Introduction
The limited success and considerable side effects of
classic chemotherapeutic agents has led to researchers attempting
to identify natural anti-tumor compounds with fewer side effects.
Umbelliprenin (Umb; C24H30O3;
molecular weight, 366), a member of 7-prenyloxycoumarins, is a
naturally occurring compound, which is isolated from Ferula
species, including Ferula szowitsiana (1,2). This
compound is able to suppress tumorigenesis through its
anti-inflammatory, anti-genotoxicity, anti-invasive and
lipoxygenase inhibitory activities (3–6). In
vitro, Umb has been reported to attenuate the proliferation of
various cancer cell lines, and is able to prevent/delay cancer in
in vivo animal models of papilloma and lung cancer (7–9). It
hasbeen stated that the anti-tumor activities of Umb are partly due
to a prenyl moiety (C7-OH) on the structure of the umbelliferone
nucleus (3).
Lung cancer may be divided into two groups based on
the size and appearance of the malignant cells: Non-small cell lung
carcinoma (NSCLC) and small cell lung carcinoma (10). In France, NSCLC accounts for >80%
of all lung tumors (10) and is
comprised of three major subtypes, including adenocarcinoma,
squamous and large-cell carcinoma. Large-cell lung carcinoma (13%
of all lung cancer cases) has the poorest prognosis of the three
subtypes, with a 4-year mortality rate of 93% compared with 88% for
adenocarcinoma and 85% for squamous cell lung carcinoma (10). The cytotoxic/anti-proliferative
effects of Umb have previously been demonstrated in lung cancer
cell lines (2). Using MTT assay in
combination with flow cytometry, it was demonstrated that Umb has
more potent cytotoxicity against QU-DB cells, a large-cell lung
cancer cell, compared withA549 adenocarcinoma cells (2). Despite the dominant cytotoxic activities
of Umb against certain cancer cell lines (2,3,8), its proliferative activities have been
observed in normal lymphocytes (2)
and at lower doses in a colorectal cancer cell line in culture
media (11), indicating that this
compound may have different levels of cytotoxicity against
different cancer cell lines in vitro.
Although a number of studies are available regarding
Umb anti-tumor activities, data regarding the molecular targets of
Umb are limited. Proteomic studies that have performed
high-throughput analyses of cell proteomes have identified
biomarkers and therapeutic targets in various forms of cancer. As
the most widely used method in proteomics, two-dimensional gel
electrophoresis (2-DE) coupled with mass spectrophotometry (MS),
has demonstrated great promise for the identification of molecular
targets of cancer drugs (12).
The present study aimed to identify the molecular
targets of Umb in two human lung cancer cell lines. To fulfill
this, the protein targets of Umb were investigatedusing2-DE coupled
with liquid chromatography (LC) -MS/MS in QU-DB large cell
carcinoma and A549 adenocarcinoma cell lines, the two cancer cell
lines whose in vitro proliferation rate were previously
investigated following treatment with Umb (2).
Materials and methods
Preparation of Umb and cell
culture
The synthesis of Umb was performed as previously
described (13), and its purity was
verified by nuclear magnetic resonance. For its addition to
culture, Umb was dissolved in dimethyl sulfoxide (DMSO) and diluted
with RPMI-1640 supplemented with 10% fetal bovine serum (Biosera,
Ringmer, UK) prior to use. Human QU-DB and A549 lung cancer cell
lines (Pasteur Institute of Iran, Tehran, Iran) were cultured at
37°C in an atmosphere of 5% CO2 and treated with either
Umb (IC30 value, ~31 µM) or DMSO (0.15%) for 48 h, as
test or control groups, respectively, as previously described
(2).
2-DE, gel staining and scanning
Sample preparations and 2-DE were performed
according to methods described previously (14). Briefly, cells were solubilized in a
lysis buffer. Following 2 h incubation at room temperature, the
supernatant was collected, aliquoted and stored at −70°C. Protein
concentration was determined using the Bradford method (15). Immobilized non-linear pH 3–10 gradient
strips (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) were used
for the first dimension, in which proteins are separated based on
their isoelectric points (14).
Isoelectric focusing (IEF) was performed on a PROTEAN®
IEF Cell system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The second dimension, in which proteins are separated based on
their molecular mass, as performed on a PROTEANII xi 2-D Cell
system (Bio-Rad Laboratories, Inc.).
Analytical silver staining (mass incompatible) and
preparative silver staining (mass compatible) were performed as
previously described (16). The gels
were scanned with a GS-800 Calibrated Densitometer (Bio-Rad
Laboratories, Inc.), and the volume of each spot was subsequently
analyzed by Prodigy SameSpots version 1.0 software (Nonlinear
Dynamics, Newcastle, UK) to quantify the protein expression
levels.
LC-MS/MS
The obtained spots were destained in 30 mM potassium
ferricyanide: 100 mM sodium thiosulfate (1:1) for 10 min. In-gel
digestion was performed as previously described (16). LC-MS/MS was performed using an Agilent
1100 Series LC/MSD Trap XCT (Agilent Technologies, Inc., Santa
Clara, CA, USA). Each sample (25 µl) was separated on a
Zorbax® 300SB-C18 column (75 µm, 150 mm; Agilent
Technologies, Inc.) Protein identification was performed using the
Agilent Spectrum MILL MS proteomics workbench (Agilent
Technologies, Inc.).
Statistical analysis
The statistical differences in protein spot
expression between treated and untreated groups were calculated
using (Prodigy Software version 1.0). The spot groups which
exhibited a >1.5 fold increase in expression after treatment
were considered as differentially expressed proteins. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were performed in triplicate.
Results
Differentially-expressed protein
targets
2-DE and LC-MS/MS were used to identify protein
targets in Umb-treated QU-DB and A549 cells in comparison with
corresponding DMSO-treated control cells. Three gels were run for
each group. In total, 30 differentially-expressed protein targets
were identified in the two Umb-treated human lung cancer cell lines
compared with their DMSO controls.
Differentially-expressed proteins in
the Umb-treated QU-DB cells
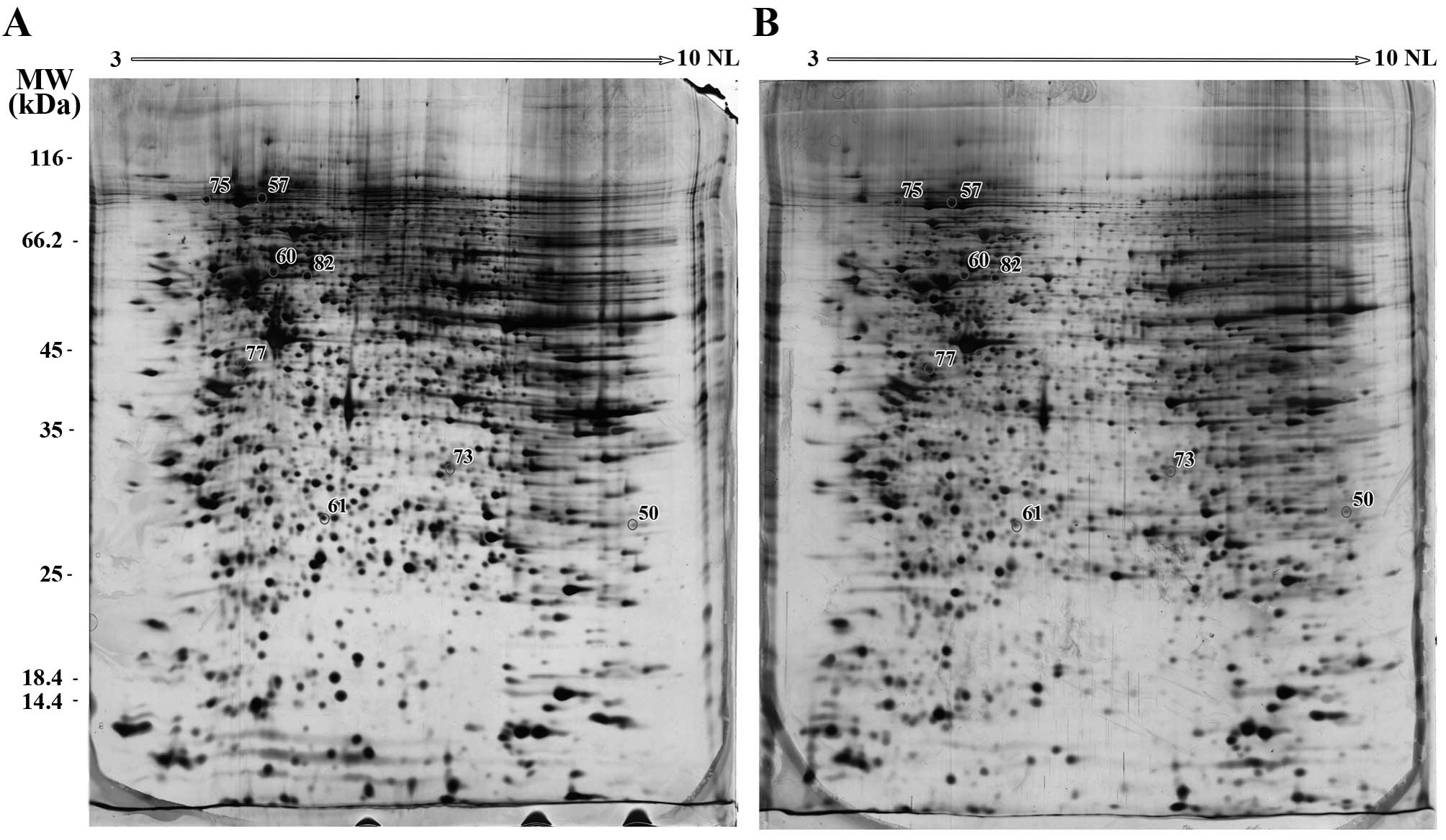
In gels derived from the QU-DB cells, collectively,
45 reproducible, distinct spots were observed to be
differentially-expressed. Due to limitation in MS availability, 20
intense spots were picked up from gels and subjected to MS. Of
these 20 spots, 8 spots were successfully identified by MS
(Table I). A number of these 8 spots
were mixture of several proteins, resulting in the identification
of 14 different proteins. A total of 12 proteins were downregulated
and two upregulated. The locations of these identified spots on the
QU-DB gels are presented in Fig.
1.
 | Table I.Descriptions of the identified
differentially-expressed protein spots in umbelliprenin-treated
QU-DB compared with dimethyl sulfoxide-treated control cells. |
Table I.
Descriptions of the identified
differentially-expressed protein spots in umbelliprenin-treated
QU-DB compared with dimethyl sulfoxide-treated control cells.
| Spot no. | Protein name | Accession
no.a | pI/MW, kDa | Scoreb | Function | Location | Expression |
|---|
| 57 | HSP90-β | P08238 | 4.97/83.26 | 92.18 | Chaperone
function | Mitochondria,
cytoplasm | ↓ |
| 57 | Transitional
endoplasmic reticulum ATPase (p97/VCP) | P55072 | 5.14/89.32 | 84.93 | ATPases, cell
cycle | Cytoplasm | ↓ |
| 60 | Vimentin | P08670 | 5.06/53.65 | 73.97 | Intermediate
filaments | Cytoskeletal
component | ↓ |
| 60 | Splicing factor 3A
subunit-3 (SF3a3) | Q12874 | 5.27/58.84 | 71.64 | Splicing
factor | Nucleus
speckle | ↓ |
| 60 | Importin α-2 | P52292 | 5.25/57.86 | 60.73 |
Nucleo-cytoplasmatic transport | Cytoplasm,
nucleus | ↓ |
| 61 | HSP27 | P04792 | 5.98/22.78 | 67.2 | Chaperone
function | Cytoplasm,
nucleus | ↓ |
| 61 | NADH dehydrogenase
[ubiquinone] iron-sulfur protein-3 (NDUFS3) | O75489 | 6.98/30.24 | 54.7 | Transfer of
electrons | Mitochondrion inner
membrane | ↓ |
| 75 | Importin β-1 | Q14974 | 4.68/97.17 | 46.4 |
Nucleo-cytoplasmatic transport | Cytoplasm,
nucleus | ↓ |
| 75 | Endoplasmin
(GRP94) | P14625 | 4.76/92.46 | 36.75 | Molecular
chaperone | Endoplasmic
reticulum lumen | ↓ |
| 77 | Heterogeneous
nuclear ribonucleoproteins C1/C2 (hnRNP C1/C2) | P07910 | 4.95/33.67 | 103.56 | Component of
ribonucleosomes | Nucleus | ↓ |
| 77 | Endoplasmin
(GRP94) | P14625 | 4.76/92.46 | 42.22 | Molecular
chaperone | Endoplasmic
reticulum lumen | ↓ |
| 82 | FK506-binding
protein (FKBP4) | Q02790 | 5.35/51.8 | 166.07 |
Co-chaperoneactivities | Cytoplasm,
nucleus | ↓ |
| 82 | Vimentin | P08670 | 5.06/53.65 | 86.48 | Intermediate
filaments | Cytoskeletal
component | ↓ |
| 82 | Tubulin α-1B
chain | P68363 | 4.94/50.15 | 70.43 | Microtubules,
motility | Cytoplasm,
cytoskeleton | ↓ |
| 50 | NipSnap1 | Q9BPW8 | 9.35/33.3 | 23.16 | Ca2+
flux | Mitochondria | ↑ |
| 73 | Glycine-tRNA ligase
(GRS) | P41250 | 6.61/83.16 | 27.91 | Translation
apparatus | Cytoplasm,
mitochondria, serum | ↑ |
The data demonstrated that Umb downregulated the
production of heat shock protein 90 kDa (HSP90), HSP27, endoplasmin
(two spots), vimentin (two spots), heterogeneous nuclear
ribonucleoproteins (hnRNP) C1/C2, transitional endoplasmic
reticulum ATPase (p97/VCP), NADH dehydrogenase [ubiquinone]
iron-sulfur protein-3 (NDUFS3), importin-α2, importin-β1, tubulin
α-1B, FK506-binding protein (FKBP4) and splicing factor 3A
subunit-3 (SF3a3) in the QU-DB cells. By contrast, Nipsnap1 and
glycine-tRNA ligase (GRS) were upregulated in the Umb-treated QU-DB
cells.
Differentially-expressed proteins in
the Umb-treated A549 cells
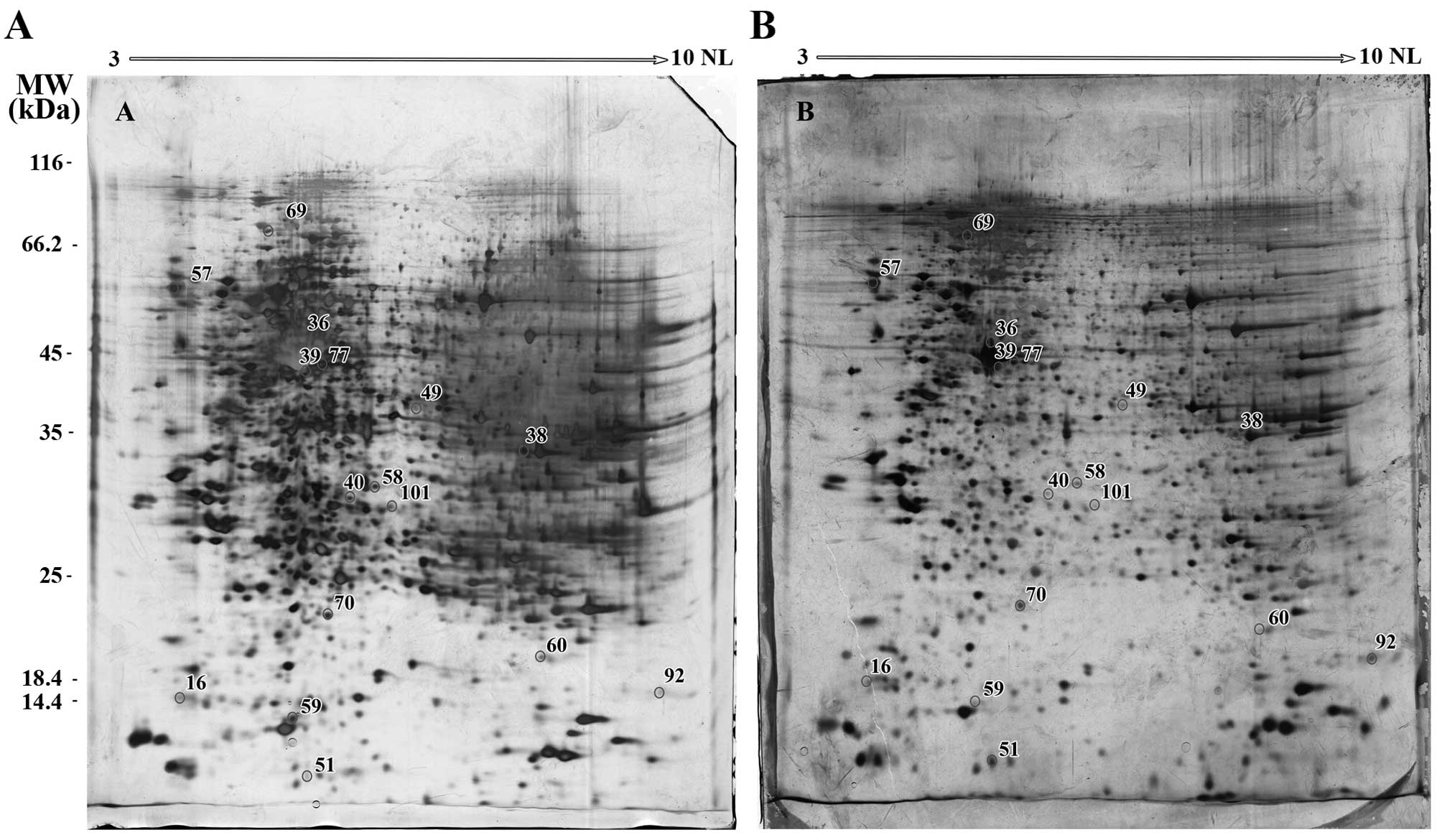
In the A549 cells collectively, 70 reproducible,
distinct spots were differentially-expressed. Of these, 42 intense
spots were picked up from gels, of which 16 were identified by MS
(Table II). The locations of these
spots on the A549 gels are presented in Fig. 2.
 | Table II.Descriptions of the 16 differentially
expressed protein spots identified in umbelliprenin-treated A549
and dimethyl sulfoxide-treated control cells. |
Table II.
Descriptions of the 16 differentially
expressed protein spots identified in umbelliprenin-treated A549
and dimethyl sulfoxide-treated control cells.
| Spot no. | Protein name | Accession
no.a | pI/MW, kDa | Scoreb | Function | Location | Expression |
|---|
| 40 | Dimethylarginine
dimethylaminohydrolase-2 (DDAH-2) | O95865 | 5.66/29.64 | 39.6 | Inhibitor of
NOS | Cytoplasm,
mitochondrion | ↓ |
| 49 | St20-like
protein kinase (MST) | P25325 | 6.13/33.17 | 28.27 | Catalytic enzyme,
antidote | Cytoplasm | ↓ |
| 51 | Keratin-1 | P04264 | 8.15/66.03 | 42.83 | Intermediate
filaments | Cell membrane | ↓ |
| 58 | Annexin A4 | P09525 | 5.84/35.88 | 25.61 | Endo- and
exocytosis | Membrane-binding
protein | ↓ |
| 59 | Proteasome α-1 | P25786 | 6.15/29.55 | 17.69 | Proteinase | Cytoplasm,
nucleus | ↓ |
| 60 | Dual specificity
protein phosphatase-3 (VHR) | P51452 | 7.66/20.47 | 23.51 | Tyrosine
phosphatase | Cytoplasm,
nucleus | ↓ |
| 70 | Adenine
phosphoribosyltransferase (APRT) | P07741 | 5.78/19.6 | 38.63 | Metabolism of
AMP | Cytoplasm | ↓ |
| 92 | Cyclophilin B | P23284 | 9.42/23.74 | 124.62 | Immunophilin
protein, protein folding | ER lumen,
melanosome | ↓ |
| 101 | Prohibitin | P35232 | 5.57/29.8 | 26.6 |
Apoptosis-associatedprotein | Mitochondrion inner
membrane | ↓ |
| 16 | Stathmin | P16949 | 5.76/17.3 | 27.57 | Cell motility | Cytoplasm,
cytoskeleton | ↑ |
| 36 | Actin, aortic
smooth muscle | P62736 | 5.24/42 | 24.24 | Cell motility | Cytoplasm,
cytoskeleton | ↑ |
| 38 | GAPDH | P04406 | 8.57/36.05 | 13.42 | Glycolysis Nuclear
functions | Cytoplasm,
nucleus | ↑ |
| 39 | Actin, α-skeletal
muscle | P68133 | 5.23/42.05 | 158.76 | Cell motility | Cytoplasm,
cytoskeleton | ↑ |
| 57 | Calreticulin | P27797 | 4.29/48.14 | 62.8 | Calcium-binding
chaperone | Endoplasmic
reticulum | ↑ |
| 69 | Glucose-regulated
protein78 kDa | P11021 | 5.07/72.33 | 60.92 | ER chaperone | Endoplasmic
reticulum | ↑ |
| 77 | Activator of heat
shock 90 kDa protein ATPase homolog-1 (AHA1) | O95433 | 5.41/38.27 | 15.5 | Co-chaperone,
Activities HSP90 | Cytoplasm, cytosol.
endoplasmic reticulum | ↑ |
Of the 16 identified spots, 9 were downregulated.
These included cyclophilin B, adenine phosphoribosyl transferase
(APRT), dimethylarginine dimethylaminohydrolase-2 (DDAH-2), dual
specificity protein phosphatase-3 (VHR), annexin A4, prohibitin,
proteasome α-1, MST and keratin-1. Data demonstrated the
upregulation of 7/16 differentially-expressed proteins, which
consisted of glucose-regulated protein (GRP) 78 kDa, aortic and
skeletal muscle α-actins, activator of HSP90, ATPase homolog-1
(AHA1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), stathmin
and calreticulin.
Discussion
To the best of our knowledge, the present study
identified molecular targets of Umb in cancer for the first time
using a high-throughput approach. The proteomes of the QU-DB
(large-cell lung carcinoma) and A549 (adenocarcinoma) cell lines
were mapped in the presence of Umb using 2-DE. The effect of Umb on
these cell lines in culture media using MTT assay and flow
cytometry has been previously investigated in vitro, with
higher cytotoxicity of Umb observed against QU-DB cells (2). Furthermore, in an in vivo mouse
model of lung cancer, a significant anti-tumor activity of Umb was
reported, in addition to deviation of the immune system in favor of
Th1 responses (7).
The present study demonstrated that Umb
downregulated the expression of HSP90, HSP27, GRP94 (two spots),
vimentin (two spots), hnRNP C1/C2, p97/VCP, NDUFS3, importin-α2,
importin-β1, tubulin α-1B, FKBP4 and SF3a3 in the QU-DB cells, and
cyclophilin B, APRT, DDAH-2, VHR, annexin A4, prohibitin,
proteasome α-1, MST and keratin-1 in the A549 cells. The pattern of
downregulated proteins suggested that Umb is an important
anti-tumor compound, particularly in QU-DB cells. The functions of
the proteins suggested that Umb is able to suppress tumorigenesis
through different pathways, including induction of apoptosis and
inhibition of cell growth, migration and angiogenesis.
In total, 12 downregulated protein spots were
identified in the QU-DB cells by MS performed in the current study.
The overexpression of chaperones, such as HSP90, HSP27 and
endoplasmin, has been identified in various forms of cancer,
including lung (17,18), and has been reported to correlate with
drug resistance (18). These
chaperones induce tumor cell survival as they inhibit caspase
activation (18), and thus we
hypothesize that Umb may induce tumor apoptosis by removing
suppression from caspases. Caspases are a family of endoproteases
that are critical in apoptosis (programmed cell death) with a wide
range of substrates, including vimentin (19). Caspase cleavage of vimentin dislocates
the cytoskeletal component of intermediate filaments and causes
nuclear fragmentation in apoptotic cells (19). Vimentin was downregulated in the QU-DB
cells treated with Umb in the present study. This intermediate
filament is a marker of epithelial-mesenchymal transition (EMT).
Vimentin overexpression has been detected in several forms of
cancer and been reported as a molecular target for cancer therapy
(20). hnRNPs have been linked to
signal transduction, cell cycle progression and regulation of
endocytosis (12). NDUFS3 is a
mitochondrial Fe-S protein in electron transport chain complex I, a
complex that is responsible for electron transfer to ubiquinone,
and its overexpression promotes apoptotic resistance to stresses,
cell proliferation, cell migration and EMT (21). VCP/p97 is an ATPase that regulates
endoplasmic reticulum-associated degradation, and its inhibition
induces cancer cell death (22).
Importin subunits (α and β1) contribute to nucleocytoplasmic
transport. Importin α, a new potential biomarker in cancer, has
been demonstrated to be a predictor of poor prognosis in different
forms of cancer (23). It has been
reported that coumarins are able to exert anti-proliferative
effects through inhibition of tubulin polymerization and induction
of cell cycle arrest at the G2/M transition of the mitotic cell
cycle (24). Umb may have a similar
effect on QU-DB cells via tubulin reduction. FKBP4 has been
associated with a poorer prognosis in several types of cancer,
including ovary, prostate and breast cancer (25), however, its role in lung cancer
requires further clarification. Finally, the function of SF3a3 in
cancer has not yet been clarified.
In the present study, 7 of the downregulated
proteins in the A549 cells were identified by MS, and with the
exception of MST, their downregulation favored the anti-tumor
activity of Umb. Cyclophilin B is a ubiquitously expressed protein
in normal cells and tissues, but protects cancer cells against
apoptosis induced by cisplatin, hypoxia and oxidative stresses
(26). The protein increases tumor
growth, angiogenesis and resistance to therapy (26). Adenine is converted to AMP by the
ubiquitous enzyme APRT (27).
Specific inhibitors of APRT induce cytotoxicity, and have been
consideredfor the treatment of cancer, arthritis, inflammation or
microbial infections (27). DDAH-2
functionsas an inhibitor of nitric oxide (NO) synthase. NO
overexpression has been reported in various tumors, including lung
cancer, and its inhibition is regarded as an anticancer therapy
(28). Cancer cells are able to evade
cell cycle arrest via VHR overexpression (29). Annexins, prohibitin and different
subunits of the proteasome complex have been associated with cancer
invasiveness (12,13). MST is a serine/threonine-specific
protein kinase and serves a conserved role as a regulator of organ
size and as a potential tumor suppressor within the Hippo pathway
(30). This proteins downregulation
may also accelerate cancer. The role of Keratin-1 in cancer
requires further clarification.
In the current study, proteins upregulated in the
QU-DB cells includedNipsnap1 and GRS, while upregulated proteins in
the Umb-treated A549 cells included GRP78, α-smooth muscle actin,
skeletal muscle α-actin, AHA1, GAPDH, stathmin and calreticulin. In
contrast to the QU-DB cells, upregulated proteins in the A549 cells
may deteriorate cancer conditions and induce tumor cell survival,
while only calreticulin overexpression appears to induce
appropriate anti-tumor and anti-proliferative responses.
Upregulated proteins in the QU-DB cells were consistent with Umb
anti-tumor activities. A previous study identified Nipsnap1 as a
tumor suppressor in prostate cancer (31). Immune stimulatory activity of GRS
indicates its therapeutic potential against tumorigenesis. GRS is a
component of the translation machinery, and is secreted from
macrophages in response to Fas ligand, which is released from tumor
cells (32). GRS binds to various
extracellular signal-regulated kinase (ERK)-activated tumor cells
and releases phosphatase 2A, which in turn suppresses ERK signaling
through dephosphorylation of ERK and induces apoptosis (32). A previous study demonstrated that
following in vivo administration of GRS, growth of tumors
with a high level of ERK activation was strongly suppressed
(32).
In the present study, 6 out of 7 upregulated
proteins in the A549 cells were in favor of tumor growth. GRP78 is
an endoplasmic reticulum (ER) chaperone, which participates in the
folding of proteins in the ER (33).
GRP78 stimulates tumor survival, metastasis and resistance to
various therapies (33). GRP78 is
considered as a novel biomarker in cancer treatment and therapeutic
regimen (33). Actins are potential
mediator of tumor development. In the current study, it was
observed that two different types of actins, α-smooth muscle actin
and skeletal muscle α-actin, were upregulated in the A549 cells.
α-smooth muscle actin overexpression is a major characteristic of
cancer-associated fibroblasts and serves a vital role in lung
cancer initiation and progression (34). Thus, this suggests that actin
polymerization and cytoskeletal rearrangement were induced in the
A549 cells in response to Umb. AHA1 (18), a co-chaperone that binds to HSP90,
GAPDH (14) and stathemin (35) are well-known for their roles in tumor
progression. Calreticulin is a Ca2+-binding chaperone
and its cell surface expression is able to induce innate immune
responses via ‘eat me’ signals, which lead to capture of dying
tumor cells by dendritic cells and macrophages (36). It has been reported that calreticulin
functions as a damage-associated molecular pattern (DAMP) following
tumor therapy (36). In tumor cells,
calreticulin in complex with cluster of differentiation 91
functions as a macrophage surface receptor for collectin and
complement factor C1q (36). This
interaction induces phagocyte binding of apoptotic cells and innate
immune response development (36).
Calreticulin also takes part in major histocompatibility class I
assembly and presentation of tumor cell antigens (36). Calreticulin expression on lung cancer
cell membranes is associated with tumor pathological classification
and grade, and is known to be a novel prognostic factor and
potential therapeutic biomarker in lung cancer (37). Despite other overexpressed proteins
identified in the Umb-treated A549 cells, it appears that only
calreticulin is able to induce A549 cell repression.
By using a proteomic approach, the present study
observed that targets of Umb were different in two different lung
cancer cell lines. In the QU-DB cells, the targets supported its
anti-tumor activities, while in the A549 cells, the targets had
contradictory effects in tumorigenesis. The proteomic data are
consistent with previous studies that demonstrated that a similar
dosage of Umb had an apoptotic effect on QU-DB cells, but not on
A549 cells, and only after increasing the Umb dose was cytotoxicity
observed in the A549 cells (2).
Notably, certain upregulated proteins observed in each cell line in
the current study (GRS in the QU-DB cells and calreticulin in the
A549 cells) are known to interact with immune cells in vivo
and boost anti-tumor immune response (32,36,37). This
may indicate that Umb has stronger in vivo anti-tumor
activities compared to in vitro results.
In conclusion, to the best of our knowledge, the
results of the present study provide the first evidence of
molecular targets of Umb in cancer obtained by high-throughput
profiling. At the molecular level, Umb affects the two cancer cell
lines differentially, reflecting different levels of cytotoxicity
against them that is consistent with previous studies. In each cell
line, immune-associated molecules were upregulated, which suggests
that Umb has stronger in vivo anti-tumor activity. In
addition, these results highlighted the possible administration of
Umb in lung cancer in an individualized manner. Whether Umb
functions in the same manner within other types of lung cancer, or
even on the same type, requires further clarification in future
studies.
Acknowledgements
The present study was funded by grants from Shiraz
University of Medical Sciences, Shiraz Iran (no. 89–5291) and
Shiraz Cancer Research Center, Shiraz, Iran (no. ICR-87-503).
References
|
1
|
Lacy A and O'Kennedy R: Studies on
coumarins and coumarin-related compounds to determine their
therapeutic role in the treatment of cancer. Curr Pharm Des.
10:3797–3811. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khaghanzadeh N, Mojtahedi Z, Ramezani M,
Erfani N and Ghaderi A: Umbelliprenin is cytotoxic against QU-DB
large cell lung cancer cell line but anti-proliferative against
A549 adenocarcinoma cells. DARU. 20:692012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iranshahi M, Askari M, Sahebkar A and
Hadjipavlou-Litina D: Evaluation of antioxidant, anti-inflammatory
and lipoxygenase inhibitory activities of the prenylated coumarin
umbelliprenin. DARU. 17:99–103. 2009.
|
|
4
|
Soltani F, Mosaffa F, Iranshahi M, Karimi
G, Malekaneh M, Haghighi F and Behravan J: Auraptene from Ferula
szowitsiana protects human peripheral lymphocytes against oxidative
stress. Phytother Res. 24:85–89. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shahverdi AR, Saadat F, Khorramizadeh MR,
Iranshahi M and Khoshayand MR: Two matrix metalloproteinases
inhibitors from Ferula persica var. persica. Phytomedicine.
13:712–717. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soltani F, Mosaffa F, Iranshahi M, Karimi
G, Malekaneh M, Haghighi F and Behravan J: Evaluation of
antigenotoxicity effects of umbelliprenin on human peripheral
lymphocytes exposed to oxidative stress. Cell Biol Toxicol.
25:291–296. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khaghanzadeh N, Samiei A, Ramezani M,
Mojtahedi Z, Hosseinzadeh M and Ghaderi A: Umbelliprenin induced
production of IFN-γ and TNF-α, and reduced IL-10, IL-4, Foxp3 and
TGF-β in a mouse model of lung cancer. Immunopharmacol
Immunotoxicol. 36:25–32. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iranshahi M, Sahebkar A, Takasaki M,
Konoshima T and Tokuda H: Cancer chemopreventive activity of the
prenylated coumarin, umbelliprenin, in vivo. Eur J Cancer Prev.
18:412–415. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barthomeuf C, Lim S, Iranshahi M and
Chollet P: Umbelliprenin from Ferula szowitsiana inhibits the
growth of human M4Beu metastatic pigmented malignant melanoma cells
through cell-cycle arrest in G1 and induction of caspase-dependent
apoptosis. Phytomedicine. 15:103–111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Blanchon F, Grivaux M, Asselain B, Lebas
FX, Orlando JP, Piquet J and Zureik M: 4-year mortality in patients
with non-small-cell lung cancer: Development and validation of a
prognostic index. Lancet Oncol. 7:829–836. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hamidinia M, Ramezani M and Mojtahedi Z:
Cytotoxic/proliferative effects of umbelliprenin on colon cancer
cell lines. Ann Colorectal Res. 1:101–105. 2013. View Article : Google Scholar
|
|
12
|
Cai XZ, Huang WY, Qiao Y, Du SY, Chen Y,
Chen D, Yu S, Che RC, Liu N and Jiang Y: Inhibitory effects of
curcumin on gastric cancer cells: A proteomic study of molecular
targets. Phytomedicine. 20:495–505. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Askari M, Sahebkar A and Iranshahi M:
Synthesis and purification of 7-prenyloxycoumarins and herniarin as
bioactivenatural coumarins. Iran J Basic Med Sci. 12:63–69.
2009.
|
|
14
|
Mojtahedi Z, Safaei A, Yousefi Z and
Ghaderi A: Immunoproteomics of HER2-positive and HER2-negative
breast cancer patients with positive lymph nodes. OMICS.
15:409–418. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okutucu B, Dinçer A, Habib O and Zihnioglu
F: Comparison of five methods for determination of total plasma
protein concentration. J Biochem Biophys Methods. 70:709–711. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sarvari J, Mojtahedi Z, Kuramitsu Y,
Malek-Hosseini SA, Shahrabadi M Shamsi, Ghaderi A and Nakamura K:
Differential expression of haptoglobin isoforms in chronic active
hepatitis, cirrhosis and HCC related to HBV infection. Oncol Lett.
2:871–877. 2011.PubMed/NCBI
|
|
17
|
Wang Q, An L, Chen Y and Yue S: Expression
of endoplasmic reticulum molecular chaperon GRP94 in human lung
cancer tissues and its clinical significance. Chin Med J (Engl).
115:1615–1619. 2002.PubMed/NCBI
|
|
18
|
Alarcon SV, Mollapour M, Lee MJ, Tsutsumi
S, Lee S, Kim YS, Prince T, Apolo AB, Giaccone G, Xu W, et al:
Tumor-intrinsic and tumor-extrinsic factors impacting hsp90-
targeted therapy. Curr Mol Med. 12:1125–1141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Byun Y, Chen F, Chang R, Trivedi M, Green
KJ and Cryns VL: Caspase cleavage of vimentin disrupts intermediate
filaments and promotes apoptosis. Cell Death Differ. 8:443–450.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target in cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng CW, Kuo CY, Fan CC, Fang WC, Jiang
SS, Lo YK, Wang TY, Kao MC and Lee AY: Overexpression of Lon
contributes to survival and aggressive phenotype of cancer cells
through mitochondrial complex I-mediated generation of reactive
oxygen species. Cell Death Dis. 4:e6812013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Magnaghi P, D'Alessio R, Valsasina B,
Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius
C, et al: Covalent and allosteric inhibitors of the ATPase VCP/p97
induce cancer cell death. Nat Chem Biol. 9:548–556. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Christiansen A and Dyrskjøt L: The
functional role of the novel biomarker karyopherin α 2 (KPNA2) in
cancer. Cancer Lett. 331:18–23. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim SN, Kim NH, Park YS, Kim H, Lee S,
Wang Q and Kim YK: 7-Diethylamino-3(2′-benzoxazolyl)-coumarin is a
novel microtubule inhibitor with antimitotic activity in multidrug
resistant cancer cells. Biochem Pharmacol. 77:1773–1779. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lawrenson K, Mhawech-Fauceglia P,
Worthington J, Spindler TJ, O'Brien D, Lee JM, Spain G, Sharifian
M, Wang G, Darcy KM, et al: Identification of novel candidate
biomarkers of epithelial ovarian cancer by profiling the secretomes
of three-dimensional genetic models of ovarian carcinogenesis. Int
J Cancer. 137:1806–1817. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim Y, Jang M, Lim S, Won H, Yoon KS, Park
JH, Kim HJ, Kim BH, Park WS, Ha J and Kim SS: Role of cyclophilin B
in tumorigenesis and cisplatin resistance in hepatocellular
carcinoma in humans. Hepatology. 54:1661–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kamal MA and Christopherson RI:
Accumulation of 5-phosphoribosyl-1-pyrophosphate in human CCRF-CEM
leukaemia cells treated with antifolates. Int J Biochem Cell Biol.
36:545–551. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burke AJ, Sullivan FJ, Giles FJ and Glynn
SA: The Yin and Yang of nitric oxide in cancer progression.
Carcinogenesis. 34:503–512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arnoldussen YJ, Lorenzo PI, Pretorius ME,
Waehre H, Risberg B, Maelandsmo GM, Danielsen HE and Saatcioglu F:
The mitogen-activated protein kinase phosphatase vaccinia
H1-related protein inhibits apoptosis in prostate cancer cells and
is overexpressed in prostate cancer. Cancer Res. 68:9255–9264.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rawat SJ and Chernoff J: Regulation of
mammalian Ste20 (Mst) kinases. Trends Biochem Sci. 40:149–156.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Malhotra A, Shibata Y, Hall IM and Dutta
A: Chromosomal structural variations during progression of a
prostate epithelial cell line to a malignant metastatic state
inactivate the NF2, NIPSNAP1, UGT2B17, and LPIN2 genes. Cancer Biol
Ther. 14:840–852. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park MC, Kang T, Jin D, Han JM, Kim SB,
Park YJ, Cho K, Park YW, Guo M, He W, et al: Secreted human
glycyl-tRNA synthetase implicated in defense against ERK-activated
tumorigenesis. Proc Natl Acad Sci USA. 109:E640–E647. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fernandez PM, Tabbara SO, Jacobs LK,
Manning FC, Tsangaris TN, Schwartz AM, Kennedy KA and Patierno SR:
Overexpression of the glucose-regulated stress gene GRP78 in
malignant but not benign human breast lesions. Breast Cancer Res
Treat. 59:15–26. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Horie M, Saito A, Mikami Y, Ohshima M,
Morishita Y, Nakajima J, Kohyama T and Nagase T: Characterization
of human lung cancer-associated fibroblasts in three-dimensional in
vitro co-culture model. Biochem Biophys Res Commun. 423:158–163.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yousefi Z, Sarvari J, Nakamura K,
Kuramitsu Y, Ghaderi A and Mojtahedi Z: Secretomic analysis of
large cell lung cancer cell lines using two-dimensional gel
electrophoresis coupled to mass spectrometry. Folia Histochem
Cytobiol. 50:368–874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Korbelik M, Zhang W and Merchant S:
Involvement of damage-associated molecular patterns in tumor
response to photodynamic therapy: Surface expression of
calreticulin and high-mobility group box-1 release. Cancer Immunol
Immunother. 60:1431–1437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu R, Gong J, Chen J, Li Q, Song C, Zhang
J, Li Y, Liu Z, Dong Y, Chen L and Jin B: Calreticulin as a
potential diagnostic biomarker for lung cancer. Cancer Immunol
Immunother. 61:855–864. 2012. View Article : Google Scholar : PubMed/NCBI
|