Introduction
Esophageal squamous cell carcinoma (ESCC) is one of
the most malignant types of cancer and is ranked as the sixth most
common cause of cancer-associated mortality in the world, with a
high incidence in China (1,2). Although diagnostic methods and cancer
therapies have been improved in recent years, the prognosis remains
poor due to widespread lymph node metastasis and frequent distant
metastasis. Therefore, the identification and understanding of
candidate target genes of genomic aberrations underlying esophageal
carcinogenesis and metastasis may provide a new treatment target in
ESCC.
ESCC is a multi-step process, and genomic aberration
is a characteristic attribute. Several studies have identified that
losses of 9p21.3, 16p13.3 and 18q22-qter and gains of 5p15, 7p22.1,
11q13.3 and 19p13.11 were the most frequent genomic aberrations in
ESCC (3–5). Functional studies identified a number of
candidate target driver genes of genomic aberration. For example,
genomic loss and hypermethylation of nc886 (noncoding RNA) were
detected in ESCC, and ectopically expressed nc886 could inhibit
proliferation of ESCC cells (6).
Togashi et al (7) also
reported that oral cancer overexpressed 1 (ORAOV1) located at 11q13
was amplified in 53% of ESCC patients, and its amplification was
significantly associated with poor differention status.
Overexpression of ORAOV1 could enhance tumorigenicity and tumor
growth (7). Thus, the identification
of candidate tumor-associated genes is crucial in revealing the
mechanism of tumorigenesis of esophageal cancer.
In the present study, we integratively analyzed the
array comparative genomic hybridization (CGH) data and observed
that homozygous deletions of cyclin-dependent kinase inhibitor
(CDKN) 2A (9p21.3) and CDKN2B (9p21.3) and gains of fascin
actin-bundling protein 1 (FSCN1) (7p22.1) and homer scaffolding
protein 3 (HOMER3) (19p13.11) occurred frequently in ESCC. We
further analyzed the correlations between the copy number changes
of CDKN2A, CDKN2B, FSCN1, HOMER3 and clinical factors, and
investigated the expression level of these genes in ESCC
samples.
Materials and methods
Patients and samples
Freshly resected tissues from 115 ESCC patients were
collected at the Department of Pathology, Cancer Hospital, Chinese
Academy of Medical Sciences, Beijing, China. All of the esophageal
cancer patients were treated with radical surgery, and none of them
received any treatment prior to surgery. Representative tumor
regions were excised by experienced pathologists and immediately
stored at −70°C until use.
Biopsy tissues were taken from symptom-free patients
during endoscopic screening for esophageal cancer in Linzhou,
China, which is a well-recognized high-risk area for ESCC. During
endoscopy, the entire esophagus was visually examined and biopsies
were taken from all focal lesions. If no focal lesions were
detected, a standard site in the mid-esophagus was sampled.
All the samples used in this study were residual
specimens following diagnostic sampling. Every patient signed
separate informed consent forms for sampling and molecular
analysis. The clinicopathological characteristics of patients are
shown in Table I. This study was
approved by the Ethics Committee of Kunming University of Science
and Technology (Kunming, Yunnan, China)
 | Table I.Correlations between copy number
changes of genes and clinical factors. |
Table I.
Correlations between copy number
changes of genes and clinical factors.
|
| CDKN2A | CDKN2B | FSCN1 | HOMER3 |
|---|
|
|
|
|
|
|
|---|
| Factors | HD | No HD | P-value | HD | No HD | P-value | Gain | No gain | P-value | Gain | No gain | P-value |
|---|
| Gender |
|
| 0.764 |
|
| 0.777 |
|
| 0.387 |
|
| 1.000 |
| Male | 20 | 43 |
| 23 | 40 |
| 25 | 38 |
| 14 | 49 |
|
|
Female | 4 | 12 |
| 5 | 11 |
| 4 | 12 |
| 3 | 13 |
|
| Age |
|
| 0.804 |
|
| 0.098 |
|
| 1.000 |
|
| 0.102 |
| ≤60 | 9 | 24 |
| 8 | 25 |
| 12 | 21 |
| 4 | 29 |
|
|
>60 | 15 | 31 |
| 20 | 26 |
| 17 | 29 |
| 13 | 33 |
|
| pT |
|
| 0.586 |
|
| 0.795 |
|
| 0.002 |
|
| 0.130 |
|
T1-T2 | 8 | 14 |
| 7 | 15 |
| 2 | 20 |
| 2 | 20 |
|
|
T3-T4 | 16 | 41 |
| 21 | 36 |
| 27 | 30 |
| 15 | 42 |
|
| pN |
|
| 0.003 |
|
| 0.011 |
|
| 0.022 |
|
| 0.013 |
| N0 | 6 | 35 |
| 9 | 32 |
| 10 | 31 |
| 4 | 37 |
|
| N1 | 18 | 20 |
| 19 | 19 |
| 19 | 19 |
| 13 | 25 |
|
| pStage |
|
| 0.207 |
|
| 0.146 |
|
| 0.000 |
|
| 0.000 |
|
I–II | 12 | 37 |
| 14 | 35 |
| 9 | 40 |
| 4 | 45 |
|
|
III | 12 | 18 |
| 14 | 16 |
| 20 | 10 |
| 13 | 17 |
|
| pGrade |
|
| 0.371 |
|
| 0.233 |
|
| 0.282 |
|
| 0.284 |
| G1 | 9 | 13 |
| 11 | 11 |
| 11 | 11 |
| 5 | 17 |
|
| G2 | 12 | 30 |
| 13 | 29 |
| 14 | 28 |
| 11 | 31 |
|
| G3 | 3 | 12 |
| 4 | 11 |
| 4 | 11 |
| 1 | 14 |
|
Microarray data analysis
We integratively analyzed the array CGH data of 79
ESCC cases using Genomic Workbench (Agilent Technologies, Santa
Clara, CA, USA) and MD-SeeGH (www.flintbox.ca). Genomic Workbench was used to
calculate the log2ratio for every probe and
identify genomic aberrations. The mean
log2ratio of all probes in a chromosome
region between 0.25 and 0.75 was classified as a genomic gain,
>0.75 as an amplification, ≤0.25 as a hemizygous loss, and ≤0.75
as homozygous deletion.
Quantitative polymerase chain reaction
(qPCR)
qPCR was used to detect the expression level of
candidate genes. The PCR reactions were performed in a total volume
of 20 µl, including 10 µl 2X Power SYBR®-Green PCR
master mix (Applied Biosystems, Warrington, UK), 2 µl cDNA (5
ng/µl) and 1 µl primer mix (10 µM each). The PCR amplification and
detection were carried out in a LightCycler 480 II (Roche Applied
Science, Manheim, Germany) as follows: initial denaturation at 95°C
for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min. The
relative expression was calculated using the comparative CT method.
The expression of the target gene normalized to an endogenous
reference (GAPDH) and relative to the calibrator was given by the
formula 2−ΔΔCq. ΔCT was calculated by
subtracting the average GAPDH CT from the average CT of the gene of
interest. The ratio defines the level of relative expression status
of the target gene to that of GAPDH. The following primer pairs
were used for the PCR assay: CDKN2A forward,
5′-CTTCCTGGACACGCTGGTG-3′, CDKN2A reverse,
5′-AATCGGGGATGTCTGAGGGA-3′; CDKN2B forward,
5′-CGGGGACTAGTGGAGAAGGTG-3′, CDKN2B reverse,
5′-CCATCATCATGACCTGGATCGC-3′; FSCN1 forward,
5′-CAAAAAGTGTGCCTTCCGTACC-3′, FSCN1 reverse,
5′-CCCATTCTTCTTGGAGGTCACA-3′; HOMER3 forward,
5′-CCAAGGACCAGGAGATTCAGAC-3′, HOMER3 reverse,
5′-AGCTCACTCAGCTCAAACAGG-3′; GAPDH forward,
5′-AAATCCCATCACCATCTTCCAG-3′, GAPDH reverse,
5′-GAGTCCTTCCACGATACCAAAGTTG-3′.
Statistical analysis
Statistical analyses were conducted using Student's
t-tests and Chi-square tests using the statistical software SPSS
version 15.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Homozygous deletion of CDKN2A and
CDKN2B in ESCC
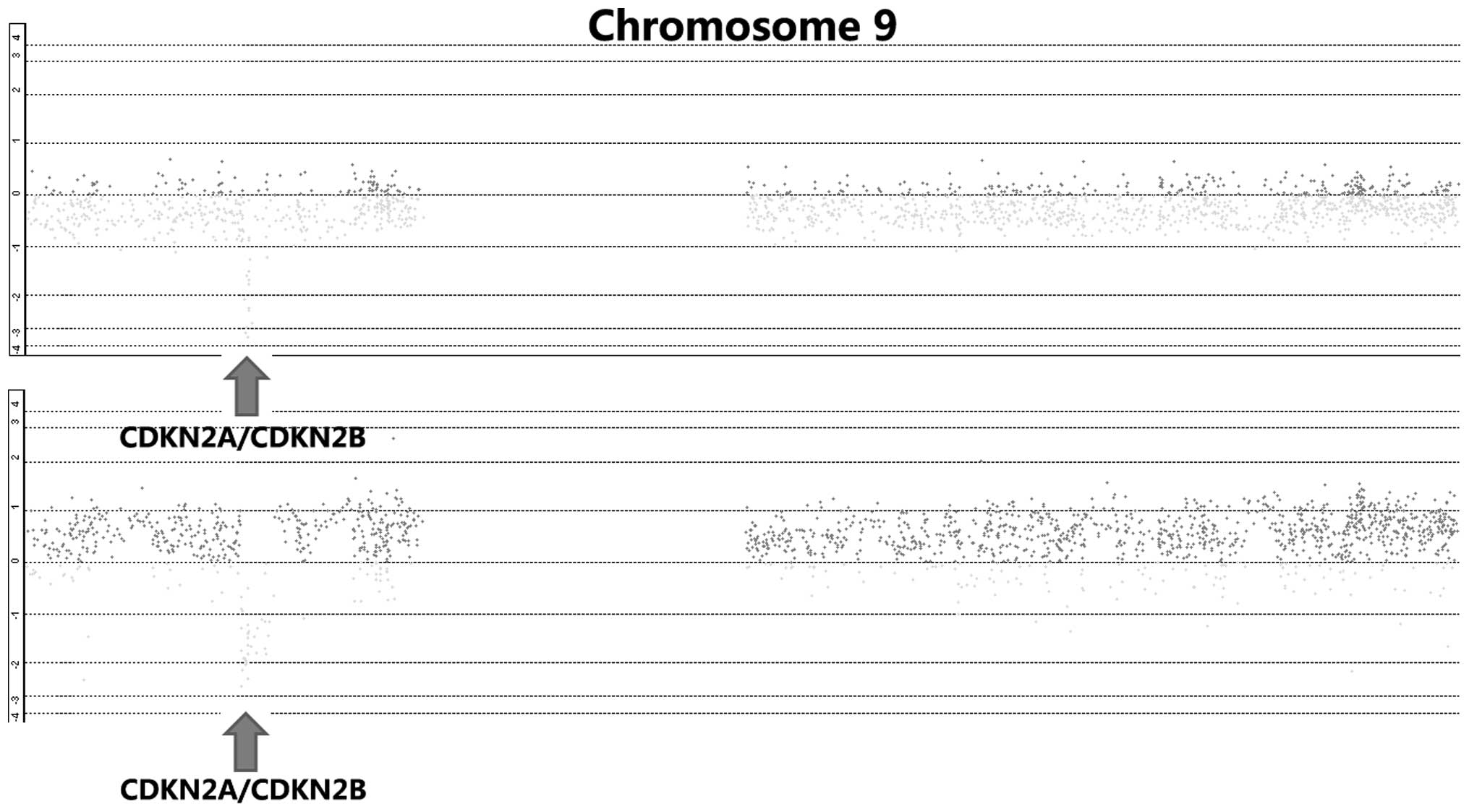
By integrative analysis of array CGH data from 79
ESCC cases, we observed that CDKN2A and CDKN2B were homozygously
deleted in 18.6% of cases (Fig. 1).
We further analyzed the correlations between copy number decreases
of CDKN2A as well as CDKN2B and clinical parameters, and observed
that homozygous deletion of CDKN2A or CDKN2B was significantly
associated with lymph node metastasis (P=0.003 and P=0011,
respectively; Table I). There were no
correlations between homozygous deletion of CDKN2A or CDKN2B and
gender, age, pT, pStage or pGrade (Table
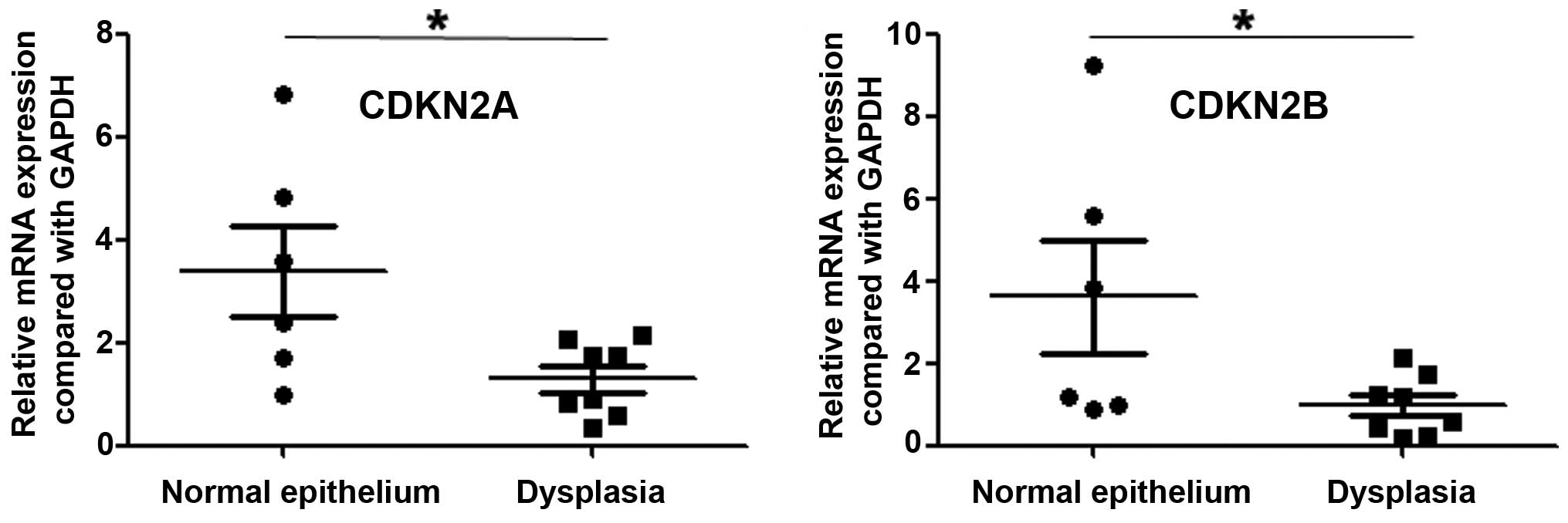
I). Most significantly, we analyzed the expression of CDKN2A
and CDKN2B in precancerous lesions, and observed that the
expression of these two genes was lower in dysplasia than in normal
esophageal epithelium (P=0.024 and P=0.048, respectively; Fig. 2).
Gains of FSCN1 and HOMER3 in ESCC
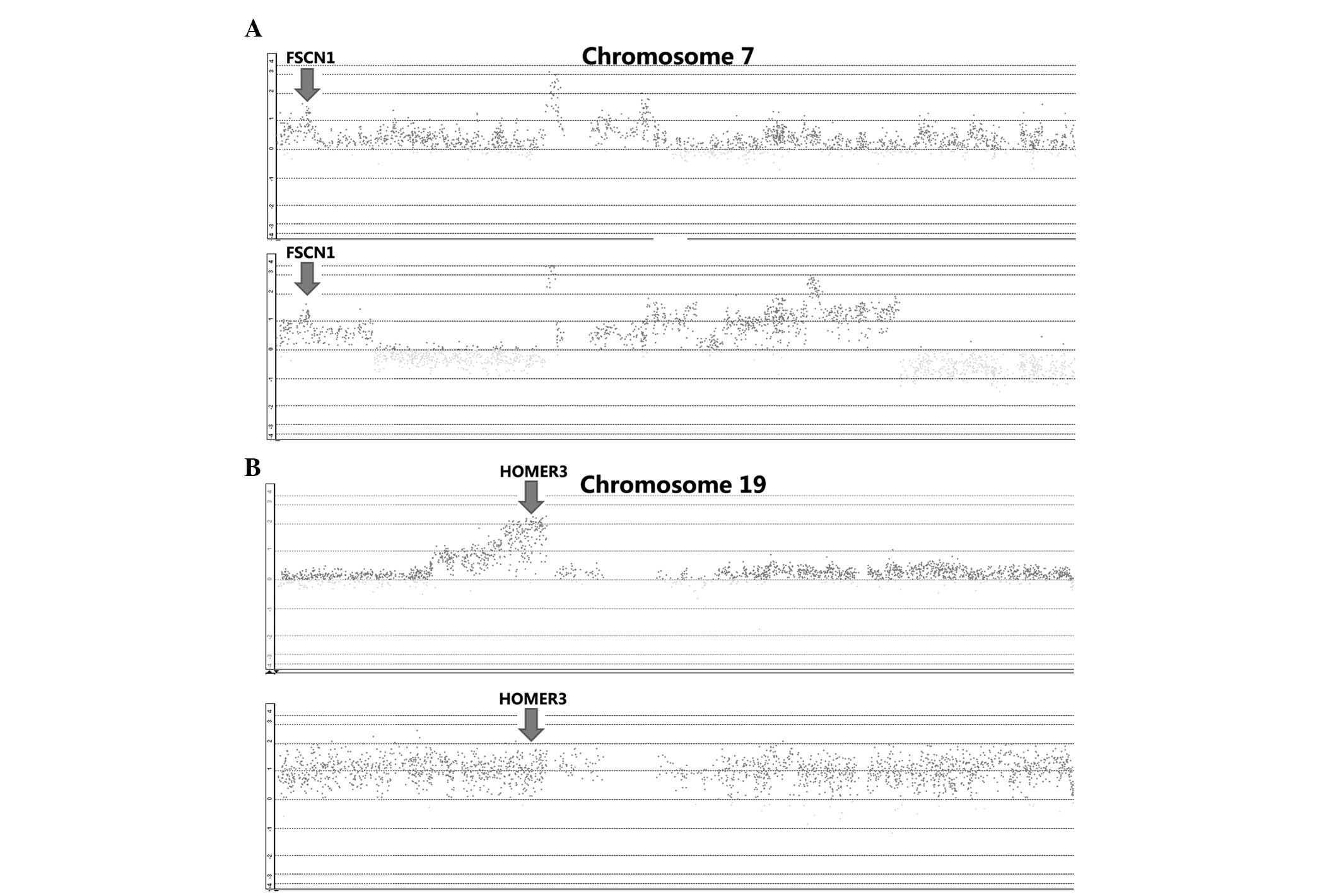
By integrative analysis of array CGH data from 79
ESCC cases, we observed that gains of FSCN1 and HOMER3 were
detected in 41 and 80% of patients, respectively (Fig. 3). We further analyzed the correlations
between gains of FSCN1 or HOMER3 and clinical factors, and observed
that gain of FSCN1 was significantly associated with pT, pN and
pStage (P=0.002, P=0.022 and P=0.000, respectively; Table I). In addition, gain of HOMER3 was
significantly linked with pN and pStage (P=0.013 and P=0.000,
respectively; Table I). We further
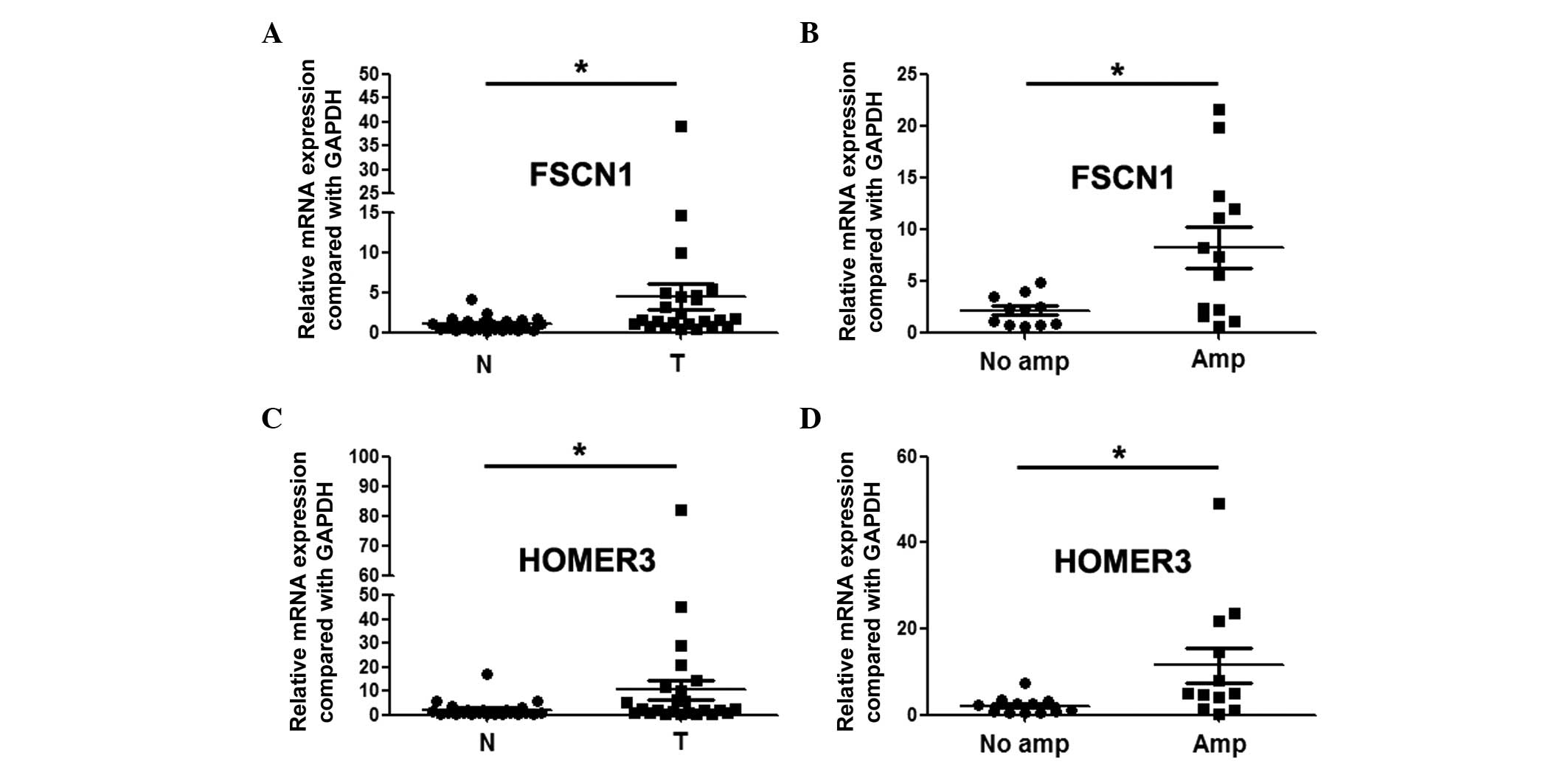
revealed through qPCR assay that FSCN1 and HOMER3 were
overexpressed in ESCC, and the expression level in patients with
gains was significantly higher than that in patients without gains
(Fig. 4).
Discussion
Genomic aberrations are one of the mechanisms
resulting in gene dysfunction, and contribute to carcinogenesis and
tumor progression. Differentially expressed genes associated with
DNA copy number changes may be candidate targets of amplifications
or homozygous deletions, and play significant roles in
tumorigenesis and the development of cancer (8).
9p21.3 is the most common loss in numerous types of
cancer, including glioblastoma, esophageal cancer and pancreatic
cancer (9,10), and is also the most frequent early
event in the tumorigenesis of Epstein-Barr virus-associated
nasopharyngeal carcinoma (11).
CDKN2A and CDKN2B were identified as the driver genes of 9p21.3
loss. CDKN2A and CDKN2B are frequently inactivated in a number of
cancer types. In lung cancer, CDKN2A is frequently inactivated via
homozygous deletions, the methylation of the promoter region, or
point mutations (12). In rectal
cancer, Kohonen-Corish et al observed that CDKN2A methlytion
occurred in 20% of patients, and that CDKN2A methlytion was
associated with poor overall survival (13). CDKN2A hypermethylation is also
associated with lymphovascular invasion, lymph node metastasis and
proximal tumor location in colorectal cancer (14). In ovarian cancer, deletion of CDKN2A
is also a common event (15). Our
study identified that homozygous deletion of CDKN2A or CDKN2B was
significantly associated with lymph node metastasis. Most
significantly, the expression of CDKN2A and CDKN2B was lower in
dysplasia than in normal esophageal epithelium. These results
indicate that CDKN2A and CDKN2B may play a significant role in the
early formation of esophageal cancer.
Our study also revealed that FSCN1 and HOMER3, which
were gained in ESCC, were overexpressed, and higher expression
levels of these two genes were associated with copy number
increase. The literature reveals that the expression of FSCN1 is an
independent poor prognostic factor according to a multivariate
analysis, and co-expression with MMP14 correlates with the poorest
overall survival in esophageal cancer. The knockdown of FSCN1
inhibits the proliferation and invasion of ESCC cells (16). Chen et al (17) reported that FSCN1 was overexpressed in
colorectal cancers, and was associated with cancer cell
progression. Hanker et al (18) observed that lower FSCN1 expression was
associated with significantly poorer overall survival in epithelial
ovarian cancer. In tumors, FSCN1 could be directly targeted and
regulated by miR-133a, miR-133b, miR-143 and miR-145 (19–22). Fuse
et al (23) further revealed
that restoration of miR-145 expression suppresses cell
proliferation, migration and invasion in prostate cancer by
targeting FSCN1. HOMER3 is a member of the cytoplasmic scaffolding
proteins family, and regulates transcription and plays an essential
role in the development and differentiation of certain tissues,
including muscle and nervous systems (24–26).
HOMER3 is overex6pressed in acute myeloid leukemia (AML), and
decreased expression of HOMER3 is associated with poor prognosis in
AML (27,28). Li et al (29) further reported that forced expression
of HOMER3 in K562 cells could inhibit proliferation, influence the
cell cycle, and affect apoptosis induced by As2O3 via inhibition of
Bcl-2 expression. However, the roles of FSCN1 and HOMER3 were
previously unclear in esophageal carcinogenesis.
In summary, our study suggested that CDKN2A, CDKN2B,
FSCN1 and HOMER3 were candidate cancer-associated genes and may
play a tumorigenic role in ESCC.
Acknowledgements
This study was funded by the National Natural
Science Foundation of China (grant no. 81460425), the Yunnan
Provincial Research Foundation for Basic Research, China (grant no.
2013FD012), the Foundation for the Talents of Kunming University of
Science and Technology (grant no. KKSY201226099) and Yunnan
Provincial Engineering Center of Translational Cancer Medicine
(grant no. 2011DH011).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yen CC, Chen YJ, Chen JT, Hsia JY, Chen
PM, Liu JH, Fan FS, Chiou TJ, Wang WS and Lin CH: Comparative
genomic hybridization of esophageal squamous cell carcinoma:
correlations between chromosomal aberrations and disease
progression/prognosis. Cancer. 92:2769–2777. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shi ZZ, Liang JW, Zhan T, Wang BS, Lin DC,
Liu SG, Hao JJ, Yang H, Zhang Y, Zhan QM, et al: Genomic
alterations with impact on survival in esophageal squamous cell
carcinoma identified by array comparative genomic hybridization.
Genes Chromosomes Cancer. 50:518–526. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hirasaki S, Noguchi T, Mimori K, Onuki J,
Morita K, Inoue H, Sugihara K, Mori M and Hirano T: BAC clones
related to prognosis in patients with esophageal squamous
carcinoma: an array comparative genomic hybridization study.
Oncologist. 12:406–417. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee HS, Lee K, Jang HJ, Lee GK, Park JL,
Kim SY, Kim SB, Johnson BH, Zo JI, Lee JS and Lee YS: Epigenetic
silencing of the non-coding RNA nc886 provokes oncogenes during
human esophageal tumorigenesis. Oncotarget. 5:3472–3481. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Togashi Y, Arao T, Kato H, Matsumoto K,
Terashima M, Hayashi H, de Velasco MA, Fujita Y, Kimura H, Yasuda
T, et al: Frequent amplification of ORAOV1 gene in esophageal
squamous cell cancer promotes an aggressive phenotype via proline
metabolism and ROS production. Oncotarget. 5:2962–2973. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability - an evolving hallmark of cancer. Nat Rev
Mol Cell Biol. 11:220–228. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Riehmer V, Gietzelt J, Beyer U, Hentschel
B, Westphal M, Schackert G, Sabel MC, Radlwimmer B, Pietsch T,
Reifenberger G, et al: Genomic profiling reveals distinctive
molecular relapse patterns in IDH1/2 wild-type glioblastoma. Genes
Chromosomes Cancer. 53:589–605. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi ZZ, Shang L, Jiang YY, Hao JJ, Zhang
Y, Zhang TT, Lin DC, Liu SG, Wang BS, Gong T, et al: Consistent and
differential genetic aberrations between esophageal dysplasia and
squamous cell carcinoma detected by array comparative genomic
hybridization. Clin Cancer Res. 19:5867–5878. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheung CC, Chung GT, Lun SW, To KF, Choy
KW, Lau KM, Siu SP, Guan XY, Ngan RK, Yip TT, et al: miR-31 is
consistently inactivated in EBV-associated nasopharyngeal carcinoma
and contributes to its tumorigenesis. Mol Cancer. 13:1842014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tam KW, Zhang W, Soh J, Stastny V, Chen M,
Sun H, Thu K, Rios JJ, Yang C, Marconett CN, et al: CDKN2A/p16
inactivation mechanisms and their relationship to smoke exposure
and molecular features in non-small-cell lung cancer. J Thorac
Oncol. 8:1378–1388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
KohonenCorish MR, Tseung J, Chan C, Currey
N, Dent OF, Clarke S, Bokey L and Chapuis PH: KRAS mutations and
CDKN2A promoter methylation show an interactive adverse effect on
survival and predict recurrence of rectal cancer. Int J Cancer.
134:2820–2828. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J
and Chen M: The prognostic value of CDKN2A hypermethylation in
colorectal cancer: a meta-analysis. Br J Cancer. 108:2542–2548.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aravidis C, Panani AD, Kosmaidou Z,
Thomakos N, Rodolakis A and Antsaklis A: Detection of numerical
abnormalities of chromosome 9 and p16/CDKN2A gene alterations in
ovarian cancer with fish analysis. Anticancer Res. 32:5309–5313.
2012.PubMed/NCBI
|
|
16
|
Akanuma N, Hoshino I, Akutsu Y, Murakami
K, Isozaki Y, Maruyama T, Yusup G, Qin W, Toyozumi T, Takahashi M,
et al: MicroRNA-133a regulates the mRNAs of two invadopodia-related
proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer.
110:189–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen MB, Wei MX, Han JY, Wu XY, Li C, Wang
J, Shen W and Lu PH: MicroRNA-451 regulates AMPK/mTORC1 signaling
and fascin1 expression in HT-29 colorectal cancer. Cell Signal.
26:102–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanker LC, Karn T, Holtrich U, Graeser M,
Becker S, Reinhard J, Ruckhäberle E, Gevensleben H and Rody A:
Prognostic impact of fascin-1 (FSCN1) in epithelial ovarian cancer.
Anticancer Res. 33:371–377. 2013.PubMed/NCBI
|
|
19
|
Wu ZS, Wang CQ, Xiang R, Liu X, Ye S, Yang
XQ, Zhang GH, Xu XC, Zhu T and Wu Q: Loss of miR-133a expression
associated with poor survival of breast cancer and restoration of
miR-133a expression inhibited breast cancer cell growth and
invasion. BMC Cancer. 12:512012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin Y, Dang X, Li W and Ma Q: miR-133a
functions as a tumor suppressor and directly targets FSCN1 in
pancreatic cancer. Oncol Res. 21:353–363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M and
Matsubara H: miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu R, Liao J, Yang M, Sheng J, Yang H,
Wang Y, Pan E, Guo W, Pu Y, Kim SJ and Yin L: The cluster of
miR-143 and miR-145 affects the risk for esophageal squamous cell
carcinoma through co-regulating fascin homolog 1. PLoS One.
7:e339872012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fuse M, Nohata N, Kojima S, Sakamoto S,
Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Naya Y, Ichikawa T
and Seki N: Restoration of miR-145 expression suppresses cell
proliferation, migration and invasion in prostate cancer by
targeting FSCN1. Int J Oncol. 38:1093–1101. 2011.PubMed/NCBI
|
|
24
|
Xiao B, Tu JC, Petralia RS, Yuan JP, Doan
A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ and Worley PF:
Homer regulates the association of group 1 metabotropic glutamate
receptors with multivalent complexes of homer-related, synaptic
proteins. Neuron. 21:707–716. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bortoloso E, Pilati N, Megighian A,
Tibaldo E, Sandonà D and Volpe P: Transition of Homer isoforms
during skeletal muscle regeneration. Am J Physiol Cell Physiol.
290:C711–C718. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ishiguro K and Xavier R: Homer-3 regulates
activation of serum response element in T cells via its EVH1
domain. Blood. 103:2248–2256. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stirewalt DL, Meshinchi S, Kopecky KJ, Fan
W, PogosovaAgadjanyan EL, Engel JH, Cronk MR, Dorcy KS, McQuary AR,
Hockenbery D, et al: Identification of genes with abnormal
expression changes in acute myeloid leukemia. Genes Chromosomes
Cancer. 47:8–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Valk PJ, Verhaak RG, Beijen MA, Erpelinck
CA, van Waalwijk van Doorn-Khosrovani S Barjesteh, Boer JM,
Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B and Delwel
R: Prognostically useful gene-expression profiles in acute myeloid
leukemia. N Engl J Med. 350:1617–1628. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Z, Qiu HY, Jiao Y, Cen JN, Fu CM, Hu
SY, Zhu MQ, Wu DP and Qi XF: Growth and differentiation effects of
Homer3 on a leukemia cell line. Asian Pac J Cancer Prev.
14:2525–2528. 2013. View Article : Google Scholar : PubMed/NCBI
|