surgery in Tongxiang People's
Hospital, Tongxiang, China, between January 2002 and December 2012.
The patients with cervical cancer underwent a hysterectomy, while
all the normal cervical samples were obtained by cervical local
excision. The study was approved by the ethics committee of
Tongxiang People's Hospital. Tissue microarray analysis of 80
specimens obtained from patients who underwent surgery was
performed by Alenabio, Inc. (Xi'an, China). All specimens were
fixed with formalin (Guangzhou Wexis Biotech Ltd., Guangzhou,
China) postoperatively, and then embedded with paraffin (Jinan
Shenghe Chemical Co., Ltd., Jinan, China)prior to being converted
into tissue microarray slides (core size, 1.50 mm; 10×8 rows). The
slides were analyzed for histological type and tumor grade by two
pathologists (Dr Fang Yu and Dr Sufang Tian, Zhongnan Hospital of
Wuhan University, Wuhan, China). Histopathological examination
revealed that the specimens consisted of 4 adenocarcinomas, 66
squamous cell carcinomas and 10 normal cases. Of the 70 tumors, 4
(5.71%) were grade I, 43 (61.43%) were grade II, 19 (27.14%) were
grade III, and 4 (5.71%) were undetermined. The clinical stage
classification of the tumors was based on the 7th edition of the
cancer staging manual published by the American Joint Committee on
Cancer (AJCC), and revealed that of the 70 tumors, 22 (31.43%) were
stage I, 20 (28.57%) were stage II and 28 (40.00%) were stage III.
The lymph node status was determined according to the
tumor-node-metastasis classification criterion, and indicated that
lymph node metastasis was present in 44 (62.86%) cases. All the
information regarding the tumor specimens is listed in Table I. Written consent was obtained from
all patients.
 | Table I.Clinicopathological characteristics
of 70 tumor cases. |
Table I.
Clinicopathological characteristics
of 70 tumor cases.
|
Characteristics | Adenocarcinoma, n
(%) | Squamous cell
carcinoma, n (%) |
|---|
| Total | 4 (5.71) | 66 (94.29) |
| Pathological
stage |
|
|
| Well
differentiated | 0 (0.00) | 4 (6.06) |
|
Moderately differentiated | 3 (75.00) | 40 (60.61) |
| Poorly
differentiated | 0 (0.00) | 19 (28.79) |
|
Unclear | 1 (25.00) | 3 (4.54) |
| Invasive
extent |
|
|
|
Confined to uterus | 1 (25.00) | 31 (46.97) |
| Beyond
the uterus | 3 (75.00) | 35 (53.03) |
| Lymph node
status |
|
|
| N0 | 3 (75.00) | 41 (62.12) |
| N1 | 0 (0.00) | 24 (36.36) |
|
Unclear | 1 (25.00) | 1 (1.52) |
| Age, years |
|
|
|
≤40 | 2 (50.00) | 18 (27.27) |
|
41–56 | 1 (25.00) | 40 (60.61) |
|
≥56 | 1 (25.00) | 8 (12.12) |
Immunohistochemistry
Tissue sections were deparaffinized with serially
decreasing concentrations of ethanol (Sinopharm Group Co., Ltd.,
Shanghai China) and then rehydrated in distilled water for 2 min.
For antigen retrieval, the slides were pretreated with 0.01 M
citrate buffer pH 6.0 (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), prior to be heated in a KJ23B microwave oven (Midea, Shunde,
China) for 15 min. To detect SMO, GLI1 and FOXM1 antigens, the
endogenous peroxidase activity was blocked with 3%
H2O2 (Nanjing Senbeijia Biological Technology
Co., Ltd., Nanjing, China) in methanol (Shanghai Macklin
Biochemical Co., Ltd., Shanghai, China) for 15 min at room
temperature, and the sections were then incubated with the
corresponding primary antibodies overnight at 4°C. Subsequently,
the sections were incubated with peroxidase-conjugated secondary
antibodies (catalogue no. PV-9000; ZSGB Biotechnology, Beijing,
China). Protein expression was visualized by the brown pigmentation
resulting from the chromogen 3,3′-diaminobenzidine (DAB)
hydrochloride (catalogue no. AR1022; Wuhan Boster Biological
Technology, Ltd., China). Next, the slides were counterstained with
hematoxylin (catalogue no. H9627, Sigma-Aldrich, St. Louis, MO,
USA), rinsed several times with phosphate-buffered saline (PBS;
Wuhan Boster Biological Technology, Ltd.) pH 7.4, dehydrated with a
series of graded ethanol solutions, cleared using xylene (Sinopharm
Group Co., Ltd.), and observed under an ECLIPSE E100 optical
microscope (Nikon Corporation, Tokyo, Japan). To detect SHh and
PTCH1 antigens, the endogenous peroxidase activity was blocked with
rabbit serum (Shanghai Beiyi Bioequip Information Co., Ltd.,
Shanghai, China) for 30 min at room temperature, prior to
incubating the tissue sections with the corresponding primary
antibodies overnight at 4°C. The primary antibodies used were as
follows: Goat anti-SHh polyclonal antibody (catalogue no. sc-1194;
1:120 dilution), goat anti-PTCH1 polyclonal antibody (catalogue no.
sc-6149; 1:120 dilution), rabbit anti-SMO polyclonal antibody
(catalogue no. sc-13943; 1:120 dilution), rabbit anti-GLI1
polyclonal antibody (catalogue no. sc-20687; 1:100 dilution) (Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) and rabbit polyclonal
anti-FOXM1 antibody (1:150 dilution; ProteinTech Group, Inc.,
Chicago, IL, USA). Next, biotinylated rabbit anti-goat
immunoglobulin G (catalogue no. SA1023; Wuhan Boster Biological
Technology) was added to the sections and incubated at 30°C for 30
min. The immunoreactivity of the tissue sections was visualized
using DAB hydrochloride. Sections were then counterstained with
hematoxylin, rinsed in PBS pH 7.4, dehydrated with ethanol, cleared
with xylene, and observed under an ECLIPSE E100 optical
microscope.
Quantitative analysis
The slides were examined under an ECLIPSE E100
microscope. Positive cells were stained as brownish granules in the
cytoplasm or in the nucleus. The staining intensity for the Hh
signaling molecules and FOXM1 protein in cervical tumors and normal
epithelia was semi-quantitatively assessed. The immunostaining
densities of each point of the tissue microarray were
quantitatively assessed with Image-Pro Plus version 6.0 (Media
Cybernetics, Inc., Rockville, MD, USA). Briefly, the sections were
placed under the microscope and photographed. The images were then
transferred via a digital camera (EOS 600D; Canon, Inc., Tokyo,
Japan) to a computer. A total of three separate positive areas at
×400 magnification were selected in each point of the
immunostaining sections, and the integrated optical density (IOD)
was measured. The IOD values obtained in three areas were averaged
and used to calculate the mean values.
Statistical analysis
Statistical analyses were performed with SPSS 20.0
software (IBM SPSS, Armonk, NY, USA). The association between the
protein expression levels (according to the IOD values) of FOXM1
and Hh signaling molecules and the clinicopathological parameters
of the tumors was evaluated with independent t-tests in
order to compare the differences between two groups. To compare
differences between ≥2 groups, one-way analysis of variance was
used for those cases of equal variance, while Kruskal-Wallis test
was used for unequal variance. The correlation between the
expression levels of each protein was analyzed with the Spearman's
rank-order correlation test. Data were reported as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Overall data
The 80 specimens analyzed in the present study
consisted of 70 cancer cases and 10 normal cases. The mean age of
the patients was 44 years (range, 15–72 years). The expression
levels of the aforementioned proteins were significantly higher in
the tumor specimens than in normal tissues. No significant
differences were detected between the squamous cell carcinoma
tissues and adenocarcinoma tissues (Table II). Tumor cases were divided into
three groups, based on patients' age (≤40, 41–56 and ≥56 years,
respectively). No significant correlation was observed between
protein expression and patient age, except for FOXM1 expression in
<41 and >55-year-old patients (P=0.033). No significant
correlation was observed between the expression of FOXM1, SHh,
PTCH1 or SMO and lymph node status (P=408, P=0.113, P=0.528 and
P=0.853, respectively).
 | Table II.Comparison of FOXM1 and Hh signaling
molecules in tumors and normal tissues. |
Table II.
Comparison of FOXM1 and Hh signaling
molecules in tumors and normal tissues.
|
| Squamous cell
carcinoma | Adenocarcinoma |
|
|---|
|
|
|
|
|
|---|
| Protein | IOD (mean ±
SD) |
P-valuea | IOD (mean ±
SD) |
P-valueb |
P-valuec |
|---|
| FOXM1 |
(2.27±1.64)×105 | <0.001 |
(1.39±1.17)×105 | 0.014 | 0.340 |
| SHh |
(3.27±0.95)×105 | <0.001 |
(3.38±0.37)×105 | 0.007 | 0.743 |
| SMO |
(1.89±1.16)×105 | <0.001 |
(1.58±0.84)×105 | 0.007 | 0.743 |
| GLI1 |
(1.33±0.96)×105 | <0.001 |
(1.41±0.73)×105 | 0.014 | 0.620 |
| PTCH1 |
(1.71±0.58)×105 | <0.001 |
(1.97±0.17)×105 | 0.007 | 0.236 |
FOXM1 expression in cervical cancer
and normal tissues
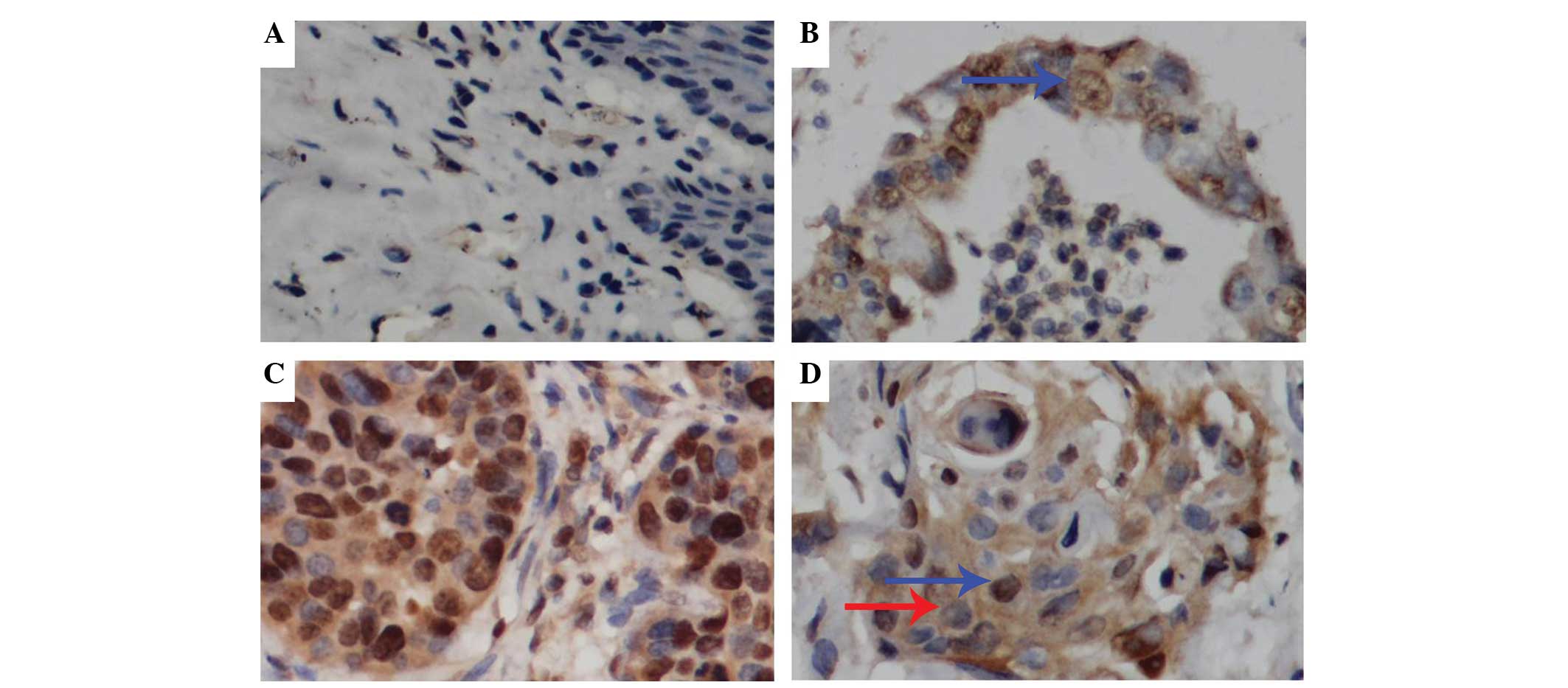
FOXM1 protein was stained as brownish granules in
the cytoplasm and particularly in the nucleus of positive cells
(Fig. 1). In the control samples, the
background was clear with no specific staining. Table III shows the expression of FOXM1 in
cervical cancer tissues. The IOD value of FOXM1 in normal cases was
(2.97±1.67)×104. According to the AJCC staging
classification system, FOXM1 protein expression differed at
different tumor stages (P=0.011), but did not exhibit any
significant correlation with pathological grade. In addition, FOXM1
expression exhibited no correlation with the lymph node metastasis
status of cervical cancer. Spearman's rank-order correlation test
indicated that FOXM1 expression correlated with GLI1 (R=0.405,
P<0.001), SHh (R=0.416, P<0.001) and PTCH1 (R=0.281, P=0.012)
expression.
 | Table III.Association between FOXM1 expression
and clinicopathological characteristics. |
Table III.
Association between FOXM1 expression
and clinicopathological characteristics.
|
Characteristics | Patients, n
(%) | IOD (mean ±
SD) | P-value |
|---|
| Pathological
stagea |
|
|
|
|
Well/moderately
differentiated | 47 (67.14) |
(2.18±1.72)×105 | 0.512 |
| Poorly
differentiated | 19 (27.14) |
(2.48±1.45)×105 |
|
| Invasive
extentb |
|
|
|
| Stage
1 | 22 (31.43) |
(1.54±1.49)×105 | 0.011c |
| Stage
2 | 20 (28.57) |
(2.34±1.36)×105 | 0.103d |
| Stage
3 | 28 (40.00) |
(2.71±1.75)×105 | 0.421e |
| Lymph node
statusf |
|
|
|
| N0 | 44 (62.86) |
(2.09±1.60)×105 | 0.408 |
| N1 | 24 (34.29) |
(2.44±1.72)×105 |
|
| Age, years |
|
|
|
|
≤40 | 20 (28.57) |
(2.72±1.71)×105 | 0.033g |
|
41–56 | 41 (58.57) |
(2.20±1.63)×105 | 0.230h |
|
≥56 | 9
(12.86) |
(1.33±0.99)×105 | 0.143i |
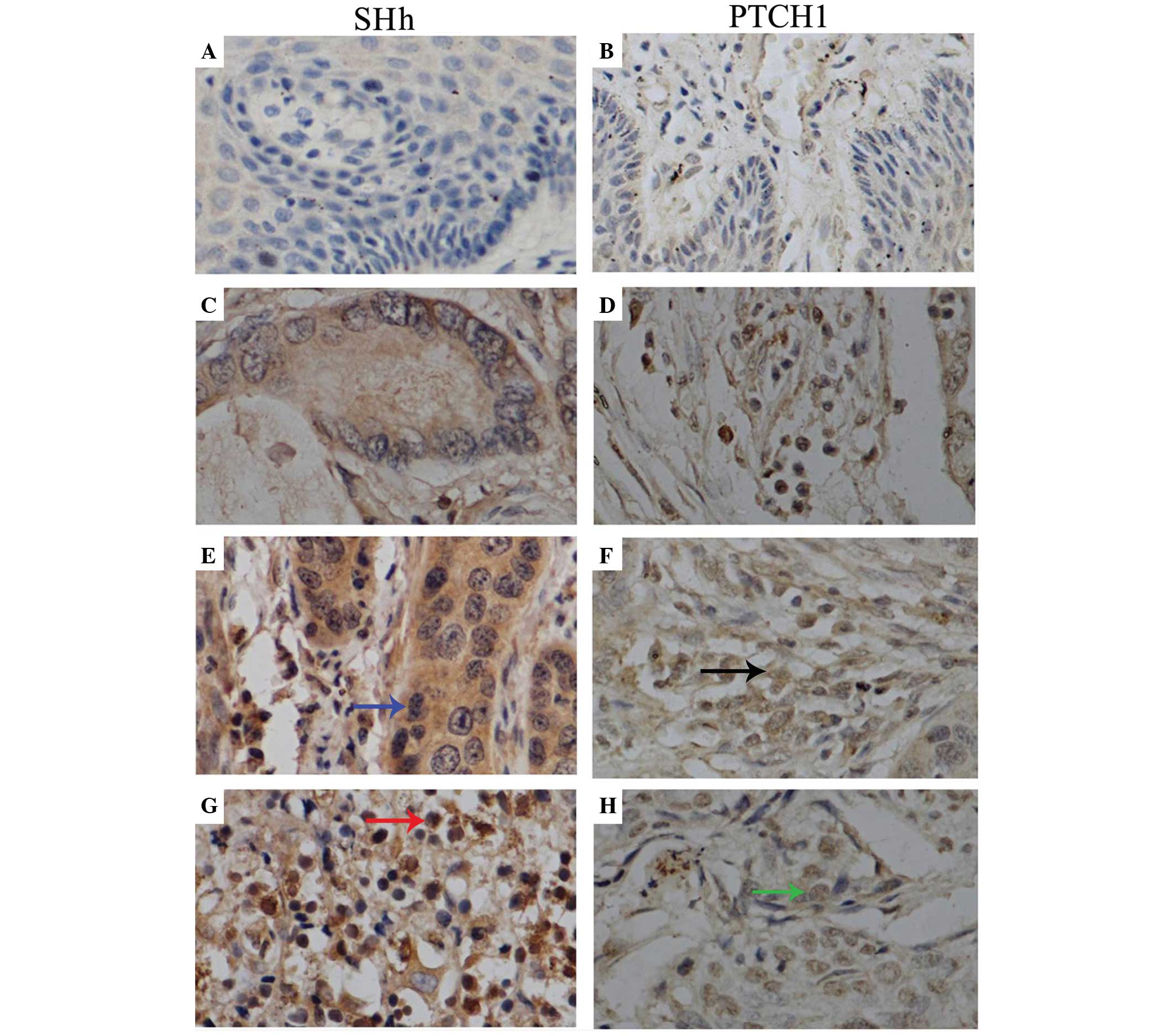
Expression levels of SHh and PTCH1 in
cervical cancer and normal tissues
Immunohistochemical staining identified SHh and
PTCH1 in the cytoplasm and nucleus of tumor cells (Fig. 2). The IOD values of SHh and PTCH1 in
normal specimens were (8.82±3.04)×103 and
(3.46±1.27)×104, respectively. The expression of SHh
significantly differed between different pathological grades, and
was significantly higher in poorly differentiated tumors than in
moderately differentiated tumors (P<0.001). No difference was
observed in terms of invasive extent or lymph node metastasis. The
expression levels of PTCH1 were not associated with invasive
extent. By contrast, the expression of PTCH1 was significantly
correlated with the tumor pathological grade, being the expression
levels of PTCH1 higher in poorly differentiated tissues than in
moderately differentiated tissues (P=0.023). The results of the
statistical analysis conducted for SHh and PTCH1 expression are
listed in Table IV.
 | Table IV.Association between SHh and PTCH1
expression and clinicopathological characteristics. |
Table IV.
Association between SHh and PTCH1
expression and clinicopathological characteristics.
|
|
| SHh | PTCH1 |
|---|
|
|
|
|
|
|---|
|
Characteristics | Patients, n
(%) | IOD (mean ±
SD) | P-value | IOD (mean ±
SD) | P-value |
|---|
| Pathological
gradea |
|
|
|
|
|
|
Well/moderately
differentiated | 47 (67.14) |
(2.98±0.82)×105 |
<0.001a |
(1.60±0.49)×105 | 0.023b |
| Poorly
differentiated | 19 (27.14) |
(3.95±0.92)×105 |
|
(1.96±0.73)×105 |
|
| Invasive
extentc |
|
|
|
|
|
| Stage
1 | 22 (31.43) |
(3.04±0.83)×105 |
0.463d |
(1.81±0.57)×105 | 0.473d |
| Stage
2 | 20 (28.57) |
(3.25±1.06)×105 |
0.413e |
(1.68±0.38)×105 | 0.911e |
| Stage
3 | 28 (40.00) |
(3.47±0.94)×105 |
0.104f |
(1.66±0.69)×105 | 0.373f |
| Lymph node
statusg |
|
|
|
|
|
| N0 | 44 (62.86) |
(3.13±0.87)×105 |
0.113 |
(1.75±0.50)×105 | 0.528 |
| N1 | 24 (34.29) |
(3.52±1.05)×105 |
|
(1.66±0.72)×105 |
|
| Age, years |
|
|
|
|
|
|
≤40 | 20 (28.57) |
(3.19±1.06)×105 |
0.601h |
(1.74±0.54)×105 | 0.802k |
|
41–56 | 41 (58.57) |
(3.32±0.96)×105 |
0.767i |
(1.67±0.44)×105 |
|
|
≥56 | 9
(12.86) |
(3.27±0.95)×105 |
0.933j |
(1.86±0.11)×105 |
|
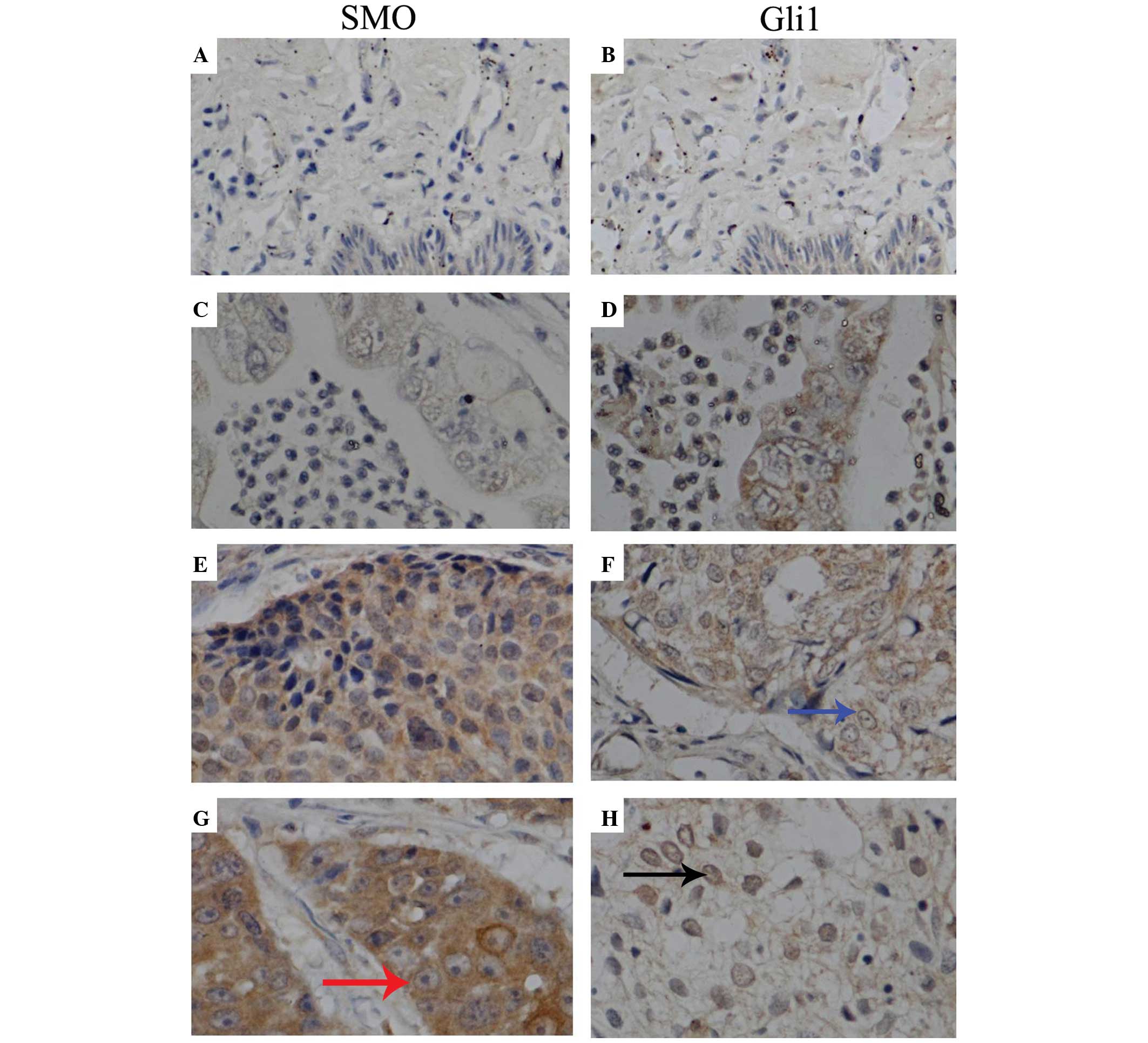
Expression levels of SMO and GLI1 in
cervical cancer and normal tissues
Immunohistochemical staining identified SMO mainly
in the cytoplasm of tumor cells, while GLI1 was expressed both in
the cytoplasm and nucleus (Fig. 3).
The IOD values of SMO and GLI1 in normal specimens were
(2.41±1.43)×104 and (2.48±2.35)×104,
respectively. The expression of SMO was not associated with the
tumor pathological grade. However, the expression levels of this
protein correlated with the tumor invasive extent, as revealed by
the results of Kruskal-Wallis test (P=0.019). The expression of
GLI1 was significantly lower in moderately differentiated cervical
cancer tissues than in poorly differentiated cervical cancer
tissues (P=0.022). In addition, the expression of GLI1 correlated
with the invasive extent of cervical tumors (P=0.005). GLI1
expression was also higher in tumors with lymph node metastasis
than in tissues without lymph node metastasis (P=0.017). The
results of the statistical analysis regarding these proteins are
listed in Table V.
 | Table V.Association between GLI1 and SMO
expression and clinicopathological characteristics. |
Table V.
Association between GLI1 and SMO
expression and clinicopathological characteristics.
|
|
| GLI1 | SMO |
|---|
|
|
|
|
|
|---|
|
Characteristics | Patients, n
(%) | IOD (mean ±
SD) | P-value | IOD (mean ±
SD) | P-value |
|---|
| Pathological
stagea |
|
|
|
|
|
|
Well/moderately
differentiated | 47 (67.14) |
(1.18±0.94)×105 | 0.022b |
(1.78±1.05)×105 | 0.322 |
| Poorly
differentiated | 19 (27.14) |
(1.77±0.90)×105 |
|
(2.10±1.36)×105 |
|
| Invasive
extentc |
|
|
|
|
|
| Stage
1 | 22 (31.43) |
(8.89±6.87)×104 | 0.005d |
(2.40±1.51)×105 |
0.019d |
| Stage
2 | 20 (28.57) |
(1.33±0.53)×105 |
|
(1.26±0.55)×105 |
|
| Stage
3 | 28 (40.00) |
(1.69±1.20)×105 |
|
(1.91±0.92)×105 |
|
| Lymph node
statuse |
|
|
|
|
|
| N0 | 44 (62.86) |
(1.15±0.73)×105 | 0.017b |
(1.92±1.26)×105 | 0.853 |
| N1 | 24 (34.29) |
(1.72±1.20)×105 |
|
(1.87±0.95)×105 |
|
| Age, years |
|
|
|
|
|
|
≤40 | 20 (28.57) |
(1.25±0.89)×105 | 0.981f |
(1.90±1.33)×105 |
0.747f |
|
41–56 | 41(58.57) |
(1.36±1.01)×105 | 0.783g |
(1.80±1.11)×105 |
0.567g |
|
≥56 | 9
(12.86) |
(1.42±0.84)×105 | 0.788h |
(2.17±0.90)×105 |
0.389h |
Discussion
Cervical cancer is the second most common malignancy
among women worldwide, and its incidence and mortality rates rank
the third and fourth, respectively, among female malignant tumors
(2). The metastasis capability of
cervical cancer contributes to its malignance (35). Therefore, elucidation of the molecular
mechanism underlying cancer formation and progression is urgently
required. In the present study, the expression patterns of FOXM1
and Hh signaling molecules in cervical cancer were characterized
via immunohistochemistry. The results indicated that FOXM1 protein
and Hh signaling molecules were overexpressed in cervical cancer
tissues. In addition, the association between FOXM1 and the Hh
signaling pathway was also analyzed. The results indicated that
FOXM1 overexpression correlated with overexpression of molecules
participating in the Hh signaling pathway. These data suggest that
FOXM1 is important in cervical cancer and that the Hh signaling
pathway participates in its modulation. Overall, the present study
suggests that therapies directed against FOXM1 and the Hh signaling
pathway are a promising novel approach for cervical cancer
treatment.
Increased expression of FOXM1 has been previously
detected in diverse cancer cell lines and tissues (36,37). In
the present study, FOXM1 expression was significantly higher in
cervical cancer tissues than in normal cervical tissues (squamous
carcinoma vs. normal cervical tissue, P<0.001; adenocarcinoma
vs. normal cervical tissue, P=0.014). This result is in accordance
with the studies by Chan et al (20) and He et al (21). The present study also revealed that
the expression of FOXM1 correlated with cancer invasion (P=0.011),
similarly to the results reported by Chan et al (20) and He et al (21), who also noted an association between
FOXM1 expression and tumor stage. In the current study, no
association was detected between FOXM1 expression and tumor
pathological grade or lymph node metastasis. However, FOXM1
expression was lower in patients older than 55 years than in
patients younger than 41 years (P=0.033). This result suggests that
FOXM1 expression decreases with age. To the best of our knowledge,
the present study is the first to explore the association between
FOXM1 expression and patient age. In a previous study, the present
authors demonstrated that RNA interference (RNAi)-mediated FOXM1
knockdown inhibited cervical cancer migration, invasion and
angiogenesis in vivo and in vitro (18). In the present study, the results of
immunohistochemistry analysis indicated that FOXM1 expression also
correlated with cancer invasive extent in human cervical cancer
tissues. These results suggest an important function of FOXM1 in
cervical cancer metastasis and invasion.
Previous studies have reported that the Hh signaling
pathway participates in various tumor processes, including
formation, development, metastasis and angiogenesis (38–40). In
the present study, the expression of the molecules involved in the
Hh signaling pathway was investigated, and the results revealed
that GLI1, PTCH1, SMO and SHh were overexpressed in cervical cancer
tissues (squamous carcinoma vs. normal cervical tissue, all
P<0.01; adenocarcinoma vs. normal cervical tissue, P=0.014,
P=0.007, P=0.007 and P=0.007, respectively). In addition, the
association between the expression levels of the above Hh signaling
molecules and the clinicopathological characteristics of patients
was also analyzed in the present study. The expression levels of
SHh, PTCH1 and GLI1 were significantly higher in the poorly
differentiated tumor cases than in the moderately differentiated
tumor cases (P<0.001, P=0.023 and P=0.022, respectively). The
expression levels of GLI1 and SMO also correlated with the invasive
extent of cervical tumors (P=0.005 and P=0.019, respectively),
while GLI1 expression additionally correlated with lymph node
metastasis (P=0.017). The overexpression of these molecules, which
are involved in the Hh signaling pathway, implies that this pathway
participates in the formation and invasion of cervical cancer. Xuan
et al (41) also studied the
expression of the aforementioned Hh signaling molecules in cervical
cancer, and observed that their expression was higher in carcinoma
tissues and cervical intraepithelial neoplasia of stage II/III than
in normal tissues. Furthermore, Samarzija and Beard (42) reported that the overexpression of
GLI1, PTCH1, SMO and SHh in cervical cancer cells (including C33-A,
SiHa, C4-1, CaSki and HeLa), and the inhibition of the Hh signaling
pathway by its inhibitor, reduced the proliferation and survival of
cervical cancer cells. Overall, the results of these studies imply
that the Hh signaling pathway is important in cervical cancer.
FOXM1 is a major oncogenic transcription factor,
since it mediates cancer cell cycle progression, apoptosis,
angiogenesis, migration, invasion and metastasis (9,43–45). Considerable efforts have been exerted
in recent years to elucidate the translation and activity of FOXM1
(46). The Hh signaling pathway may
modulate the transcription of FOXM1 in human transitional cell
carcinoma of the bladder (47). Teh
et al (22) indicated that
GLI1 overexpression in primary keratinocytes and other cell lines
significantly elevated the mRNA levels and transcriptional activity
of FOXM1. Thus, FOXM1 may be the target of GLI1 in basal cell
carcinomas. In addition, FOXM1 overexpression in non-small cell
lung carcinoma has been demonstrated to correlate with PTCH1, SMO
and GLI1 expression (48). In present
study, the association between FOXM1 and Hh signaling molecules was
analyzed, and the results indicated that FOXM1 expression
significantly correlated with GLI1 (R=0.405, P<0.001), SHh
(R=0.416, P<0.001) and PTCH1 (R=0.281, P=0.012) expression.
These data indicate that FOXM1 may be the downstream of the Hh
signaling pathway in cervical cancer. To the best of our knowledge,
the present study is the first to describe an association between
FOXM1 and the Hh signaling pathway in cervical cancer. Considering
previous and present results, it is possible to conclude that FOXM1
is a downstream target of the Hh signaling pathway in human
tumors.
The mechanism responsible for the regulation of
metastasis and invasion in cervical cancer is not thoroughly
understood at present. MMP-2 and MMP-9 principally function in
tumor invasion and migration (49),
whereas VEGF is a key factor in angiogenesis (50,51). In
epithelial ovarian cancer cells, FOXM1 downregulation led to
reduced expression of MMP-2, MMP-9 and VEGF (52). In a previous study, the present
authors used RNAi to inhibit the expression of FOXM1 in cervical
cancer cells, which consequently suppressed the activity and
expression of MMP-2, MMP-9 and VEGF (18). Previous studies have reported that the
inhibition of the Hh signaling pathway in glioma (53) and liver cancer (54) suppressed the expression of MMP-2,
MMP-9 and VEGF. FOXM1 may act as the downstream element of the Hh
signaling pathway, and both FOXM1 and the Hh signaling pathway may
modulate the expression and activity of MMP-2, MMP-9 and VEGF
(18,54–56). Thus,
it is possible to hypothesize that the Hh signaling pathway may
regulate the expression and activity of MMP-2, MMP-9 and VEGF
through FOXM1. Further studies are required to explore the exact
mechanism underlying this regulation.
In conclusion, FOXM1 may act as the downstream
target of the Hh signaling pathway. GLI1, as a translational
activator of this pathway, may be responsible for the translation
and activation of FOXM1. In the present study, the expression of
FOXM1, GLI1 and SMO correlated with cervical cancer clinical stage,
whereas the expression of GLI1, SHh and PTCH1 correlated with tumor
pathological grade. The present results were mainly based on the
analysis of cervical cancer tissues. Therefore, further experiments
using cervical cancer cells and cervical cancer orthotopic
implantation models should be conducted in order to understand the
exact mechanism underlying the regulation of cervical cancer.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
Statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giraldi G, Martinoli L and De Lucad'
Alessandro E: The human papillomavirus vaccination: A review of the
cost-effectiveness studies. Clin Ter. 165:e426–e432.
2014.PubMed/NCBI
|
|
4
|
Yang SH, Kong SK, Lee SH, Lim SY and Park
CY: Human papillomavirus 18 as a poor prognostic factor in stage
I–IIA cervical cancer following primary surgical treatment. Obstet
Gynecol Sci. 57:492–500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ludmir EB, Palta M, Zhang X, Wu Y, Willett
CG and Czito BG: Incidence and prognostic impact of high-risk HPV
tumor infection in cervical esophageal carcinoma. J Gastrointest
Oncol. 5:401–407. 2014.PubMed/NCBI
|
|
6
|
Buitrago-Pérez A, Garaulet G,
Vázquez-Carballo A, Paramio JM and García-Escudero R: Molecular
signature of HPV-induced carcinogenesis: pRb, p53 and gene
expression profiling. Curr Genomics. 10:26–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saha SK and Khuda-Bukhsh AR: Berberine
alters epigenetic modifications, disrupts microtubule network, and
modulates HPV-18 E6-E7 oncoproteins by targeting p53 in cervical
cancer cell HeLa: A mechanistic study including molecular docking.
Eur J Pharmacol. 744:132–146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jaiswal N, John R, Chand V and Nag A:
Oncogenic human papillomavirus 16E7 modulates SUMOylation of
FOXM1b. Int J Biochem Cell Biol. 58:28–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang C, Qiu Z, Wang L, Peng Z, Jia Z,
Logsdon CD, Le X, Wei D, Huang S and Xie K: A novel FOXM1-caveolin
signaling pathway promotes pancreatic cancer invasion and
metastasis. Cancer Res. 72:655–665. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Visnovsky J, Kudela E, Farkasova A,
Balharek T, Krkoska M and Danko J: Amplification of TERT and TERC
genes in cervical intraepithelial neoplasia and cervical cancer.
Neuro Endocrinol Lett. 35:518–522. 2014.PubMed/NCBI
|
|
11
|
Halasi M, Pandit B, Wang M, Nogueira V,
Hay N and Gartel AL: Combination of oxidative stress and FOXM1
inhibitors induces apoptosis in cancer cells and inhibits xenograft
tumor growth. Am J Pathol. 183:257–265. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Laoukili J, Stahl M and Medema RH: FOXM1:
At the crossroads of ageing and cancer. Biochim Biophys Acta.
1775:92–102. 2007.PubMed/NCBI
|
|
13
|
Zhao F, Siu MK, Jiang L, Tam KF, Ngan HY,
Le XF, Wong OG, Wong ES, Gomes AR, Bella L, et al: Overexpression
of forkhead box protein M1 (FOXM1) in ovarian cancer correlates
with poor patient survival and contributes to paclitaxel
resistance. PLoS One. 9:e1134782014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khongkow P, Karunarathna U, Khongkow M,
Gong C, Gomes AR, Yagüe E, Monteiro LJ, Kongsema M, Zona S, Man EP,
et al: FOXM1 targets NBS1 to regulate DNA damage-induced senescence
and epirubicin resistance. Oncogene. 33:4144–4155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bergamaschi A, MadakErdogan Z, Kim YJ,
Choi YL, Lu H and Katzenellenbogen BS: The forkhead transcription
factor FOXM1 promotes endocrine resistance and invasiveness in
estrogen receptor-positive breast cancer by expansion of stem-like
cancer cells. Breast Cancer Res. 16:4362014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang L, Wang P, Chen L and Chen H:
Down-regulation of FOXM1 by thiostrepton or small interfering RNA
inhibits proliferation, transformation ability and angiogenesis,
and induces apoptosis of nasopharyngeal carcinoma cells. Int J Clin
Exp Pathol. 7:5450–5460. 2014.PubMed/NCBI
|
|
17
|
Inoguchi S, Seki N, Chiyomaru T, Ishihara
T, Matsushita R, Mataki H, Itesako T, Tatarano S, Yoshino H, Goto
Y, et al: Tumour-suppressive microRNA-24-1 inhibits cancer cell
proliferation through targeting FOXM1 in bladder cancer. FEBS Lett.
588:3170–3179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen H, Zou Y, Yang H, Wang J and Pan H:
Downregulation of FOXM1 inhibits proliferation, invasion and
angiogenesis of HeLa cells in vitro and in vivo. Int J Oncol.
45:2355–2364. 2014.PubMed/NCBI
|
|
19
|
Guan P, Chen H, Li HJ, Duan J and Chen JY:
Expression and significance of FOXM1 in human cervical cancer: A
tissue micro-array study. Clin Invest Med. 34:E1–E7.
2011.PubMed/NCBI
|
|
20
|
Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW,
Cheung AN and Ngan HY: Over-expression of FOXM1 transcription
factor is associated with cervical cancer progression and
pathogenesis. J Pathol. 215:245–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He SY, Shen HW, Xu L, Zhao XH, Yuan L, Niu
G, You ZS and Yao SZ: FOXM1 promotes tumor cell invasion and
correlates with poor prognosis in early-stage cervical cancer.
Gynecol Oncol. 127:601–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Teh MT, Wong ST, Neill GW, Ghali LR,
Philpott MP and Quinn AG: FOXM1 is a downstream target of GLI1 in
basal cell carcinomas. Cancer Res. 62:4773–4780. 2002.PubMed/NCBI
|
|
23
|
Huang C, Du J and Xie K: FOXM1 and its
oncogenic signaling in pancreatic cancer pathogenesis. Biochim
Biophys Acta. 1845:104–116. 2014.PubMed/NCBI
|
|
24
|
Katoh Y and Katoh M: Hedgehog target
genes: Mechanisms of carcinogenesis induced by aberrant hedgehog
signaling activation. Curr Mol Med. 9:873–886. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shigemura K and Fujisawa M: Hedgehog
signaling and urological cancers. Curr Drug Targets. 16:258–271.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nüsslein-Volhard C and Wieschaus E:
Mutations affecting segment number and polarity in Drosophila.
Nature. 287:795–801. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Varjosalo M and Taipale J: Hedgehog:
Functions and mechanisms. Genes Dev. 22:2454–2472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hooper JE and Scott MP: Communicating with
Hedgehogs. Nat Rev Mol Cell Biol. 6:306–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang J and Hui CC: Hedgehog signaling in
development and cancer. Dev Cell. 15:801–812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Laurendeau I, Ferrer M, Garrido D, D'Haene
N, Ciavarelli P, Basso A, Vidaud M, Bieche I, Salmon I and Szijan
I: Gene expression profiling of the hedgehog signaling pathway in
human meningiomas. Mol Med. 16:262–270. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rimkus TK, Carpenter RL, Qasem S, Chan M
and Lo HW: Targeting the sonic hedgehog signaling pathway: Review
of smoothened and GLI inhibitors. Cancers (Basel). 8:E222016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Callahan BP and Wang C: Hedgehog
cholesterolysis: Specialized gatekeeper to oncogenic signaling.
Cancers (Basel). 7:2037–2053. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rovida E and Stecca B: Mitogen-activated
protein kinases and Hedgehog-GLI signaling in cancer: A crosstalk
providing therapeutic opportunities? Semin Cancer Biol. 35:154–167.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mathew E, Zhang Y, Holtz AM, Kane KT, Song
JY, Allen BL and di Magliano M Pasca: Dosage-dependent regulation
of pancreatic cancer growth and angiogenesis by hedgehog signaling.
Cell Reports. 9:484–494. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X,
Jin X and Tian X: MicroRNA-183 functions as the tumor suppressor
via inhibiting cellular invasion and metastasis by targeting MMP-9
in cervical cancer. Gynecol Oncol. S0090-8258(16): 300322016.(Epub
ahead of print).
|
|
36
|
Halasi M and Gartel AL: FOX(M1) news - it
is cancer. Mol Cancer Ther. 12:245–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kalin TV, Ustiyan V and Kalinichenko VV:
Multiple faces of FOXM1 transcription factor: Lessons from
transgenic mouse models. Cell Cycle. 10:396–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shin K, Lim A, Zhao C, Sahoo D, Pan Y,
Spiekerkoetter E, Liao JC and Beachy PA: Hedgehog signaling
restrains bladder cancer progression by eliciting stromal
production of urothelial differentiation factors. Cancer Cell.
26:521–533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sabol M, Trnski D, Uzarevic Z, Ozretic P,
Musani V, Rafaj M, Cindric M and Levanat S: Combination of
cyclopamine and tamoxifen promotes survival and migration of mcf-7
breast cancer cells - interaction of hedgehog-gli and estrogen
receptor signaling pathways. PLoS One. 9:e1145102014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kai K, Aishima S and Miyazaki K:
Gallbladder cancer: Clinical and pathological approach. World J
Clin Cases. 2:515–521. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xuan YH, Jung HS, Choi YL, Shin YK, Kim
HJ, Kim KH, Kim WJ, Lee YJ and Kim SH: Enhanced expression of
hedgehog signaling molecules in squamous cell carcinoma of uterine
cervix and its precursor lesions. Mod Pathol. 19:1139–1147.
2006.PubMed/NCBI
|
|
42
|
Samarzija I and Beard P: Hedgehog pathway
regulators influence cervical cancer cell proliferation, survival
and migration. Biochem Biophys Res Commun. 425:64–69. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Koo CY, Muir KW and Lam EW: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819:28–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Laoukili J, Alvarez M, Meijer LA, Stahl M,
Mohammed S, Kleij L, Heck AJ and Medema RH: Activation of FOXM1
during G2 requires cyclin A/Cdk-dependent relief of autorepression
by the FOXM1 N-terminal domain. Mol Cell Biol. 28:3076–3087. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wierstra I: Cyclin D1/Cdk4 increases the
transcriptional activity of FOXM1c without phosphorylating FOXM1c.
Biochem Biophys Res Commun. 431:753–759. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xue J, Zhou A, Tan C, Wu Y, Lee HT, Li W,
Xie K and Huang S: Forkhead box M1 is essential for nuclear
localization of glioma-associated oncogene homolog 1 in
glioblastoma multiforme cells by promoting importin-7 expression. J
Biol Chem. 290:18662–18670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pignot G, Vieillefond A, Vacher S, Zerbib
M, Debre B, Lidereau R, AmsellemOuazana D and Bieche I: Hedgehog
pathway activation in human transitional cell carcinoma of the
bladder. Br J Cancer. 106:1177–1186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gialmanidis IP, Bravou V, Amanetopoulou
SG, Varakis J, Kourea H and Papadaki H: Overexpression of hedgehog
pathway molecules and FOXM1 in non-small cell lung carcinomas. Lung
Cancer. 66:64–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bourboulia D and Stetler-Stevenson WG:
Matrix metalloproteinases (MMPs) and tissue inhibitors of
metalloproteinases (TIMPs): Positive and negative regulators in
tumor cell adhesion. Semin Cancer Biol. 20:161–168. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bourdeanu L and Luu T: J Adv Pract Oncol.
5:246–260. 2014.PubMed/NCBI
|
|
51
|
Wainberg ZA and Drakaki A: The importance
of optimal drug sequencing in metastatic colorectal cancer:
Biological rationales for the observed survival benefit conferred
by first-line treatment with EGFR inhibitors. Expert Opin Biol
Ther. 15:1205–1220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wen N, Wang Y, Wen L, Zhao SH, Ai ZH, Wang
Y, Wu B, Lu HX, Yang H, Liu WC and Li Y: Overexpression of FOXM1
predicts poor prognosis and promotes cancer cell proliferation,
migration and invasion in epithelial ovarian cancer. J Transl Med.
12:1342014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cui D, Chen X, Yin J, Wang W, Lou M and Gu
S: Aberrant activation of Hedgehog/GLI1 pathway on angiogenesis in
gliomas. Neurol India. 60:589–596. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen JS, Huang XH, Wang Q, Huang JQ, Zhang
LJ, Chen XL, Lei J and Cheng ZX: Sonic hedgehog signaling pathway
induces cell migration and invasion through focal adhesion
kinase/AKT signaling-mediated activation of matrix
metalloproteinase (MMP)-2 and MMP-9 in liver cancer.
Carcinogenesis. 34:10–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hwang J, Kang MH, Yoo YA, Quan YH, Kim HK,
Oh SC and Choi YH: The effects of sonic hedgehog signaling pathway
components on non-small-cell lung cancer progression and clinical
outcome. World J Surg Oncol. 12:2682014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Moeini A, Cornellà H and Villanueva A:
Emerging signaling pathways in hepatocellular carcinoma. Liver
Cancer. 1:83–93. 2012. View Article : Google Scholar : PubMed/NCBI
|