Introduction
Non-small cell lung cancer (NSCLC) is the most
common respiratory system tumor clinically, accounting for ~80% of
lung carcinoma cases (1). It is
clinically characterized by rapid progression, strong invasiveness
and a high mortality rate. Surgical resection is the current
optimal therapy for NSCLC, but <50% of NSCLC patients can
receive surgical treatment due to late diagnosis and advanced stage
of disease (2). Therefore, with the
exception of surgery, chemotherapy is also an essential strategy in
the treatment of NSCLC, particularly for molecular targeted therapy
(3,4).
Epidermal growth factor receptor tyrosine kinase inhibitors
(EGFR-TKIs) are the most well known molecular targeted anti-cancer
drugs and are particularly effective in patients with activating
EGFR mutations (5). Gefitinib is a
specific EGFR-TKI, which is extensively applied in the treatment of
non-small lung cells in clinical practice (6,7). Clinical
studies (8,9) have reported that as first-line therapy,
gefitinib significantly improves the survival in advanced EGFR
mutation-positive NSCLC patients. However, for various reasons,
such as EGFR gene mutant and tumor microenvironment changes, the
sensitivity of NSCLC patients to gefitinib differs (10). In addition, previous studies
demonstrated that certain patients develop resistance to gefitinib
within a year (11). Thus, evaluating
and predicting the effectiveness of gefitinib is crucial for
adjusting therapeutic regimens and avoiding futile or
low-efficiency treatment.
MicroRNA is a type of short-chain non-coding RNA
that has been confirmed to be closely associated with the
incidence, progression and drug resistance of tumors. For example,
Fu et al found that microRNA-93 was significantly
upregulated in cisplatin-resistant ovarian cancer cells (12). Dong et al found that
microRNA-31 inhibits cisplatin-induced apoptosis in NSCLC (13). In addition, microRNAs can present
stably in the serum or plasma and the techniques of quantitative
detection for microRNA are ready for clinical practice. Thus,
microRNAs have potential to become biomarkers that can be used for
evaluating the sensitivity to chemotherapy. In previous years,
extensive studies on microRNA-200b induced drug-resistance have
been performed. Previous studies showed that microRNA-200b
contributed to multi-drug resistance of small cell lung cancer
(14), but at present, studies on the
effect of microRNA-200b on NSCLC chemotherapy have been rarely
reported. In particular, whether microRNA-200b level may affect the
gefitinib treatment of NSCLC remains unclear.
Based on the aforementioned considerations, the
present study was designed to investigate the association between
microRNA-200b and the sensitivity of NSCLC patients to gefitinib.
Furthermore, in vitro, the present study used A549 cells to
confirm the results from clinical data and explore its potential
mechanism involved with the factor-1 receptor pathway, as previous
studies have shown that insulin-like growth factor-1 receptor
(IGF-1R) is associated with acquired resistance of gefitinib
(15).
Materials and methods
Patients and specimens
In total, 100 patients (43 males and 57 females;
median age, 63 years) with pathologically confirmed advanced NSCLC
admitted to the First Hospital of Lanzhou University (Lanzhou,
China) between March 2012 and May 2014 were enrolled in the present
study. Additional demographic data, including age, gender,
histological type of tumor and clinical stage, are shown in
Table I. All patients were
administered with gefitinib orally (250 mg/day) and the effect of
gefitinib was evaluated according to the Response Evaluation
Criteria in Solid Tumors (RECIST) guidelines. In brief, the
diameter of the tumors were calculated and analyzed, and patients
were assessed for complete response (CR), partial response (PR),
stable disease (SD) or progressive disease (PD). The tumor tissue
samples were collected and stored in liquid nitrogen for additional
analysis. Plasma samples were collected prior to and subsequent to
therapy. The experimental protocols were approved by the Ethics
Committee of The First Hospital of Lanzhou University, and all
patients provided informed consent.
 | Table I.Clinical characteristics of 100
patients with non-small cell lung cancer. |
Table I.
Clinical characteristics of 100
patients with non-small cell lung cancer.
| Characteristic | CR+PR | SD+PD | P-value |
|---|
| Gender |
|
| 0.424 |
| Male | 14 | 29 |
|
|
Female | 23 | 34 |
|
| Median age,
years | 60 | 63 | 0.546 |
| Histological
subtypes |
|
| 0.884 |
|
Adenocarcinoma | 20 | 35 |
|
|
Squamous carcinoma | 8 | 15 |
|
|
Large-cell carcinoma | 1 | 3 |
|
|
Bronchioloalveolar
carcinoma | 8 | 10 |
|
| Clinical
stages |
|
| 0.002 |
|
IIIA+IIIB | 25 | 22 |
|
| IV | 12 | 41 |
|
| Smoking
history |
|
| 0.207 |
|
Yes | 28 | 40 |
|
| No | 9 | 23 |
|
| Chemotherapy
history |
|
| 0.855 |
|
Yes | 30 | 52 |
|
| No | 7 | 11 |
|
| Radiotherapy
history |
|
| 0.259 |
|
Yes | 27 | 39 |
|
| No | 10 | 24 |
|
Cell culture
The human lung adenocarcinoma A549 cell line was
obtained from the American Type Culture Collection (Manassas, VA,
USA) and cultured at 37°C in a 5% CO2 atmosphere. All
cells were maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Reverse transcription quantitative PCR
(RT-qPCR) for microRNA-200b detection
The microRNA of plasma specimens and tissue samples
were extracted with the MiRcute miRNA Extraction-Separation kit
[Tiangen Biotech (Beijing) Co., Ltd, Beijing, China], according to
the manufacturer's protocol. Following measurement of the purity
and concentration, reverse transcription of microRNA to cDNA was
assessed using One Step PrimeScript miRNA cDNA Synthesis kit
[Takara Biotechnology (Dalian) Co., Ltd., Dalian, China]. The
reverse transcription system and conditions were also followed in
accordance with the manufacturer's protocol; a no cDNA control and
a no reverse transcription control were also used in the same
reaction system. The obtained cDNA was amplified by RT-qPCR using
SYBR® Premix Ex Taq™ II [Takara Biotechnology (Dalian)
Co., Ltd.]. The reaction system and conditions were in accordance
with the manufacturer's protocol. The microRNA-200b upstream primer
sequence was 5′-GCGGCTAATACTGCCTGGTAA-3′. For normalizing the data
of plasma microRNA, microRNA-16 was used as reference (upstream
primer, 5′-CGCGCTAGCAGCACGTAAAT-3′). For normalizing the data of
tissue microRNA, U6 small nuclear RNA (snRNA) was used as a
reference (upstream primer, 5′-CTCGCTTCGGCAGCACA-3′). All
downstream primers were universal 3′ miRNA primers. The relative
expression level of microRNA-200b was calculated by the
2−ΔΔCq method (16,17),
ΔCq=Cqtarget-Cqreference.
MicroRNA-200b upregulation
The microRNA-200b mimic was designed and synthesized
by Invitrogen (Thermo Fisher Scientific, Inc.). Prior to
transfection, A549 cells were routinely maintained and grew to 70%
confluency. Subsequently, 100 nM microRNA-200b mimic and microRNA
negative controls (NC) were transfected into cells by
N-[1-(2,3-Dioleoyloxy)propyl]-N,N,N-trimethylammonium
methyl-sulfate reagent (Roche Applied Science, Mannheim, Germany),
according to the manufacturer's protocol. The transfection
efficiency was assessed by RT-qPCR in a preliminary experiment
using a similar protocol to the aforementioned protocol. For
RT-qPCR analysis, the data in cells were normalized by U6 snRNA
with the upstream primer sequence 5′-CTCGCTTCGGCAGCACA-3′.
Cell Counting Kit-8 (CCK-8) assay
Transfected and non-transfected A549 cells were
plated at a density of 1.0×105 cells/well into a 96-well
plate and incubated for 24 h at 37°C in a 5% CO2
atmosphere. Subsequent to being maintained for 24 h, all cells were
treated with 0.1 µM gefitinib for 6, 12, 24 and 48 h. Subsequent to
gefitinib treatment, 10 µl CCK-8 reagent (Dojindo Laboratories,
Kumamoto, Japan) was immediately added to each well. After 4 h, the
intensity of staining was monitored by SpectraMax i3 microtiter
plate spectrophotometer (Molecular Devices, LLC, Sunnyvale, CA,
USA) at 450 nm and the cell viability was estimated.
Flow cytometry analysis
Transfected and non-transfected A549 cells were
treated with 0.1 µM gefitinib for 24 h and harvested. For cell
cycle analysis, cells were fixed in 70% ethanol for 12 h and then
incubated with bromodeoxyuridine (BrdU) and 7-amino-actinomycin D
(7-AAD), according to the detailed protocol provided by BD
Pharmingen BrdU Flow Kits (BD Biosciences, Heidelberg, Germany).
The cell cycle was analyzed using FACSCalibur flow cytometer system
(BD Biosciences). For apoptosis analysis, double staining (Annexin
V-fluorescein isothiocyanate and propidium iodide) was performed
and cells were also analyzed using the FACSCalibur flow cytometry
system. The flow cytometry data were analyzed by CellQuest Pro
software (BD Biosciences).
Transwell migration assay
Using 24-well Transwell chambers (Corning
Incorporated, Corning, NY, USA) the migration of A549 cells was
observed. According to the manufacturer's protocol, the lower
chambers were filled with 500 µl DMEM supplemented with 10% FBS,
and 1×104 cells and 100 µl DMEM without FBS were placed
into the upper chambers. As routinely, Cells were cultured for 24 h
at 37°C in the presence of 5% CO2 and then the cells
that had invaded the lower surface of the microporous membrane of
the Transwell chamber were fixed using methanol. Subsequent to
being stained with crystal violet, the cells were quantified.
Quantitative PCR for IGF-1R, protein
kinase B (AKT) and extracellular signal-related kinases (ERK)
Transfected and non-transfected A549 cells were
cultured for 24 h; the total RNA in cells was extracted by TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was
reverse-transcribed to cDNA using PrimeScript® RT
reagent kit [Takara Biotechnology (Dalian) Co., Ltd.]. The reverse
transcription system was in accordance with the protocols of the
kit. The synthesized cDNA was amplified by SYBR® Premix
Ex Taq™ II kit [Takara Biotechnology (Dalian) Co., Ltd.]. Primers
are shown in Table II, The reaction
system and program was in accordance with the protocols of the kit.
The relative expression of target IGF-1 mRNA was analyzed by the
2−ΔΔCq method
(ΔCq=Cqtarget-CqGAPDH).
 | Table II.Primer sequences of IGF-1R, AKT and
ERK. |
Table II.
Primer sequences of IGF-1R, AKT and
ERK.
| Gene | Forward primer | Reverse primer |
|---|
| IGF-1R |
5′-CGGCATGGCATACCTCAAC-3′ |
5′-CGTCATACCAAAATCTCCGA-3′ |
| AKT |
5′-GCGACGTGGCTATTGTGA-3′ |
5′-CAGTCTGGATGGCGGTTG-3′ |
| ERK |
5′-CGCTACACGCAGTTGCCAGTACA-3′ |
5′-AAGCGCAGCAGGATCTGGA-3′ |
| GAPDH |
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ |
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ |
Western blot assay
Cells were lysed in radioimmunoprecipitation assay
buffer on ice for 30 min and total proteins were extracted. In
total, 25 µg of protein was boiled for 5 min, separated using 12%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred to a polyvinylidene fluoride membrane. Subsequent to
being blocked with 5% nonfat dry milk in Tris-buffered saline and
Tween 20, membranes were probed with anti-IGF-1R (dilution, 1:100;
catalog no. ab39398), anti-AKT (dilution, 1:500; catalog no.
ab8805), anti-ERK (dilution, 1:1,000; catalog no. ab196883),
anti-phospho-AKT (dilution, 1:500; catalog no. ab8933), phospho-ERK
(dilution, 1:100; catalog no. ab214362) and anti-β-actin (dilution,
1:10,000; ab8227) antibodies (Abcam, Cambridge, UK) overnight at
4°C. Subsequent to washing, membranes were incubated with Goat
Anti-Rabbit IgG H&L (HRP) secondary antibody (dilution,
1:2,000; catalog no. ab97051) for 1 h. Incubated membranes were
treated with ECL reagent (Applygen Technologies, Inc., Beijing,
China). Please state the host and target animals, clonality,
dilution and catalog number for the antibodies.
Statistical analysis
All data are reported as the mean ± standard
deviation. The statistical significance of the differences was
analyzed by one-way analysis of variance or least significant
difference-t test using SPSS 15.0 software (SPSS, Inc., Chicago,
IL, USA). For non-parametric statistics, Mann-Whitney U test was
performed for comparison between the two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
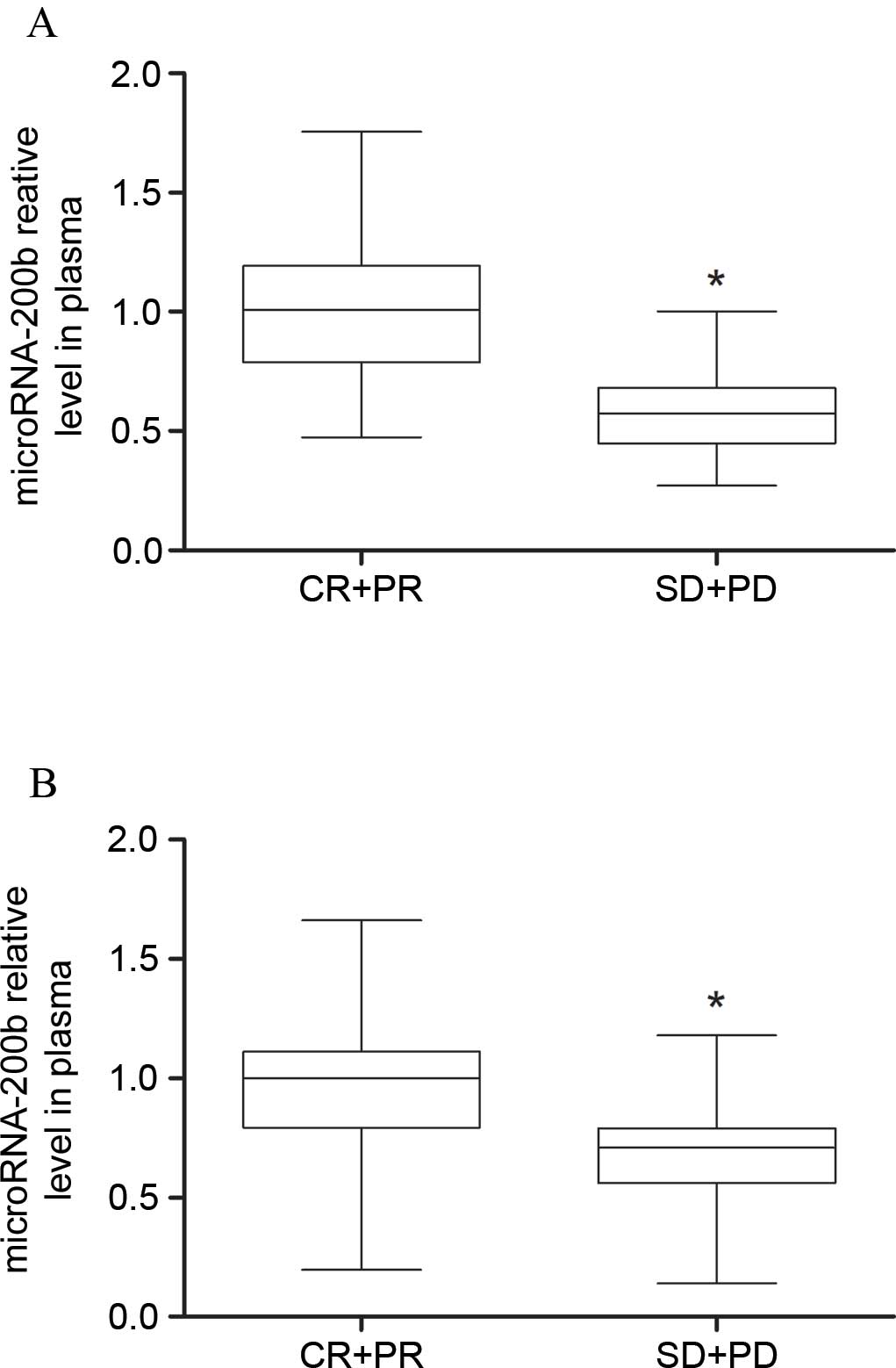
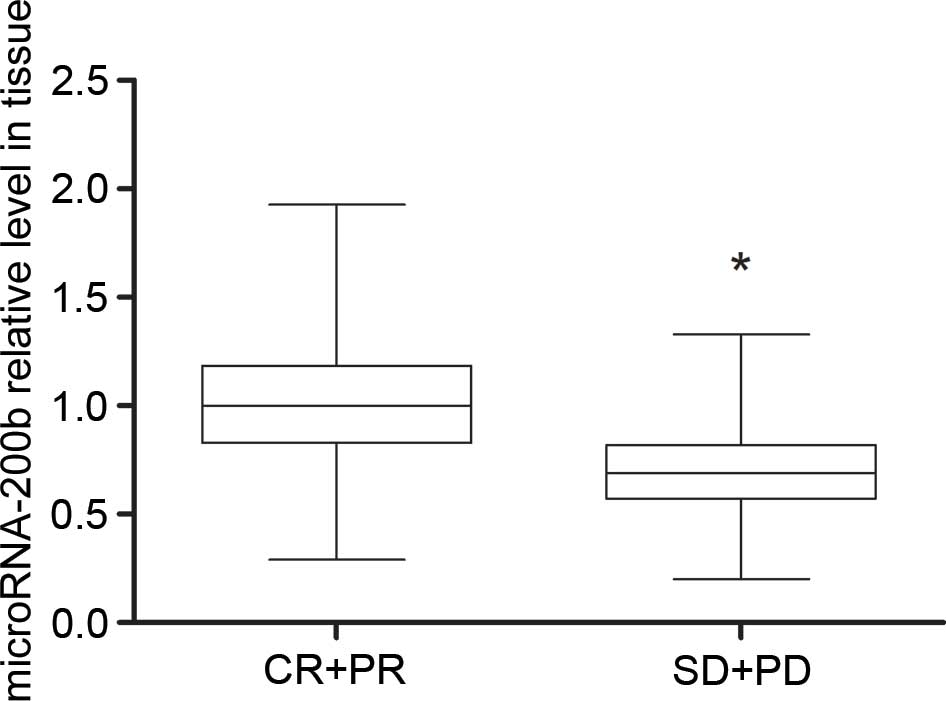
Low levels of microRNA-200b expression
were detected in gefitinib-insensitive patients
According to the RECIST guidelines, 7 patients were
classified as CR, 30 as PR, 28 as SD and 35 as PD. The present
study defined CR+PR patients as gefitinib-sensitive patients (n=37)
and SD+PD patients as gefitinib-insensitive patients (n=63). As
shown in Table I, the age, gender,
clinical stage and history of treatments showed no statistical
difference between CR+PR patients and SD+PD patients. RT-qPCR
results showed that the plasma microRNA-200b levels in SD+PD
patients were lower compared with the CR+PR patients (P=0.0068),
and the microRNA-200b levels in tissue samples of
gefitinib-insensitive patients were also lower, compared with
gefitinib-sensitive patients (P=0.0000075) (Figs. 1 and 2).
It is suggested that detecting microRNA-200b level may reflect the
sensitivity of NSCLC patients to gefitinib.
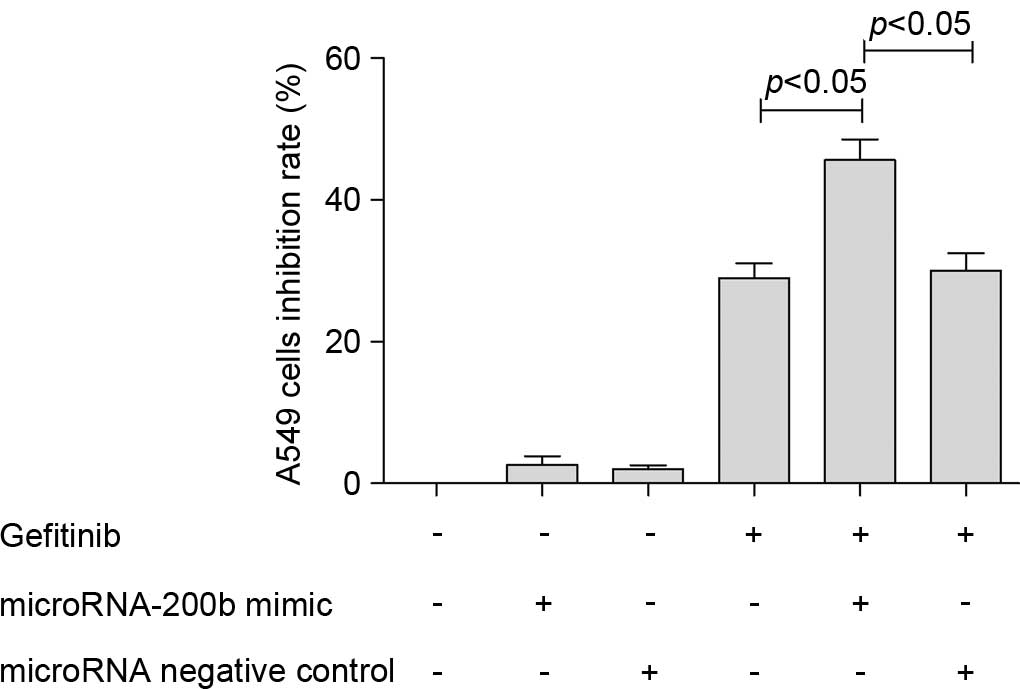
Upregulation of microRNA-200b
increased the anti-proliferative effect of gefitinib
Subsequent to upregulating the microRNA-200b level
of A549 cells through transfection with a microRNA-200b mimic, the
present study found that the anti-proliferative effect of gefitinib
on A549 cells increased. For the non-transfected A549 cells 24 h
subsequent to treatment with 0.1 µM gefitinib, the cell inhibition
rate was 20.8±3.2%, and for the A549 cells transfected with
microRNA-200b mimic, the inhibition rate increased to 45.6±4.3%,
showing a statistically significant difference (P=0.0071). However,
transfection with microRNA NC did not affect the inhibiting effect
of gefitinib on A549 cells (Fig.
3).
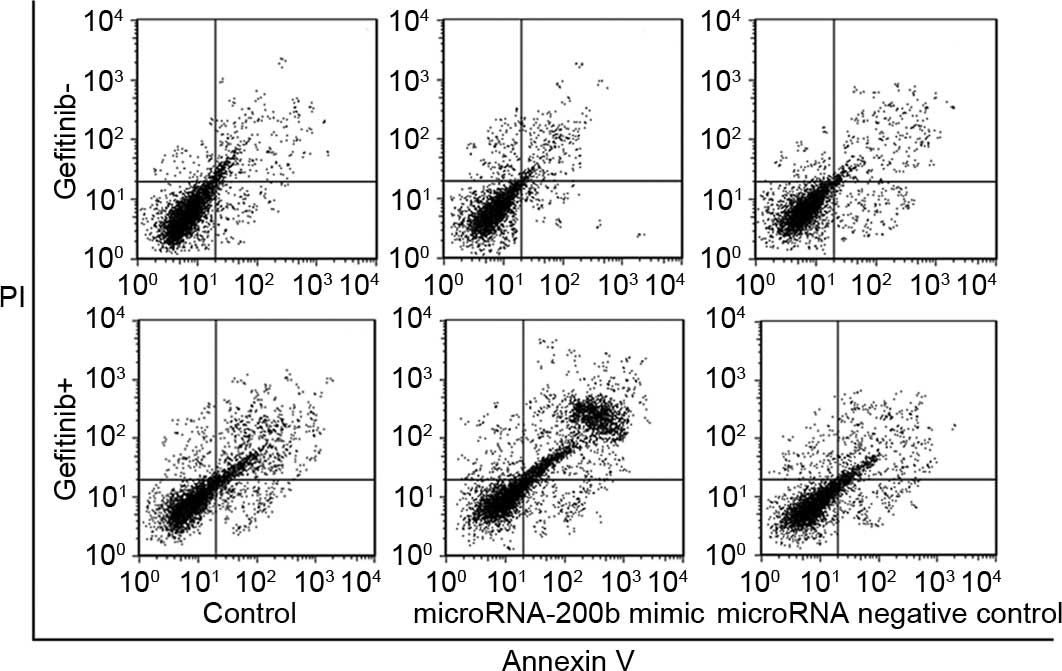
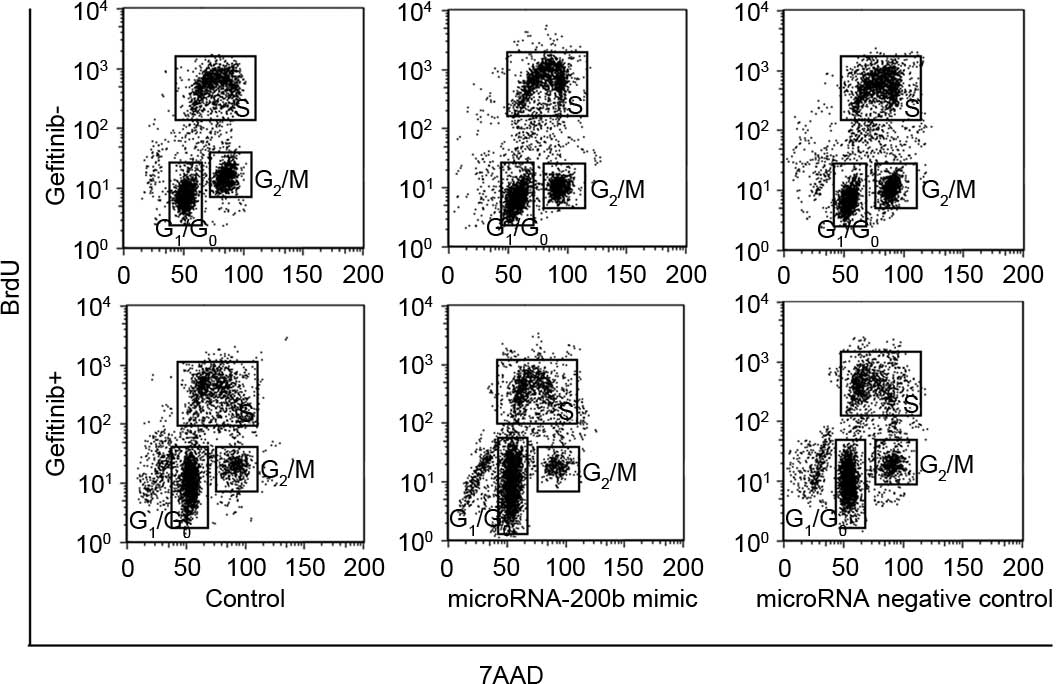
Upregulating microRNA-200b enhanced
apoptosis induction and cycle arrest ability of gefitinib
Following a time period of 24 h subsequent to
treatment of A549 cells using 0.1 µM gefitinib, the A549 cell
apoptosis rate without transfection treatment was 18.3±2.9%, while
the A549 cell apoptosis rate with upregulation of microRNA-200b was
29.2±3.1%, which was significantly increased compared with
non-transfected A549 cells (P=0.032), as shown in Fig. 4. Cell cycle analysis showed that,
subsequent to treatment with 0.1 µM gefitinib, the
G0/G1 percentage of untransfected A549 cells
was 38.6±4.2% and the G2/M percentage was 15.6±2.3%,
suggesting that gefitinib could cause G0/G1
arrest of A549 cells. Following treatment of the A549 cells with
upregulation of microRNA-200b with gefitinib, the
G0/G1 percentage was 70.4±7.9% and the
G2/M rate was 8.2±0.9%, indicating that upregulation of
microRNA-200b could enhance arrest of G0/G1
phase induced by gefitinib. Similarly, the effect of microRNA NC on
the cell cycle was not observed (Fig.
5).
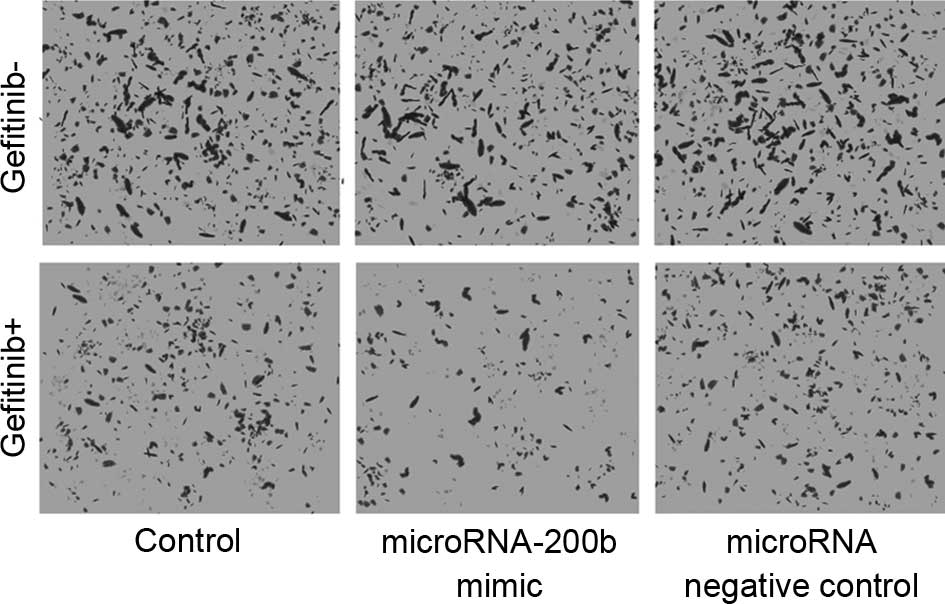
Upregulating microRNA-200b increased
the migration inhibition ability of gefitinib
As shown in Fig. 6,
the Transwell chamber assay revealed that, following upregulation
of microRNA-200b, gefitinib exhibited a markedly increased
migration inhibition on A549 cells, which was manifested by a
significantly reduced number of cells invading the lower surface of
the polycarbonate membranes. The microRNA NC did not affect the
migration inhibition ability of gefitinib.
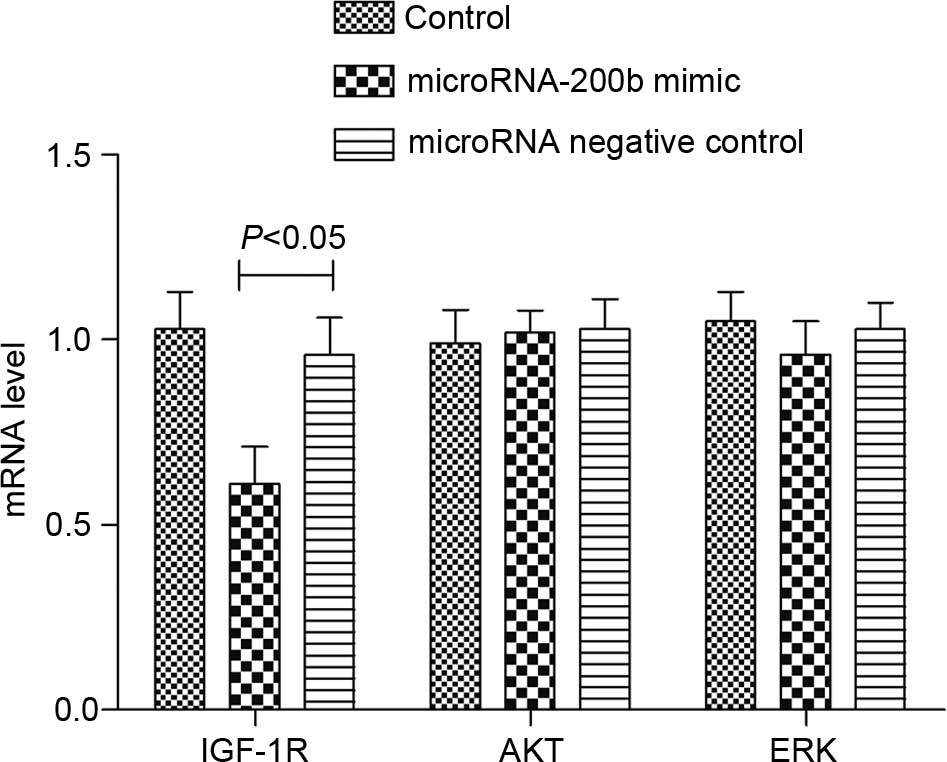
Upregulating microRNA-200b affected
the expression of IGF-1R and phosphorylation of AKT
RT-qPCR analysis revealed that, following the
upregulation of the microRNA-200b level of A549 cells, the IGF-1R
mRNA level in the cells was significantly reduced (P=0.012), but
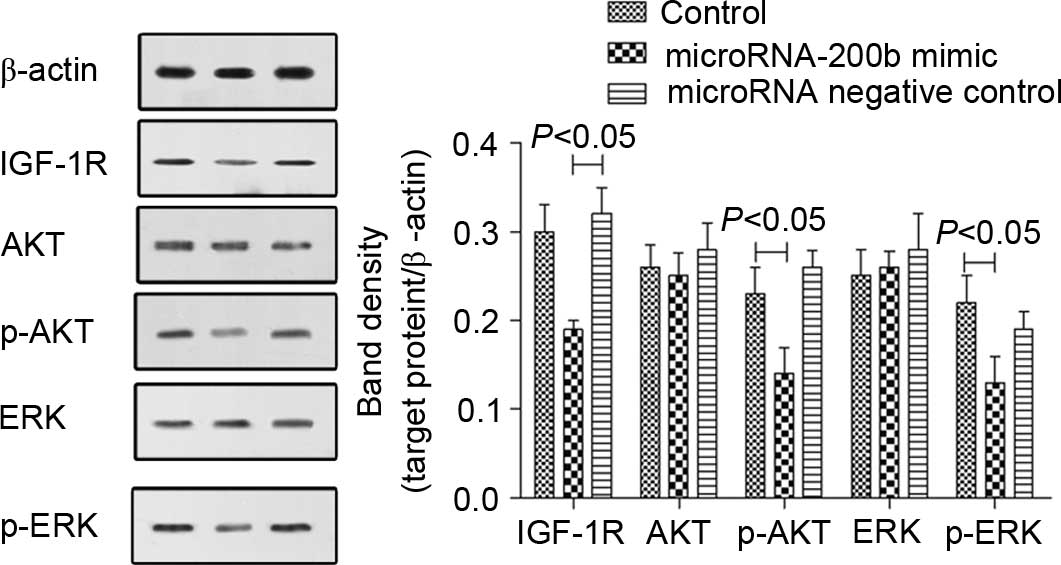
the mRNA levels of AKT and ERK were not changed (Fig. 7). The western blot analysis results
were similar to those of RT-qPCR. The IGF-1R protein level was
reduced, and AKT and ERK protein levels were not changed, but the
levels of phosphorylated AKT and ERK were reduced (Fig. 8). The microRNA NC showed no effect on
IGF-1R expression and AKT or ERK phosphorylation in A549 cells.
Discussion
The promotion and progression of NSCLC involves
signal transduction abnormality of a variety of growth factors.
Studies have confirmed that the growth of NSCLC is associated with
EGF, vascular endothelial growth factor, fibroblast growth factor
and IGF (18–20). Therefore, molecular targeted drugs can
suppress the growth of NSCLC by inhibiting or activating the
receptors or associated pathways of the aforementioned growth
factors. EGFR is the specific EGF receptor, and studies have shown
that overexpression of EGFR exists in more than half of NSCLC
patients (18,21). EGFR has tyrosine kinase activity, and
subsequent to binding with its ligand it can cause phosphorylation
of downstream proteins and activate downstream signaling pathways,
including the mitogen-activated protein kinase (MAPK) and
phosphoinositide 3-kinase (PI3K)/AKT pathways (22,23). These
pathways regulate tumorigenesis and tumor cell metastasis,
including control over cell proliferation and apoptosis. Studies
have shown that sustained activation of the MAPK and PI3K/AKT
pathways can promote tumor cell division, inhibit cell apoptosis
and promote invasiveness (24,25). Thus,
EGFR is a crucial treatment target in NSCLC. The application of
EGFR-TKI drugs can block the activation of EGFR downstream
pathways, for example, inhibiting the MAPK and PI3K/AKT pathway, to
play the role of inhibiting tumor cells (26). Gefitinib is one of the first
generation EGFR-TKIs, which can effectively improve the management
of NSCLC by inhibiting EGFR autophosphorylation and downstream
signaling. However, the resistance to gefitinib limits its
development and the majority of patients develop resistance within
a year (12–16,18–27).
Therefore, it is necessary to evaluate and predict the sensitivity
of patients to gefitinib. Previous studies showed that microRNAs
could regulate the response to gefitinib. Zhong et al
(28) found that let-7a, hsa-miR-126
and hsa-miR-145 may enhance cytotoxicity induced by gefitinib in
NSCLC. Another study showed that miR-34a rescues hepatocyte growth
factor-induced gefitinib resistance in EGFR mutant NSCLC cells
(29).
The present study revealed that the microRNA-200b
levels were lower in those patients that experienced a poor
curative effect of gefitinib. This indicated that detecting the
microRNA-200b level may reflect the sensitivity of NSCLC patients
to gefitinib. Furthermore, the current in vitro studies
showed that upregulating the expression of microRNA-200b could
increase the sensitivity of A549 cells to gefitinib. These results
suggest that the inhibitory effect of gefitinib was associated with
the expression of microRNA-200b. Previous studies have reported the
association between microRNA-200b and chemotherapy resistance and
its involved mechanism. For example, in a docetaxel-resistant human
lung adenocarcinoma cell line, microRNA-200b was identified as the
most downregulated microRNA. Additionally, a decreased
microRNA-200b level was detected in lung adenocarcinoma patients
treated with docetaxel-based chemotherapy and was associated with
decreased sensitivity to docetaxel (14). The mechanism of microRNA-200b-induced
chemotherapy resistance is complicated. Generally, the association
between microRNA-200b dysregulation and cancer chemoresistance can
be explained through the aspects of epithelial-mesenchymal
transition, cancer stem cell maintenance, angiogenesis, apoptosis
and cell cycle distribution (14,30).
However, the present study focused on the IGF-1R pathway
specifically.
IGF-1R is a transmembrane tyrosine protein receptor
with tyrosine kinase activity, when binding with a ligand, it can
activate the downstream EGFR pathways independent of EGFR, such as
the PI3K-AKT pathway, to reduce the efficacy of EGFR-TKI drugs and
even cause drug resistance (24,31).
Therefore, to avoid the activation of pathways downstream of the
EGFR pathway, such as the MAPK and PI3K/AKT pathways, it is
particularly important to maintain the efficacy of gefitinib. In
the present study, it was shown that upregulating the expression of
microRNA-200b could reduce IGF-1R in the cells and inhibit
phosphorylation of ERK and AKT. Therefore, it is considered that
microRNA-200b may increase the inhibitory effect of gefitinib on
NSCLC cells by inhibiting the expression of IGF-1R and reducing the
activation of MAPK and PI3K/AKT pathways caused by IGF-1R.
The majority of previous studies reported that the
target genes of the microRNA-200 family in tumors are the zinc
finger E-box binding homeobox 1 and 2 with transcriptional
regulation function (32). This study
revealed that the increased level of microRNA-200b could reduce the
expression of IGF-1R, but it could not identify that IGF-1R is the
direct target gene of microRNA-200b. In fact, the prediction of
bioinformatics tools does not confirm that IGF-1R is the direct
target gene of microRNA-200b. Therefore, the pathway for IGF-1R
expression affected by microRNA-200b remains to be further
studied.
In conclusion, low expression of microRNA-200b
exists in gefitinib-insensitive patients with NSCLC. Detecting the
level of microRNA-200b may be useful to evaluate the effect of
gefitinib on NSCLC patients. The increased microRNA-200b level can
inhibit the activation of MAPK and PI3K/AKT pathways caused by
IGF-1R, which aids improvement of the inhibitory effect of
gefitinib.
References
|
1
|
Wang YW, Yin CL, Zhang HY, Hao J, Yang YY,
Liao H and Jiao BH: High expression of forkhead box protein C2 is
related to poor prognosis in human gliomas. Asian Pac J Cancer
Prev. 15:10621–10625. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Islam KM Monirul, Shostrom V, Kessinger A
and Ganti AK: Outcomes following surgical treatment compared to
radiation for stage I NSCLC: A SEER database analysis. Lung Cancer.
82:90–94. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gridelli C, Peters S, Sgambato A, Casaluce
F, Adjei AA and Ciardiello F: ALK inhibitors in the treatment of
advanced NSCLC. Cancer Treat Rev. 40:300–306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roemans P, Emami B, Cox J, Pacagnella A,
Holsti L, Monteau M, Helle P, Comis R and Schaake C: Quality
control in NSCLC treatment: A consensus report. Lung Cancer.
7:19–20. 1991. View Article : Google Scholar
|
|
5
|
Pircher A, Manzl C, Fiegl M, Popper H,
Pirker R and Hilbe W: Overcoming resistance to first generation
EGFR TKIs with cetuximab in combination with chemotherapy in an
EGFR mutated advanced stage NSCLC patient. Lung Cancer. 83:408–410.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han SY, Zhao MB, Zhuang GB and Li PP:
Marsdenia tenacissima extract restored gefitinib sensitivity in
resistant non-small cell lung cancer cells. Lung Cancer. 75:30–37.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Koizumi T, Agatsuma T, Ikegami K, Suzuki
T, Kobayashi T, Kanda S, Yoshikawa S, Kubo K, Shiina T, Takasuna K,
et al: Prospective study of gefitinib readministration after
chemotherapy in patients with advanced non-small-cell lung cancer
who previously responded to gefitinib. Clin Lung Cancer.
13:458–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grigoriu B, Berghmans T and Meert AP:
Management of EGFR mutated nonsmall cell lung carcinoma patients.
Eur Respir J. 45:1132–1141. 2001. View Article : Google Scholar
|
|
9
|
Dhillon S: Gefitinib: A review of its use
in adults with advanced non-small cell lung cancer. Target Oncol.
10:153–170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manegold C: New perspectives in the
management of non-small cell lung cancer (NSCLC): Gefitinib
(Iressa, ZD 1839). European Journal of Cancer Supplements. 2:34–39.
2004. View Article : Google Scholar
|
|
11
|
Jeong CH, Park HB, Jang WJ, Jung SH and
Seo YH: Discovery of hybrid Hsp90 inhibitors and their
anti-neoplastic effects against gefitinib-resistant non-small cell
lung cancer (NSCLC). Bioorg Med Chem Lett. 24:224–227. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu X, Tian J, Zhang L, Chen Y and Hao Q:
Involvement of microRNA-93, a new regulator of PTEN/Akt signaling
pathway, in regulation of chemotherapeutic drug cisplatin
chemosensitivity in ovarian cancer cells. FEBS Lett. 586:1279–1286.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong Z, Zhong Z, Yang L, Wang S and Gong
Z: MicroRNA-31 inhibits cisplatin-induced apoptosis in non-small
cell lung cancer cells by regulating the drug transporter ABCB9.
Cancer Lett. 343:249–257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
MacDonagh L, Gray SG, Finn SP, Cuffe S,
O'Byrne KJ and Barr MP: The emerging role of microRNAs in
resistance to lung cancer treatments. Cancer Treat Rev. 41:160–169.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tandon R, Kapoor S, Vali S, Senthil V,
Nithya D, Venkataramanan R, Sharma A, Talwadkar A, Ray A, Bhatnagar
PK and Dastidar SG: Dual epidermal growth factor receptor
(EGFR)/insulin-like growth factor-1 receptor (IGF-1R) inhibitor: A
novel approach for overcoming resistance in anticancer treatment.
Eur J Pharmacol. 667:56–65. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Tang N, Hui T, Wang S, Zeng X, Li
H and Ma J: Identification of endogenous reference genes for
RT-qPCR analysis of plasma microRNAs levels in rats with
acetaminophen-induced hepatotoxicity. J Appl Toxicol. 33:1330–1336.
2013.PubMed/NCBI
|
|
18
|
Grossi F and Tiseo M: Granulocyte growth
factors in the treatment of non-small cell lung cancer (NSCLC).
Crit Rev Oncol Hematol. 58:221–230. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mukohara T, Kudoh S, Yamauchi S, Kimura T,
Yoshimura N, Kanazawa H, Hirata K, Wanibuchi H, Fukushima S, Inoue
K and Yoshikawa J: Expression of epidermal growth factor receptor
(EGFR) and downstream-activated peptides in surgically excised
non-small-cell lung cancer (NSCLC). Lung Cancer. 41:123–130. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pallis AG and Syrigos KN: Epidermal growth
factor receptor tyrosine kinase inhibitors in the treatment of
NSCLC. Lung Cancer. 80:120–130. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han X, Liu M, Wang S, Lv G, Ma L, Zeng C
and Shi Y: An integrative analysis of the putative
gefitinib-resistance related genes in a lung cancer cell line model
system. Curr Cancer Drug Targets. 15:423–434. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Van Emburgh BO, Sartore-Bianchi A, Di
Nicolantonio F, Siena S and Bardelli A: Acquired resistance to
EGFR-targeted therapies in colorectal cancer. Mol Oncol.
8:1084–1094. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hwang KE, Kwon SJ, Kim YS, Park DS, Kim
BR, Yoon KH, Jeong ET and Kim HR: Effect of simvastatin on the
resistance to EGFR tyrosine kinase inhibitors in a non-small cell
lung cancer with the T790M mutation of EGFR. Exp Cell Res.
323:288–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao Z, Liu LZ, Dixon DA, Zheng JZ,
Chandran B and Jiang BH: Insulin-like growth factor-I induces
cyclooxygenase-2 expression via PI3K, MAPK and PKC signaling
pathways in human ovarian cancer cells. Cell Signall. 19:1542–1553.
2007. View Article : Google Scholar
|
|
25
|
Kim MN, Lee KE, Hong JY, Heo WI, Kim KW,
Kim KE and Sohn MH: Involvement of the MAPK and PI3K pathways in
chitinase 3-like 1-regulated hyperoxia-induced airway epithelial
cell death. Biochem Biophys Res Commun. 421:790–796. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang CY, Kuan YH, Ou YC, Li JR, Wu CC,
Pan PH, Chen WY, Huang HY and Chen CJ: Autophagy contributes to
gefitinib-induced glioma cell growth inhibition. Exp Cell Res.
327:102–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu R, Shen H, Guo R, Sun J, Gao W and Shu
Y: Combine therapy of gefitinib and fulvestrant enhances antitumor
effects on NSCLC cell lines with acquired resistance to gefitinib.
Biomed Pharmacother. 66:384–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhong M, Ma X, Sun C and Chen L: MicroRNAs
reduce tumor growth and contribute to enhance cytotoxicity induced
by gefitinib in non-small cell lung cancer. Chem Biol Interact.
184:431–438. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou JY, Chen X, Zhao J, Bao Z, Chen X,
Zhang P, Liu ZF and Zhou J: MicroRNA-34a overcomes HGF-mediated
gefitinib resistance in EGFR mutant lung cancer cells partly by
targeting MET. Cancer Lett. 351:265–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng B, Wang R and Chen LB: Review of
MiR-200b and cancer chemosensitivity. Biomed Pharmacother.
66:397–402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ochi N, Takigawa N, Harada D, Yasugi M,
Ichihara E, Hotta K, Tabata M, Tanimoto M and Kiura K: Src mediates
ERK reactivation in gefitinib resistance in non-small cell lung
cancer. Exp Cell Res. 322:168–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu Q, Guo R, Lin M, Zhou B and Wang Y:
MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration
and invasion by targeting E-cadherin repressor ZEB2. Gynecol Oncol.
122:149–154. 2011. View Article : Google Scholar : PubMed/NCBI
|