Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide, and the second cause of cancer related
deaths in developed countries (1,2). CRC is
the 3rd and the 4th commonly diagnosed cancer in Iranian men and
women, respectively (3). The lack of
clinical manifestations in CRC patients until the late stages of
cancer is a common disease characteristic, which results in poor
prognosis and high mortality. The process of carcinogenesis in
primary adenomas, which are precursor lesions of colon cancer that
eventually develop into colorectal carcinomas, is slow, and is a
cause of the late diagnosis (4).
Eighty percent of early-diagnosed patients are referred for tissue
resection and are eventually cured (5). In order to reduce the morbidity and
mortality of the disease, early diagnosis and treatment of CRC
appears to be of critical importance, since there is a large
preclinical asymptomatic stage in CRC patients (6).
There are several CRC screening methods, such as
fecal occult blood testing (FOBT), barium enema, flexible
sigmoidoscopy and colonoscopy. FOBT and colonoscopy are commonly
used clinically; however, they have some technical restrictions and
disadvantages (7). FOBT, while a
simple method in practice, does not have high sensitivity and
specificity. Considering that colonoscopy is the ‘gold standard’
method for CRC screening, its invasiveness nature and complicated
required preparation procedure make patients reluctant to choose it
as an acceptable screening method (7). Therefore, developing new and useful
screening methods is a high priority (8).
Sporadic CRC occurrence is due to accumulation of
genetic and epigenetic changes, which cause normal epithelial cells
to transform into the adenocarcinoma cells (9). There is increasing evidence that
widespread epigenetic alterations are the key features of most of
cancer types (10,11), and these changes may be important in
the pathogenesis of CRC. Aberrant DNA methylation is one of the
best known and well-defined epigenetic changes in tumors, and is a
frequent mechanism for inappropriate gene silencing among tumor
suppressor genes (12–14). DNA methylation is of particular
interest, as it occurs in the primary stages of carcinogenesis, and
hence, can be used as a marker for early detection of CRC (15).
Septin 9 (SEPT9) is a member of the septin
gene family, which are highly conserved and encode GTP-binding
proteins. Septins are multidomain proteins which together form
filamentous structures that form part of cytoskeleton (16,17).
Furthermore, septins belong to P-loop GTPases superclass and were
first identified in yeast as key genes in cell division (18). These proteins have prominent roles in
multiple cellular processes, including cell membrane rigidity,
establishment of separate cellular domains by creating membrane
diffusion barriers, providing scaffold for localization of proteins
to certain subcellular regions and determination of cell polarity
(16,18,19). The
precise mechanism of SEPT9 molecular function in colon
cancer pathogenesis has not yet been clearly described (20), however, several studies suggest
possible roles in different malignancies, including leukemia
(21), breast and ovarian cancer
(22–24), brain tumors (25) and CRC (19,26–28).
The methylation status of the SEPT9 gene has
been examined previously in CRC patients, as well as cases with
precancerous lesions including adenomas in several studies
(29,30). In a recent study conducted by Ahmed
et al (31), a methylation
panel including the SEPT9 gene was analyzed in CRC patients,
and concluded that SEPT9 promoter methylation is a promising
biomarker with the ability to discriminate CRC tissues as well as
adenomas, from normal mucosa.
The NTRK neurotrophin receptor family
includes NTRK1 (TrkA), NTRK2 (TrkB) and
NTRK3 (TrkC), which, in conjunction with their
ligands (NGF, BDNF and NT4/5,
NT-3, respectively) are important in development of the
nervous system (32). It has been
demonstrated previously that NTRKs have oncogenic effects in
some cancer types, such as breast cancer and liver cancer (33). Recent studies have demonstrated that
NTRK1 and NTRK3 may be dependent receptors, which
depend on availability of their ligands to select their specific
cell signals; these receptors are defined as having the ability to
induce opposite effects in the presence or absence of their ligands
(34,35). The availability of the ligand leads to
the transduction of a positive cellular survival or differentiation
signal, whereas the induction of apoptosis is a result of the
absence of ligand (36). Observations
which have demonstrated NTRK3 is a beneficial prognostic
factor in certain cancers, including melanoma and medulloblastomas,
indirectly support that NTRK3 has a dependence receptor role
and is a conditional tumor suppressor gene (37,38).
Therefore, these findings suggest that NTRK3 may act as a
conditional tumor suppressor gene in CRC.
Specific somatic missense mutations in NTRK3,
which probably inhibit its function, have been identified in
colorectal cancer, as well as breast, lung, and pancreatic cancers
(39,40). Considering the possibility of
NTRK3 as a CRC tumor suppressor gene, based on the discovery
of its mutant and methylated forms in CRC, Luo et al
(41) conducted a study in order to
define the effect of aberrant methylation on NTRK3
expression, and also to define whether NTRK3 has oncogenic
or tumor suppressor functions in CRC cell lines. The authors
concluded that aberrant methylation of NTRK3 is prevalent in
CRC and adenomas that consequently silences its expression, which
suggests its tumor suppressor role. The results exhibited
NTRK3's function as a dependent receptor which means that
binding its ligand, NT-3, it can induce proliferation while
the absence of NT-3 leads to NTRK3-mediated
apoptosis. Overall, those findings suggested NTRK3 as a
novel conditional tumor suppressor gene in CRC.
The present study aimed to analyze the methylation
status of SEPT9 and NTRK3 gene promoters in order to
examine their ability to differentiate CRC tissues from normal
mucosa, and to assess the validity of NTRK3 as a methylation
marker in clinical CRC samples.
Materials and methods
Study design
The present cross-sectional study was undertaken as
a collaboration between the Immunology Research Center of Tabriz
University of Medical Sciences, Imam Reza Hospital (Tabriz, Iran),
Amiralmomenin Hospital (Tabriz, Iran) and Pasteur Institute of
Tehran. Written informed consent was obtained from all patients
participating in this study. The ethical protocol of the study was
proved by the Ethical Committee of Tabriz University of Medical
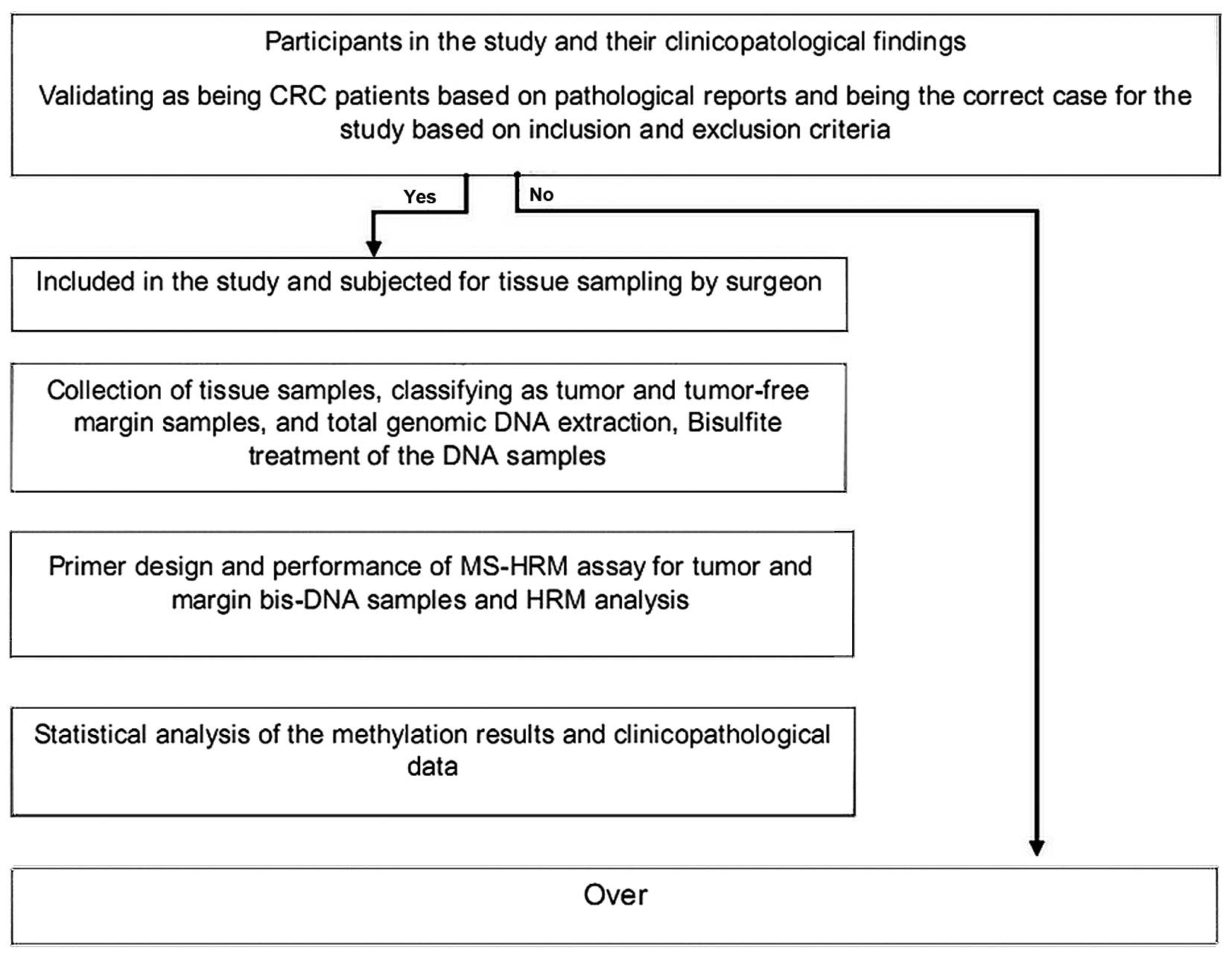
Sciences. A scheme of the study design is presented in Fig. 1.
Study population
Participants in this study were Iranians with the
same ethnicity and geographical residency. Tumor and matched
tumor-free margin samples were obtained from 45 colorectal cancer
patients during surgery, which was a part of the routine treatment
of patients. All patients were precisely identified as CRC patients
on the basis of clinicopathological findings, all were candidates
for cancer surgery, and underwent appropriate surgery at Imam Reza
Hospital and Amiralmomenin Hospital, between 2014 and 2015. All the
samples were referred to the laboratory under certain conditions
with the complete patient information including clinicopathological
and demographic data. The patients comprised of 19 males (42.2%)
and 26 females (57.8%). None of the patients were undergone
preoperative chemotherapy and/or radiotherapy and have no other
malignancies. Tissue samples were separated into two distinct 45
tumor samples and 45 margin samples, which had been validated
according to the pathological analysis.
Samples collection
Fresh tumor tissue and tumor-free margin tissue
samples were collected during surgery in the Imam Reza Hospital,
Tabriz and Amiralmomenin Hospital. Following resection, the tissue
samples were immediately snap-frozen in liquid nitrogen and stored
at −80°C in the laboratory until further sample processing.
DNA extraction and sodium bisulfite
modification
Total genomic DNA was extracted from tissue samples
using CinnaPure-DNA kit (Cinna Colon, Iran) according to the
manufacture's protocol. DNA concentrations were measured using a
NanoDrop spectrophotometer, and then stored at −20°C until the next
step. Extracted DNA samples were excluded from the further analysis
if the final concentration was <100 ng/µl, or the A260/A280
ratio was outside the range of 1.7–1.9. In the next step, total
genomic DNA samples underwent sodium bisulfite conversion using a
EZ DNA methylation-Gold kit (Zymo Research Corp. Irvine, CA, USA),
according to the instructions provided by the company. The modified
DNA samples were stored immediately at −20°C.
Methylation specific-high resolution
melting (MS-HRM)
HRM primer pairs were designed for specific GC rich
islands in promoter sequences of each gene according to the HRM
primer design guidelines (Table I).
Two specific locations were analyzed within the promoter region of
SEPT9 gene, and one location within the promoter region of
NTRK3 gene by MS-HRM assay. For each reaction, 2 µl of
bis-DNA template was added to 10 µl of master mix (SYBR Premix Ex
Taq™ II), and 2 µl of specific primer pairs with 6 µl double
distilled water, then placed in Real Time PCR (Applied bio system,
step one plus) with the following conditions: initial denaturation
at 95°C for 30 sec; and then 40 cycles at 95°C of denaturation for
5 sec, appropriate annealing temperature for each primer set
(Table I) for 30 sec, and extension
at 72°C for 30 sec. HRM analysis was done at a temperature range
from 60°C to 95°C with the ramp rate of 0.3°C/15 sec. The standard
curves were included in each assay with DNA samples with known
methylation ratios and then used to deduce the methylation ratio of
each unknown sample. Using the HRM v.2.2 software (Applied
Biosystems, Thermofisher Scientific, Waltham, MA, USA), melting
curves were normalized relative to two normalization regions before
and after the major decrease of fluorescence indicating the melting
region of the PCR product. The output plots were in the shape of
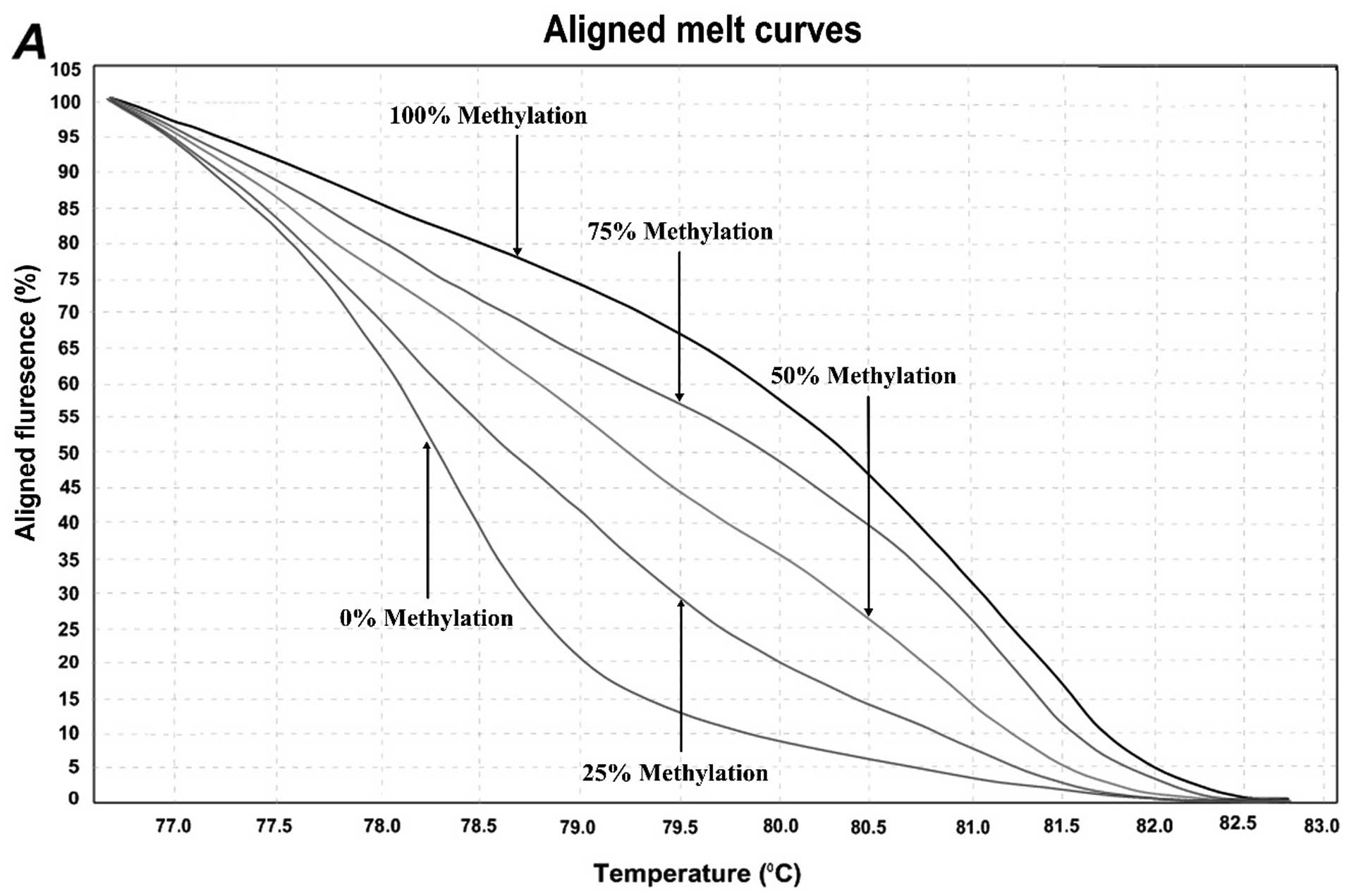
normalized melting curves. Considering the standard curves (0, 25,
50, 75, and 100%), HRM data for each unknown sample were classified
into different ranges of methylation by three independent
observers.
 | Table I.Primers used for MS-HRM assay. |
Table I.
Primers used for MS-HRM assay.
|
| Amplified
region |
|---|
|
|
|
|---|
| Primer | Sequences
(5′-3′) | Ta°C | Product size
(bp) | Number of GC
dinucleotide | Accession no. | Nucleotide
numbers |
|---|
| SEPT9 (1st
location) | F:
CGGTGATAGAGAATTTTGTTTGGT | 60 | 178 | 11 | NC_000017.11 |
77372911–77373089 |
|
| R:
CGACCTCAACCCCTCCC |
|
| SEPT9 (2nd
location) | F:
GACGTGTTGGAGAGGATTTTG | 60 | 181 | 24 | NC_000017.11 |
77373582–77373763 |
|
| R:
CGAATACCCCTAACAAAATCCC |
|
| NTRK3 | F:
TGGTTCGGGAGATGTTTTT | 58 | 164 | 16 | NC_000015.10 |
88255602–88255765 |
|
| R:
AAACGAACCAACAACTAATTAAA |
|
Statistical analysis
The analyzed data were found not to be normally
distributed, therefore, nonparametric tests were used. Statistical
analysis was performed in each group using the Mann-Whitney U and
Kruskal-Wallis tests. The Spearman correlation coefficient test was
used to analyze any correlation between clinicopathological
findings of patients and gene specific methylation. Sensitivity and
specificity of test were examined using ROC curve analysis. In all
tests, P<0.05 was considered to indicate a statistically
significant difference. SPSS version 22 was used for all
statistical analyses (SPSS Inc., Chicago, IL, USA).
Results
Clinicopathological findings of
patients
The mean age of the patients was 58.28 years (range
29–83 years), the median weight and height of patients were 71.11
kg (range 50–94 kg) and 165.51 cm (range, 150–196 cm),
respectively. The tumor samples comprised of all CRC stages,
however, the most prevalent stage was IIB. The pathological
features of samples are displayed in Table II. The mean size of tumors was 5.5 cm
(range 3–18 cm). Only 7 out of 45 patients were smokers.
 | Table II.Clinicopathological findings of
patients and their correlations with SEPT9 and NTRK3
methylation. |
Table II.
Clinicopathological findings of
patients and their correlations with SEPT9 and NTRK3
methylation.
| Clinicopathological
features | Frequency | SEPT9
methylation (first location) P-value | SEPT9
methylation (second location) P-value | NTRK3
methylation P-value |
|---|
| Age |
| 0.31 | 0.17 | 0.18 |
|
<50 | 8 |
|
|
|
|
>50 | 37 |
|
|
|
| Gender |
| 0.76 | 0.73 | 0.60 |
|
Male | 19 |
|
|
|
|
Female | 25 |
|
|
|
| Tumor location |
| 0.04 | 0.62 | 0.38 |
| Right
colon | 12 |
|
|
|
|
Transverse colon | 4 |
|
|
|
| Left
colon | 5 |
|
|
|
| Sigmoid
colon | 12 |
|
|
|
|
Cecal | 3 |
|
|
|
|
Rectosigmoid | 8 |
|
|
|
| Tumor size
(cm) |
| 0.51 | 0.34 | 0.06 |
|
<5 | 25 |
|
|
|
|
>5 | 20 |
|
|
|
| Tumor grade |
| 0.27 | 0.29 | 0.97 |
| G1 | 20 |
|
|
|
| G2 | 23 |
|
|
|
| G3 | 2 |
|
|
|
| Tumor stage |
| 0.61 | 0.81 | 0.11 |
| Stage
I | 8 |
|
|
|
| Stage
II | 17 |
|
|
|
| Stage
III | 14 |
|
|
|
| Stage
IV | 6 |
|
|
|
| Smoking |
| 0.73 | 0.63 | 0.87 |
| No | 38 |
|
|
|
|
Yes | 7 |
|
|
|
| Pre-operative
hemoglobin (g/dl) |
| 0.41 | 0.38 | 0.41 |
|
<12 | 26 |
|
|
|
|
>12 | 19 |
|
|
|
Quantification of DNA methylation by
MS-HRM assay
In order to determine the methylation level of
SEPT9 and NTRK3 gene promoters, the MS-HRM assay, a
semi-quantitative sensitive method was used. The assay optimized by
using control dilution series including 0, 25, 50, 75, and 100%
methylation controls. Representative results are shown in Fig. 2. For the first location of
SEPT9 gene, methylation was observed in 5/45 (11.11%) of
normal adjacent samples and 25/45 (55.55%) of CRCs. In the second
location of SEPT9 gene, methylation was observed in 18/45
(40%) of normal adjacent samples and 42/45 (93.33%) of CRC samples.
For NTRK3 gene, the overall methylation level was high in
both normal adjacent samples and CRC samples. The methylation
status of the NTRK3 gene promoter in the analyzed location
was observed in 43/45 (95.5%) of normal samples and 45/45 (100%)
CRC sampless, however, the mean methylation levels in tumor samples
were much higher than those of normal adjacent ones.
The median methylation levels of SEPT9 first
location, SEPT9 sec location and NTRK3 in tumor samples was
24.16% (range 0 to 100%), 60.27% (range 0 to 100%), and 70.83%
(range 25 to 100%) respectively, and in adjacent normal tissue was
3.61% (range 0 to 75%), 11.38% (range 0 to 75%), and 40% (range 0
to 75%) respectively. After doing statistical analysis it was shown
that the methylation levels of SEPT9 gene in both locations
and NTRK3 gene between tumor and matched normal adjacent
tissue was significantly different (P<0.001; U Mann-Whitney
test). Interestingly there was not any correlation between clinical
and pathological features of patients with methylation levels of
SEPT9 gene in both locations and NTRK3 gene, except
for the first location of SEPT9, which was in correlation
with primary tumor site. Methylation levels of SEPT9 gene
first location were different in distinct tumor sites. Moreover,
there was no significant correlation between the two analyzed
locations of SEPT9 gene. Their methylation level was
completely independent of each other.
In order to assess the applicability of SEPT9
and NTRK3 methylation as diagnostic biomarkers for CRC, the
sensitivity and specificity of tests were analyzed using receiver
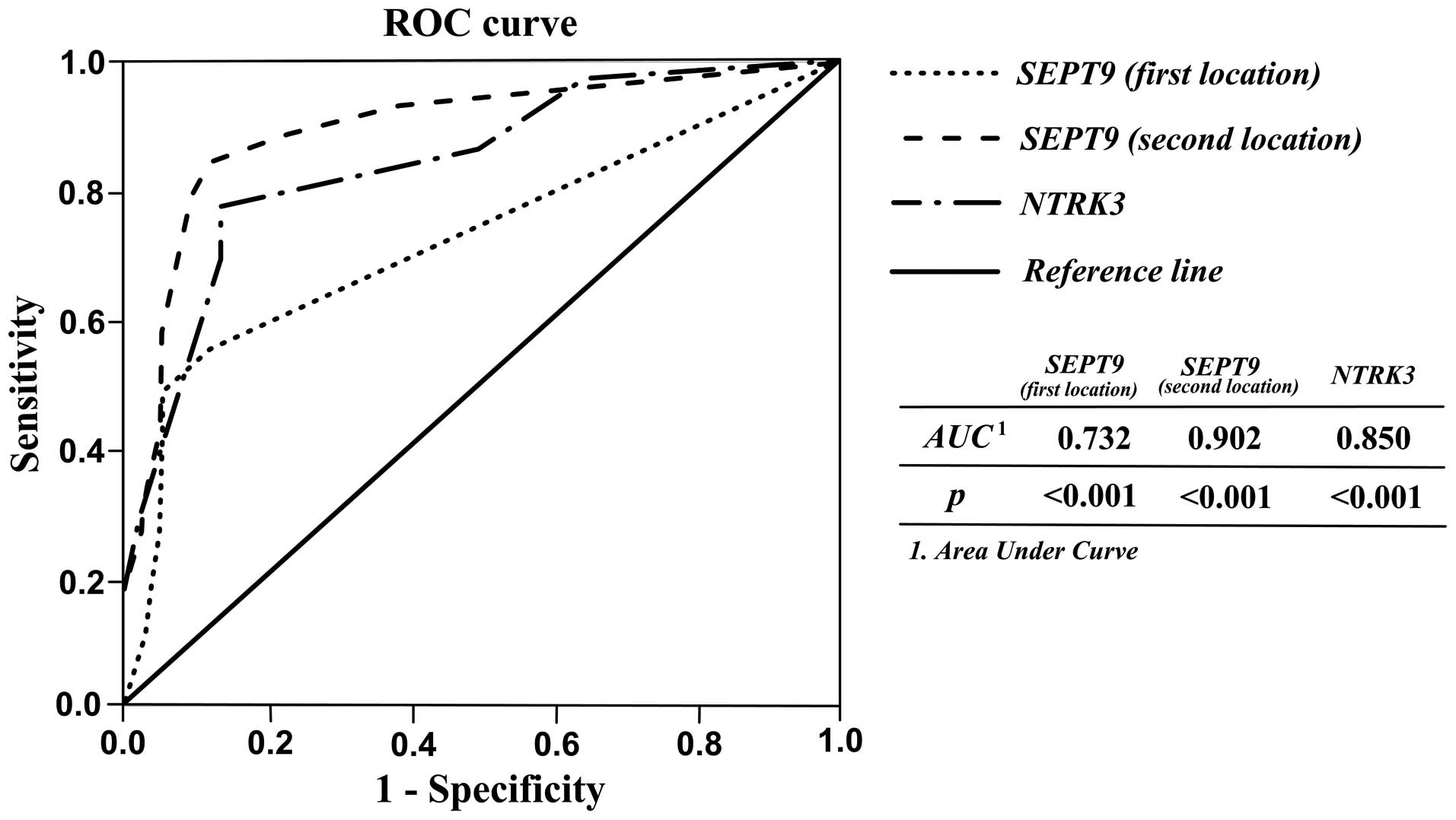
operating characteristic (ROC) curve analysis. As shown in Table III and Fig. 3, acceptable sensitivity and
specificity for SEPT9 and NTRK3 were resulted. When
the cutoff of SEPT9 sec location methylation percentage was 31.25,
the sensitivity, specificity, PPV, NPV, and accuracy were 84.40,
99, 90.36, 98.27, and 87.77% respectively. The results showed that
the second location of SEPT9 gene methylation could be
better diagnostic marker for discriminating CRC tissues from
matched normal adjacent with high accuracy. Likewise, high
percentages for these parameters indicating diagnostic ability of
NTRK3 and SEPT9 first location were also observed
(Table III).
 | Table III.Diagnostic performance of
SEPT9 and NTRK3 methylation. |
Table III.
Diagnostic performance of
SEPT9 and NTRK3 methylation.
| Gene | AUC (95% CI) | Cutoff value
(%)a | Sensitivity
(%) | Specificity
(%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|
| SEPT9 (first
location) | 0.732
(0.626–0.839) |
6.25 | 55.6 | 99 | 85.53 | 95.29 | 72.22 |
| SEPT9 (second
location) | 0.902
(0.835–0.970) | 31.25 | 84.40 | 99 | 90.36 | 98.27 | 87.77 |
| NTRK3 | 0.850
(0.771–0.929) | 56.25 | 77.80 | 86.7 | 39.39 |
97.223 | 66.66 |
Discussion
The aberrant methylation in the promoter regions of
genes is a prevalent event in a number of cancers including CRC
(42). There are several methods to
identify the methylation status of genomic DNA in specific sites;
of those, MS-HRM is a highly sensitive semi-quantitative method,
which can detect methylation level of specific region of bis-DNA
precisely.
The MS-HRM assay was performed in order to identify
the methylation status of SEPT9 and NTRK3 gene
promoters. The methylation levels of SEPT9 and NTRK3
genes in CRC were high compared with matched normal tissue
(P<0.001). There was not any significant association between
clinical features and pathological findings of patients and
hyper-methylation of analyzed genes in this study. The results of
the present study demonstrated that the first and second studied
locations of SEPT9 promoter region were not associated with
age, gender, TNM stage, and grade of tumor, while the first
location was just associated with tumor location. Similarly,
NTRK3 methylation was not associated with such clinical and
pathological parameters. Previous studies have reported that the
proportion of methylated SEPT9 genes augmented with the
progression of CRC (30,42), whereas our results demonstrate that
this proportion of SEPT9 was similar between different
stages and tumor grades of CRC cases. These results are in
agreement with the results of other similar studies such as Su
et al (43). However, these
findings may be biased due to the small number of cases included in
the study. Statistical analysis indicated that there is no
association between methylation level of first and second location
of SEPT9 gene, which suggests that their methylation process
is independent of each other, however, the overall methylation in
either tumor or normal matched adjacent tissue was higher in the
second location in comparison to the first location, which may be
due to the importance of this location in methylation-mediated
silencing of SEPT9. In addition, the sensitivity and
specificity of the second analyzed location for detection of CRC
tissues form normal ones was higher than the first location. This
finding suggests that this location may be more significant in
SEPT9 hypermethylation and the development of CRC, and can
detect tumor tissues more precisely then first location. In an
study established by Wassekort et al (44) it has been shown that hypermethylation
in a specific CpG island of SEPT9 promoter (including our
studied locations) is probably an early event in adenocarcinoma
progression. Furthermore, Wassekort et al (44) have proved that there is a direct link
between this region and the region cross-examined by EpiproColon
test that detects methylation of SEPT9 in cell free DNA. Our
results, in concordance with the published data, supports that
SEPT9 methylation in this CpG island can be a useful
biomarker in CRC diagnosis. Tóth et al (20) analyzed SEPT9 methylation in
both tissue and plasma of healthy, adenoma and CRC cases
quantitatively, and detected methylated SEPT9 in all tissue
samples at different levels regardless of the group. Methylated
SEPT9 levels in CRC and adenoma tissue samples were not
significantly different; however, its levels in healthy tissue
samples were much lower and considerably distinct from either
adenoma or CRC (20). Overall, our
findings supported previous studies for SEPT9 gene being a
promising marker for detection of CRC.
The NTRK3 gene has been recently demonstrated
to become hypermethylated and silenced in CRC cell lines, which
suggests its role as a tumor suppressor gene in colorectal cancer
(41). The present study analyzed
NTRK3 promoter methylation in order to determine whether it
can act as a biomarker in diagnosis of CRC or not. Our results
showed that the overall mean of methylation level in either normal
tumor free tissue or CRC tumor tissue samples was high in which
there were just two samples with 0% methylation. Although our
results are similar to those of Luo et al (41) and obviously NTRK3 promoter
methylation is able to discriminate the tumoral samples from normal
tumor free samples with acceptable sensitivity and specificity,
however, its high level methylation in tumor adjacent tissue
suggests that it may start methylation process far before the
appearance of any pathological features in cancerous cells.
However, this hypothesis should indeed be further analyzed with
samples in very initial stages of carcinogenesis.
In conclusion, based on our findings, SEPT9
methylation can be used as a diagnostic marker independent of
different clinicopathological features of CRC patients. In
particular, the second location is a promising candidate,
considering its high sensitivity and specificity and also the
accuracy of the test. For NTRK3 gene, we suggest further
analysis with large sample size and specially samples in very
initial stages of colorectal carcinogenesis in order to define its
potential as a diagnostic biomarker in CRC.
References
|
1
|
Perez-Carbonell L, Balaguer F, Toiyama Y,
Egoavil C, Rojas E, Guarinos C, Andreu M, Llor X, Castells A, Jover
R, et al: IGFBP3 methylation is a novel diagnostic and predictive
biomarker in colorectal cancer. PLoS One. 9:e1042852014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carmona FJ, Azuara D, Berenguer-Llergo A,
Fernández AF, Biondo S, de Oca J, Rodriguez-Moranta F, Salazar R,
Villanueva A, Fraga MF, et al: DNA methylation biomarkers for
noninvasive diagnosis of colorectal cancer. Cancer Prev Res
(Phila). 6:656–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mahmodlou R, Mohammadi P and Sepehrvand N:
Colorectal cancer in northwestern Iran. ISRN Gastroenterol.
2012:9685602012.PubMed/NCBI
|
|
4
|
Wang X, Kuang YY and Hu XT: Advances in
epigenetic biomarker research in colorectal cancer. World J
Gastroenterol. 20:4276–4287. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith RA, Cokkinides V and Eyre HJ:
American Cancer Society guidelines for the early detection of
cancer, 2006. CA Cancer J Clin. 56:11–25; quiz 49–50. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jenkinson F and Steele R: Colorectal
cancer screening-methodology. Surgeon. 8:164–171. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu H, Huang S, Zhang X, Wang D, Zhang X,
Yuan X, Zhang Q and Huang Z: DNA methylation analysis of SFRP2,
GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in
fecal DNA. Oncol Lett. 8:1751–1756. 2014.PubMed/NCBI
|
|
9
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Timp W and Feinberg AP: Cancer as a
dysregulated epigenome allowing cellular growth advantage at the
expense of the host. Nat Rev Cancer. 13:497–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome-biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kondo Y and Issa JP: Epigenetic changes in
colorectal cancer. Cancer Metastasis Rev. 23:29–39. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ushijima T: Detection and interpretation
of altered methylation patterns in cancer cells. Nat Rev Cancer.
5:223–231. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Petko Z, Ghiassi M, Shuber A, Gorham J,
Smalley W, Washington MK, Schultenover S, Gautam S, Markowitz SD
and Grady WM: Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon
polyps and in fecal DNA from patients with colorectal polyps. Clin
Cancer Res. 11:1203–1209. 2005.PubMed/NCBI
|
|
15
|
Draht MX, Riedl RR, Niessen H, Carvalho B,
Meijer GA, Herman JG, van Engeland M, Melotte V and Smits KM:
Promoter CpG island methylation markers in colorectal cancer: The
road ahead. Epigenomics. 4:179–194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Estey MP, Kim MS and Trimble WS: Septins.
Curr Biol. 21:R384–R387. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sandrock K, Bartsch I, Bläser S, Busse A,
Busse E and Zieger B: Characterization of human septin
interactions. Biol Chem. 392:751–761. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hall PA and Russell S: The pathobiology of
the septin gene family. J Pathol. 204:489–505. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wasserkort R, Kalmar A, Valcz G, Spisak S,
Krispin M, Toth K, Tulassay Z, Sledziewski AZ and Molnar B:
Aberrant septin 9 DNA methylation in colorectal cancer is
restricted to a single CpG island. BMC Cancer. 13:3982013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tóth K, Wasserkort R, Sipos F, Kalmár A,
Wichmann B, Leiszter K, Valcz G, Juhász M, Miheller P, Patai ÁV, et
al: Detection of methylated septin 9 in tissue and plasma of
colorectal patients with neoplasia and the relationship to the
amount of circulating cell-free DNA. PloS One. 9:e1154152014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kojima K, Sakai I, Hasegawa A, Niiya H,
Azuma T, Matsuo Y, Fujii N, Tanimoto M and Fujita S: FLJ10849, a
septin family gene, fuses MLL in a novel leukemia cell line CNLBC1
derived from chronic neutrophilic leukemia in transformation with
t(4; 11)(q21; q23). Leukemia. 18:998–1005. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Connolly D, Yang Z, Castaldi M, Simmons N,
Oktay MH, Coniglio S, Fazzari MJ, Verdier-Pinard P and Montagna C:
Septin 9 isoform expression, localization and epigenetic changes
during human and mouse breast cancer progression. Breast Cancer
Res. 13:R762011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Scott M, McCluggage WG, Hillan KJ, Hall PA
and Russell SE: Altered patterns of transcription of the septin
gene, SEPT9, in ovarian tumorigenesis. Int J Cancer. 118:1325–1329.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burrows JF, Chanduloy S, McIlhatton MA,
Nagar H, Yeates K, Donaghy P, Price J, Godwin AK, Johnston PG and
Russell SE: Altered expression of the septin gene, SEPT9, in
ovarian neoplasia. J Pathol. 201:581–588. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim DS, Hubbard SL, Peraud A, Salhia B,
Sakai K and Rutka JT: Analysis of mammalian septin expression in
human malignant brain tumors. Neoplasia. 6:168–178. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Church TR, Wandell M, Lofton-Day C, Mongin
SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T,
Osborn N, et al: Prospective evaluation of methylated SEPT9 in
plasma for detection of asymptomatic colorectal cancer. Gut.
63:317–325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tóth K, Sipos F, Kalmár A, Patai ÁV,
Wichmann B, Stoehr R, Golcher H, Schellerer V, Tulassay Z and
Molnár B: Detection of methylated SEPT9 in plasma is a reliable
screening method for both left- and right-sided colon cancers. PLoS
One. 7:e460002012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tóth K, Galamb O, Spisák S, Wichmann B,
Sipos F, Valcz G, Leiszter K, Molnár B and Tulassay Z: The
influence of methylated septin 9 gene on RNA and protein level in
colorectal cancer. Pathol Oncol Res. 17:503–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tänzer M, Balluff B, Distler J, Hale K,
Leodolter A, Röcken C, Molnar B, Schmid R, Lofton-Day C, Schuster T
and Ebert MP: Performance of epigenetic markers SEPT9 and ALX4 in
plasma for detection of colorectal precancerous lesions. PLoS One.
5:e90612010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Warren JD, Xiong W, Bunker AM, Vaughn CP,
Furtado LV, Roberts WL, Fang JC, Samowitz WS and Heichman KA:
Septin 9 methylated DNA is a sensitive and specific blood test for
colorectal cancer. BMC Med. 9:1332011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ahmed D, Danielsen SA, Aagesen TH,
Bretthauer M, Thiis-Evensen E, Hoff G, Rognum TO, Nesbakken A,
Lothe RA and Lind GE: A tissue-based comparative effectiveness
analysis of biomarkers for early detection of colorectal tumors.
Clin Transl Gastroenterol. 3:e272012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luther JA and Birren SJ: Neurotrophins and
target interactions in the development and regulation of
sympathetic neuron electrical and synaptic properties. Auton
Neurosci. 151:46–60. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nakagawara A: Trk receptor tyrosine
kinases: A bridge between cancer and neural development. Cancer
Lett. 169:107–114. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bouzas-Rodriguez J, Cabrera JR,
Delloye-Bourgeois C, Ichim G, Delcros JG, Raquin MA, Rousseau R,
Combaret V, Bénard J, Tauszig-Delamasure S and Mehlen P:
Neurotrophin-3 production promotes human neuroblastoma cell
survival by inhibiting TrkC-induced apoptosis. J Clin Invest.
120:850–858. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nikoletopoulou V, Lickert H, Frade JM,
Rencurel C, Giallonardo P, Zhang L, Bibel M and Barde YA:
Neurotrophin receptors TrkA and TrkC cause neuronal death whereas
TrkB does not. Nature. 467:59–63. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Goldschneider D and Mehlen P: Dependence
receptors: A new paradigm in cell signaling and cancer therapy.
Oncogene. 29:1865–1882. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu X, Tahan SR, Pasha TL and Zhang PJ:
Expression of neurotrophin receptor Trk-C in nevi and melanomas. J
Cutan Pathol. 30:318–322. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Segal RA, Goumnerova LC, Kwon YK, Stiles
CD and Pomeroy SL: Expression of the neurotrophin receptor TrkC is
linked to a favorable outcome in medulloblastoma. Proc Natl Acad
Sci USA. 91:12867–12871. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bardelli A, Parsons DW, Silliman N, Ptak
J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler
KW, et al: Mutational analysis of the tyrosine kinome in colorectal
cancers. Science. 300:9492003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wood LD, Calhoun ES, Silliman N, Ptak J,
Szabo S, Powell SM, Riggins GJ, Wang TL, Yan H, Gazdar A, et al:
Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum
Mutat. 27:1060–1061. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Luo Y, Kaz AM, Kanngurn S, Welsch P,
Morris SM, Wang J, Lutterbaugh JD, Markowitz SD and Grady WM: NTRK3
is a potential tumor suppressor gene commonly inactivated by
epigenetic mechanisms in colorectal cancer. PLoS Genet.
9:e10035522013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
deVos T, Tetzner R, Model F, Weiss G,
Schuster M, Distler J, Steiger KV, Grützmann R, Pilarsky C,
Habermann JK, et al: Circulating methylated SEPT9 DNA in plasma is
a biomarker for colorectal cancer. Clinical chemistry.
55:1337–1346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su XL, Wang YF, Li SJ, Zhang F and Cui HW:
High methylation of the SEPT9 gene in Chinese colorectal cancer
patients. Genet Mol Res. 13:2513–2520. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wasserkort R, Kalmar A, Valcz G, Spisak S,
Krispin M, Toth K, Tulassay Z, Sledziewski AZ and Molnar B:
Aberrant septin 9 DNA methylation in colorectal cancer is
restricted to a single CpG island. BMC Cancer. 13:3982013.
View Article : Google Scholar : PubMed/NCBI
|