Colorectal cancer (CRC) is one of the most commonly
diagnosed cancers worldwide, ranking as the third most prevalent
and fourth most frequent cause of cancer-related mortality
(1). In the early era of modern
molecular biology research, CRC was considered to be a disease
caused by genetic alterations, which influenced the gene product by
changing the amino acid sequence of a protein. With advances in
cancer research, however, more studies have focused on the
alterations in the regulation of gene expression that are
independent from the DNA sequence of the cell and are referred to
as ‘epigenetic’ changes. Epigenetic changes are likely to be a
major feature of cancer, and the most prominent epigenetic
modifications involve the aberrant methylation of CpG islands
(CGIs) and deacetylation or methylation of histone proteins
(2).
MicroRNAs (miRNAs or miRs) are non-coding,
endogenous small RNAs that bind the 3′ untranslated regions (UTRs)
of their target messenger RNA (mRNA) transcripts to suppress
protein translation or cause mRNA degradation (3). These newly identified small molecules
have been classified as epigenetic since the regulation of mRNAs
does not directly involve changing the DNA sequence. In the last
decades, emerging evidence has indicated that miRNAs are
deregulated in CRC and are directly connected with the epigenetic
machinery, playing a significant role in the occurrence and
progression of CRC.
In this review, we summarise recent findings on
miRNAs in CRC, in particular the reciprocal regulation between
miRNAs and other epigenetic machinery, to better understand the
epigenetic mechanisms that contribute to CRC carcinogenesis and to
identify new avenues for the diagnosis and treatment of this
disease.
miRNAs are short 18–24 nucleotide single-stranded
RNAs that mediate post-transcriptional gene repression (4). The miRNA family is comprised of gene
miRNA clusters, which have high homogeneity. In animals, primary
miRNA genes are transcribed by RNA polymerase II and are
subsequently processed to precursor hairpin miRNAs (pre-miRNAs)
that are ~70 nt long by Drosha in the nucleus. The pre-miRNAs are
then transported to the cytoplasm, where they are recognised and
processed by the RNAse III enzyme Dicer into mature miRNAs of ~22
nt. The mature miRNAs induce the degradation of mRNAs and
downregulate the target genes through the imperfect binding of the
RNA-induced silencing complex to the 3′ UTR of target mRNAs
(5,6).
With the application of high-throughput screening
technology, including microarray-based miRNA profiling platforms
and next-generation sequencing (NGS) approaches, more studies have
focused on searching for biomarkers by identifying different miRNA
expressions in different types of cancer. To date, a number of
aberrantly expressed miRNAs and their gene targets have been
identified in CRC. The first study to investigate a miRNA
alteration in CRC was performed in 2003, in which miR-143 and
miR-145 were expressed at reduced steady-state levels at the
adenomatous and cancer stages of CRC (13). In another study, 37 miRNAs with
various expression levels were identified in CRC using
comprehensive array-based analyses in 84 CRC and matched normal
colonic tissues (14). As a new
emerging throughput screening platform, NGS technology easily
enriches, detects and analyses miRNAs using a genome-wide scale.
Moreover, the combination of miRNA and transcriptome sequencing
enables the prediction of miRNA target genes, which aids in the
identification of new and known miRNAs from a systematic and
functional perspective. Using NGS technology in normal, tumour and
metastasis tissue samples from the same patients with CRC, an
earlier study investigated the complete set of miRNAs and their
potential downstream regulated genes as well as the signalling
network, and explored the power of miRNA-1 response prediction in
individual patients (15).
Despite the use of various detection methods,
including microarray, sequencing, real-time polymerase chain
reaction-based approaches and in situ hybridisation, a high
consistency of miRNA expression profiles exists among these
studies, which indicates that these miRNAs are essential elements
in cancer progression (16–19).
As each miRNA has several different mRNA targets,
miRNA genes are predicted to represent ~3% of the human genome,
whereas ~30% of the genes are regulated by miRNAs (20). Evidence has demonstrated that miRNAs
act either as tumour suppressors by suppressing the expression of
target oncogenes, or as proto-oncogenes by inhibiting the
expression of tumour suppressor genes (TSGs) (21–24). Both
effects correlate with cancer development and its progression in
CRC (11,25,26).
The deregulation of the Wnt/β-catenin pathway is one
of the earliest events during CRC development. In this pathogenic
pathway, β-catenin acts as a transcriptional activator and
upregulates the expression of Wnt target genes. Overexpression of
constitutively active β-catenin may result in colorectal
tumourigenesis (27). Through in
vitro and in vivo experiments, Ma et al proved
that miRNA-17–92 increases the expression of β-catenin indirectly
by targeting P130, and subsequently promotes the tumourigenesis and
progression of CRC (28). Strillacci
et al revealed that miR-101 regulates Wnt/β-catenin
signalling in CRC through the strong impairment of β-catenin
nuclear accumulation and β-catenin-driven transcriptional activity,
following the control of downstream target gene expression and
malignant phenotype in cancer cells (29).
CSCs are a group of heterogeneous cells that are
vital for the initiation and progression of cancers, including CRC.
The Wnt pathway plays an essential role in the induction of the
symmetrical cell division (SCD) of CSCs, which disturbs the
homeostasis of the stem cell pool and leads to carcinogenesis
(30,31). miRNA-146a was identified by Hwang
et al as an activator of the Wnt pathway in CRCSCs by
stabilising β-catenin, which directs SCD to promote CRC
progression. Notably, the study uncovered an upstream regulatory
mechanism of miR-146a, in which Snail activates miR-146a
transcription via a β-catenin-TCF4 complex. A feedback circuit of
Snail-miR-146a-β-catenin loops exists in CRCSCs to maintain Wnt
activity using a miRNA-dependent regulation method (32).
EMT is a cellular process of converting polarised
epithelial cells into mesenchymal cells. During this process,
cancer cells acquire malignant, migratory and invasive capabilities
(33). The activation of EMT in CRC
enables the cancer cells to detach, migrate and disseminate through
the blood or lymphatic vessels, suggesting a mechanistic role of
EMT in CRC metastasis.
A range of miRNAs were identified as regulators in
this CRC process. For example, miR-29c expression in primary CRC
and distant liver metastases tissues was observed to be negatively
related to the mRNA levels of E-cadherin and vimentin, which are
known to be specifically upregulated and established as markers of
EMT (34). Moreover, ectopic miR-29c
expression suppresses the expression of these EMT markers and
induces morphological transformations to reverse EMT in CRC cells
(34). Additional mechanistic
research revealed that GNA13 and PTP4A are direct targets of
miR-29c that act through the ERK/GSK3β/β-catenin and
AKT/GSK3β/β-catenin pathways, respectively, to regulate EMT
(34).
Using an azoxymethane and dextran sodium
sulfate-induced colitis-associated tumour mouse model, Rokavec
et al demonstrated that exposure of CRC cells to the
cytokine IL-6 represses the miR-34a gene by activating the STAT3
transcription factor. Furthermore, the IL-6 receptor, which
mediates IL-6-dependent STAT3 activation, was identified as a
direct miR-34a target. Such miRNA-related self-stabilising circuits
consisting of IL-6R/STAT3/miR-34a are crucial for the induction of
EMT and capacitation of metastasis in CRC cells (35).
Human EGFR comprises an extracellular ligand-binding
domain and an intracellular domain with tyrosine kinase activity.
The selective induction of EGFR activates two major downstream
signalling pathways, namely KRAS/RAF/ERK and PI3K/AKT, and is
responsible for the progression of a broad spectrum of solid
tumours, including CRC.
KRAS is described as a major driver of CRC
progression and may be regulated directly by a number of miRNAs
with tumour suppressor functions, including miR-143 (36,37),
miR-145 (37), miR-18a* (38), miR-30b (39) and let-7 (40). In contrast, KRAS upregulates the
expression of oncogenic miR-200c, miR-221/222, miR-181a and miR-210
in CRC cells (41). In an
inflammatory bowel disease-associated CRC study, researchers
identified a correlation between miR-133a, miR-143, miR-145 and
miR-223 dysregulation and alteration of PI3K/Akt signalling.
Furthermore, the authors observed that miR-223 directly targeted
IGF1R to regulate this pathway, representing molecular links
between inflammation and cancer (37).
Since aberrant miRNAs are well recognised as
representing the hallmark of and playing a key role in several
biological processes of CRC, increasing numbers of studies have
focused on exploring the mechanisms responsible for disturbed miRNA
expression. Dysregulated miRNAs are known as epigenetic
alterations. Notably, a previous study has suggested that gene
encoding for miRNAs may be affected by the same epigenetic
regulatory mechanisms that modulate any other protein-coding genes
(48). We summarise the altered
miRNAs regulated by epigenetic factors in Table I, including promoter methylation,
histone acetylation and chromatin changes in CRC. The data are
derived from various array platforms and supported by validation
experiments and function assays (49–72).
The chemical modification of DNA methylation is
mediated by DNA methyltransferases (DNMTs), which catalyse the
covalent attachment of a methyl group to the 5′ carbon of cytosine
in CpG dinucleotides. The majority of CpGs are maintained in a
methylated state, whereas the CpGs located in CGIs, local
enrichments of CpG sequences, are normally unmethylated. The
methylation of CGIs in promoters and start sites inhibits
interactions with transcription factors and induces a reduction in
gene expression (73). A
comprehensive analysis of miRNA gene sequences has revealed that
~50% are associated with CGIs, suggesting that they could be
subjected to a DNA methylation mechanism of regulation (74).
Evidence has confirmed that DNA methylation may be
responsible for the altered expression of miRNAs in CRC by
silencing potential tumour suppressor miRNAs. The DNA demethylating
agent 5-aza-2′-deoxycytidine (5-AZA) reduces the methylation levels
of genes by covalently binding to DNMTs and inhibiting their
function (75), and is widely applied
in the search for methylation-regulated miRNAs. By combining the
treatment of CRC cells using 5-AZA and high-throughput analysis,
eight miRNAs were identified as novel miRNAs regulated by DNA
methylation (59). The authors
subsequently investigated the functions of these
hypermethylation-silenced miRNAs by ectopically expressing select
candidates, which resulted in the repression of proliferation and
the migration of cancer cells (59).
After treating the CRC cells with 5-AZA and the histone deacetylase
(HDAC) inhibitor trichostatin A, Grady et al demonstrated
that the expression of miR-342 is commonly suppressed by
hypermethylation in CRC and that the reconstitution of miR-342 in
the HT-29 cells induced apoptosis, implying its function as a
pro-apoptotic tumour suppressor. More significantly, the methylated
CGI located in the promoter of EVL, which has been proven to be the
host gene of miR-342, has a higher frequency of methylation in
cancer and adenoma samples compared with normal samples. This study
addressed a novel mechanism for silencing intronic miRNAs by
epigenetic alterations of cognate host genes in CRC (57).
A similar approach aimed at identifying
epigenetically regulated miRNAs was represented by the miRNA
profiling of CRC cells genetically deficient for the DNA
methyltransferase enzymes. DNA hypomethylation induces a release of
miRNA silencing in cancer cells. One of the primary targets is
miRNA-124a, which is embedded in a large CGI and is capable of
targeting cyclin D kinase 6 (CDK6); CDK6 has an impact on the
phosphorylation status of downstream effector retinoblastoma TSG
(51). To identify
methylation-silenced miRNAs and to clarify their role in CRC, our
group performed a microarray analysis to screen for miRNAs in CRC
cells using 5-AZA treatment or a genetic knockdown of DNA
methyltransferases. We observed that miR-149 was epigenetically
silenced in CRC and that the downregulation of miR-149 was
associated with the hypermethylation of the neighbouring CGI. The
transfection of miR-149 inhibited the cell growth and invasion of
CRC cells in vitro by targeting specificity protein 1
(67).
Methylation-regulated miRNAs have been observed to
be closely linked with the progression of CRC. Balaguer et
al reported that methylation of the miR-137 CGI was a
cancer-specific event that was observed in virtually all CRC cell
lines, in 82% of adenomas, in 82% of CRCs but in only 14% of normal
mucosae, suggesting that the results constituted an early event in
colorectal carcinogenesis (58).
Other studies in CRC samples identified five miRNAs that were
downregulated and located around or on a CGI, and 5-AZA-restored
expression of three miRNAs (namely miR-9, miR-129 and miR-137) in
three CRC cell lines. Methylation of the miR-9-1, miR-129-2 and
miR-137 CGIs was observed in CRC cell lines and in primary CRC
tumours, but not in normal colonic mucosae. Furthermore, the
methylation of miR-9-1 occurred more frequently in the advanced
stages of CRC and was associated with nodal invasion (P=0.008),
vascular invasion (P=0.004) and distant metastasis (P=0.016)
(52). Using a case-control study
containing 94 primary colon cancer samples with and without liver
metastases, Siemens et al determined CGI methylation
frequencies of miR-34 promoters and prognostic values. Their
results revealed that miR-34a methylation was strongly associated
with metastasis to the liver (P=0.003) and lymph nodes (P=0.006).
In a confounder-adjusted multivariate regression model, miR-34a
methylation provided the most significant prognostic information
concerning metastasis to the liver (P=0.014) (61).
Inactivation of miR-200 members by DNA methylation
has been described in various cancers (76–78). In a
previous study, the authors demonstrated that the 5′-CGIs of both
miR-200 loci are unmethylated and are associated with the
expression of the corresponding miRNAs with epithelial features,
whereas CGI hypermethylation-associated silencing is observed in
transformed cells with mesenchymal characteristics (78). Additional studies revealed that
miR-200ba429 and miR-200c141 transcripts undergo a dynamic
epigenetic regulation linked to EMT or mesenchymal-to-epithelial
transition (MET) phenotypes in tumour progression. These findings
confirmed that the epigenetic silencing plasticity of the miR-200
family contributes to EMT and MET, and the development of CRC
tumourigenesis (79).
Inflammatory bowel disease (IBD), including
ulcerative colitis (UC) and Crohn's disease, is one of the most key
high-risk conditions for CRC (80).
Although it is known that epithelial dysplasia occurring in
long-standing IBD is a typical precancerous lesion, the
pathogenesis of colon cancer in IBD is poorly understood. Several
studies have demonstrated the crucial role of altered miRNAs in the
carcinogenesis of IBD-associated CRC (56,81–83), which
includes the detection of epigenetic regulation of miRNAs. By
comparing the expression and promoter methylation status of
miR-124a in tissue samples from UC patients with or without CRC,
patients with sporadic CRC and healthy volunteers, Ueda et
al observed significantly decreased expression and higher
methylation levels of miR-124a in UC-associated CRC (56). Downregulated miR-124 by
hypermethylation was also identified in another study on UC, which
promoted inflammation and the pathogenesis of UC by targeting STAT3
in paediatric patients (84).
Compared with silencing TSGs through DNA
hypermethylation, hypomethylation activates oncogenes and promotes
cancer progression. As a member of the let-7 miRNA family, let-7a
has been reported as a tumour suppressor in CRC (85,86).
However, its expression was reactivated by DNA hypomethylation in
lung cancer and was associated with enhanced tumour phenotypes and
oncogenic changes (87). In a
previous study of metastasis-related miRNAs in CRC, the authors
observed a higher expression of miR-200c in liver metastases
tissues than in primary CRCs (70). A
functional analysis indicated that the overexpression of miR-200c
induces MET in liver metastasis by targeting ZEB1, ETS1 and FLT1.
More significantly, the increased expression of miR-200c was
significantly associated with hypomethylation of the promoter
region and was proven to be reactivated by the demethylation in CRC
(70).
Methylation is not the only epigenetic mechanism
that affects miRNA expression. Histone modifications may also lead
to either activation or repression of miRNAs depending on which
residues are modified and which type of modification is present.
The N-terminal tails of histones undergo several different covalent
modifications, including acetylation, methylation, ubiquitylation,
sumoylation and phosphorylation on specific residues (88).
Several studies have confirmed that HDAC inhibitor
alters miRNA expression in CRC cells (60,88–90).
Acetylated lysine in histone tails was correlated with a more
relaxed chromatin state and gene-transcription activation, whereas
histone deacetylation led to a more compact chromatin structure and
suppressed gene transcription (91).
By combining the treatment of CRC cells with an HDAC inhibitor
suberoylanilide hydroxamic acid (SAHA) and a microarray assay, Shin
et al revealed that 32 of 275 human miRNAs were upregulated
in HCT-116 cells, with the potential target genes related to
apoptosis, cell cycle and differentiation of the cancer cells
(90). Using another HDAC inhibitor,
butyrate, which may decrease gene transcription by reducing histone
H3 and H4 acetylation near the transcription start sites (92), Hu et al downregulated the
expression of miR-106b in HCT-116 cells, restored p21 protein
expression and decreased cell proliferation (89). Similarly, the expression of the
oncogenic miR-17–92 cluster was inhibited by butyrate in HT29 and
HCT116 CRC cells, with a corresponding increase in the target TSGs,
including PTEN, BCL2L11 and CDKN1A (60).
In addition to performing their individual roles,
DNA methylation and histone modifications interact with each other
at manifold levels to influence chromatin organisation and the
expression status of genes, including miRNAs. By comparing miRNA
expression and histone modifications (H3K4me3, H3K27me3 and
H3K79me2) before and after DNA demethylation, 47 miRNAs, including
miR-1-1, were identified to be potential targets of epigenetic
silencing and may act as a tumour suppressors in early and advanced
CRC. DNA demethylation induced the upregulation of H3K4me3 and
H3K27me3, but not H3K79me2, at the promoters of these miRNAs. The
conclusion provided additional insight into the association between
hypermethylation, chromatin modifications and miRNA dysregulation
in cancer (54).
The significance of inhibitory signals that
contribute to epigenetic gene silencing, including DNA methylation,
histone modification and miRNAs, has been increasingly recognised
in recent years. However, the crosstalk between these epigenetic
regulators is not fully understood. An increasing amount of data
exists to support miRNA regulation of the expression of components
of the epigenetic machinery, creating a highly controlled feedback
mechanism.
Methylation changes to the genome are controlled by
DNMTs. To date, three catalytically active DNMTs, namely DNMT1,
DNMT3A and DNMT3B, have been identified (93). miR-143 has been confirmed to be a
tumour suppressor miRNA and to decrease in CRC cells and tissues
according to several studies (47,94–97).
Moreover, DNMT3A, which represents an essential controller of
methylation changes to the genome and is frequently increased in
various malignancies including CRC, was defined as a direct target
of miR-143 (98). Specifically,
miR-143 was inversely correlated with mRNA and protein expression
of DNMT3A in CRC. In silico prediction illustrated the
binding of miR-143 with the DNMT3A 3′ UTR, which was confirmed by a
luciferase reporter assay. Furthermore, the restoration of miR-143
expression in colon cell lines downregulated DNMT3A expression at
the mRNA and protein level.
In addition to DNA methyltransferases, miRNA has
been proven to regulate histone deacetylases in CRC. As a
nicotinamide adenine dinucleotide-dependent deacetylase, silent
information regulator 1 (SIRT1) may function as an oncogene and
play a role in cancers including CRC (100–102).
Yamakuchi et al demonstrated that miR-34a inhibits SIRT1
expression through a special binding site within the 3′ UTR of
SIRT1. miR-34a inhibition of SIRT1 leads to an increase in
acetylated p53 and apoptosis in CRC cells. Furthermore, miR-34a is
a transcriptional target of p53, suggesting a positive feedback
loop between p53 and miR-34a through SIRT1-dependent regulation
(103). Notably, another study
reported that HDAC1 also exhibits a robust decrease in protein
levels following the induction of miR-34a in CRC cells (104). In a previous study evaluating the
role of miRNAs in the antitumour action of calcitriol, miRNA
expression profiles were examined in colon cancer cells treated
with calcitriol (105). The results
revealed that calcitriol selectively induces the expression of
miR-627. Moreover, histone demethylase JMJD1A was identified as the
direct target of miR-627, suggesting that miR-627-mediated JMJD1A
downregulation is an essential intracellular target for calcitriol
and mediates epigenetic activities of calcitriol. The series of
studies showing miRNAs regulating DNMTs or HDACs in CRC are listed
in Table II.
The same miRNA that provides epigenetic regulation
may in turn affect the epigenetic machinery. miR-143, targeting
DNMT3A in CRC cells, was hypermethylated and decreased in acute
lymphoblastic leukaemia (ALL) cells. Demethylation restored
functional endogenous miR-143 expression and significantly
inhibited proliferation and induced apoptosis in ALL (106). On the one hand, CGI methylation of
miR-148a and miR-152 genes regulated by DNMT1 silenced the TSG-like
miRNAs in CRC (107–109). On the other hand, the levels of
DNMT1, a direct target of miR-148a and miR-152, were inhibited by
miR-148a and miR-152 overexpression. A negative feedback loop
between DNMT1 and miR-148a/152 promoted carcinogenesis in cancer
cells (110,111), implying that miRNAs mediate the
crosstalk between epigenetic regulators.
miR-137 has been identified as another significant
mediator of crosstalk between epigenetic regulators in CRC.
Balaguer et al revealed that miR-137 is constitutively
expressed in the normal colonic epithelium and is silenced by CGI
methylation of the promoter in neoplastic tissues (58). Restoring miR-137 expression in CRC
cells resulted in a significant decrease in proliferation,
suggesting its potential as a tumour suppressor miRNA in CRC. An
additional analysis of candidate targets was performed using a
combination of bioinformatic and transcriptomic approaches and
identified that LSD1, an HDAC, is a target of miR-137. Notably, as
a new class of histone demethylating enzymes, LSD1 was essential
for the maintenance of global DNA methylation through the
demethylation of a non-histone substrate, DNMT1, by increasing its
stability. The results implied that miR-137 mediates the interplay
between epigenetic regulators in CRC (58). All of these observations suggested a
reciprocal crosstalk between miRNAs and epigenetic regulators. In
this scenario, miRNAs functioned as a crucial factor in the valid
transmission of various patterns of epigenetic modulation and
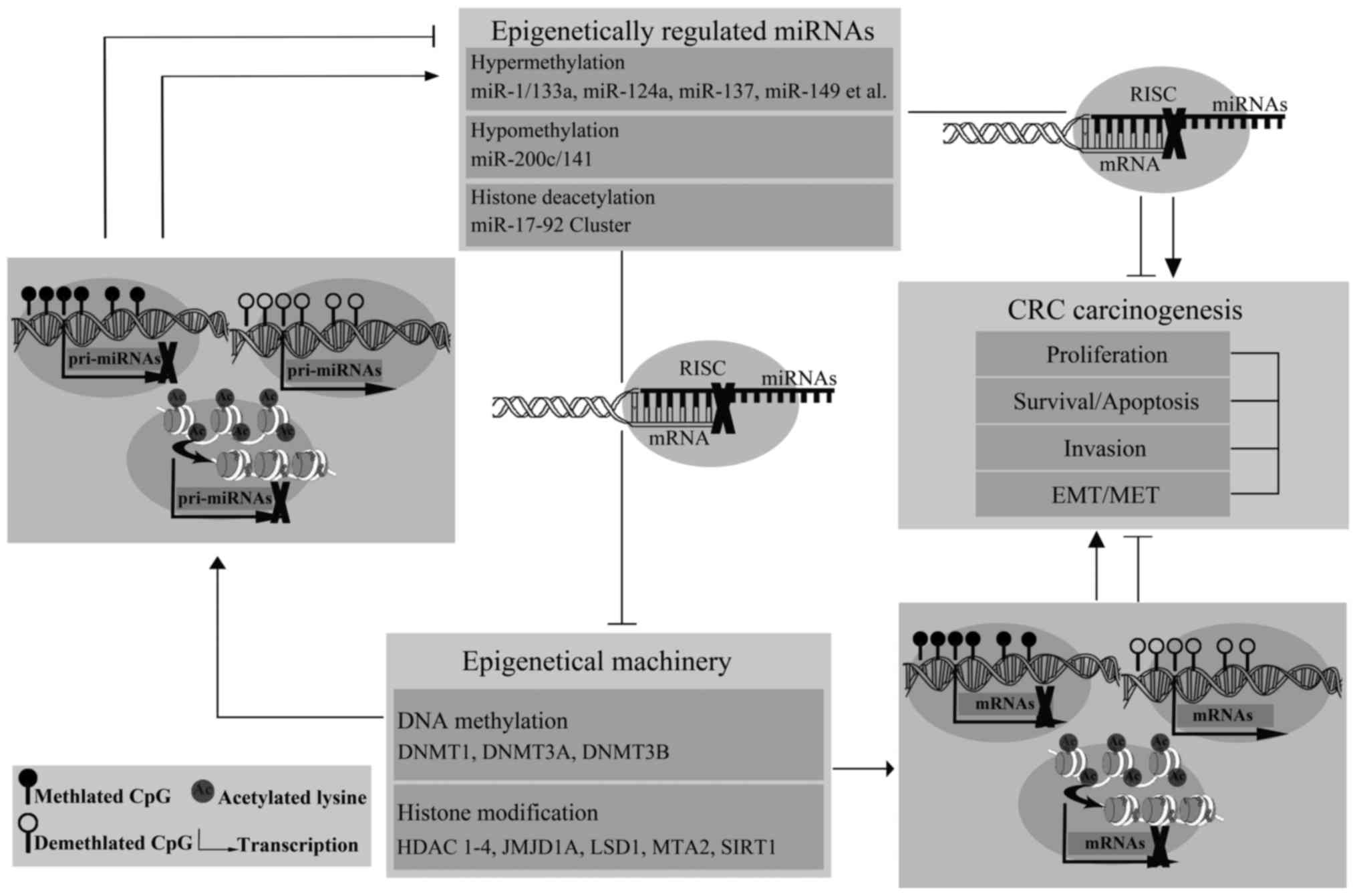
thereby interfered with CRC carcinogenesis (Fig. 1).
The evidence discussed here indicates a strong
interplay between miRNAs and epigenetic regulators in CRC.
Epigenetically regulated miRNAs by DNA methylation or histone
modifications are capable of silencing specific target molecules at
the post-transcriptional level, including members of the epigenetic
machinery, thus contributing to the conversation between other
epigenetic events. These results have extended our comprehension of
the pathogenesis and pathophysiology of CRC.
Additional exploration is warranted to annotate the
regulatory loop involving miRNAs and the epigenetic machinery, and
to explore how to translate these findings into clinical
applications. First, it is crucial to identify new avenues for
anticancer therapy based on the epigenetic regulation of miRNAs. To
date, therapy using several epigenetic-based synthetic agents has
been based on conventional protein-coding TSGs. With the
identification of a greater number of epigenetically silenced
tumour-suppressor miRNA genes in CRC, restoring these miRNAs using
epigenetic agents is likely to become a potentially powerful
approach to the remedy. Second, endeavours utilising miRNAs to
modulate the expression levels of oncogenes and suppressor genes or
to control the effector enzymes using epigenetic machinery and thus
affecting the expression of a broad range of modulated molecules
would permit the development of novel preventative therapies or
adjunctive therapeutic approaches in CRC.
This study was supported by grants from the Shanghai
Natural Science Foundation (grant no. 12ZR1447500), the National
Natural Science Foundation of China (grant no. 81301753), the
Specialized Research Fund for the Doctoral Program of Higher
Education (grant no. 20130072120046), and the State Key Program of
the National Natural Science Foundation of China (grant no.
81230057).
|
1
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kozomara A and Griffiths-Jones S: miRBase:
annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:(Database Issue). D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grady WM and Tewari M: The next thing in
prognostic molecular markers: microRNA signatures of cancer. Gut.
59:706–708. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goel A and Boland CR: Epigenetics of
colorectal cancer. Gastroenterology. 143:1442.e1–1460.e1. 2012.
View Article : Google Scholar
|
|
12
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
14
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rohr C, Kerick M, Fischer A, Kühn A,
Kashofer K, Timmermann B, Daskalaki A, Meinel T, Drichel D, Börno
ST, et al: High-throughput miRNA and mRNA sequencing of paired
colorectal normal, tumor and metastasis tissues and bioinformatic
modeling of miRNA-1 therapeutic applications. PLoS One.
8:e674612013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pencheva N and Tavazoie SF: Control of
metastatic progression by microRNA regulatory networks. Nat Cell
Biol. 15:546–554. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qian B, Nag SA, Su Y, Voruganti S, Qin JJ,
Zhang R and Cho WC: MiRNAs in cancer prevention and treatment and
as molecular targets for natural product anticancer agents. Curr
Cancer Drug Targets. 13:519–541. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suzuki HI, Katsura A, Matsuyama H and
Miyazono K: MicroRNA regulons in tumor microenvironment. Oncogene.
34:3085–3094. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berindan-Neagoe I, Pdel C Monroig,
Pasculli B and Calin GA: MicroRNAome genome: a treasure for cancer
diagnosis and therapy. CA Cancer J Clin. 64:311–336. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hermeking H: MicroRNAs in the p53 network:
micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Giordano S and Columbano A: MicroRNAs: new
tools for diagnosis, prognosis and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weisenberger DJ: Characterizing DNA
methylation alterations from the cancer Genome Atlas. J Clin
Invest. 124:17–23. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xue B and He L: An expanding universe of
the non-coding genome in cancer biology. Carcinogenesis.
35:1209–1216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ballestrero A, Garuti A, Cirmena G, Rocco
I, Palermo C, Nencioni A, Scabini S, Zoppoli G, Parodi S and
Patrone F: Patient-tailored treatments with anti-EGFR monoclonal
antibodies in advanced colorectal cancer: KRAS and beyond. Curr
Cancer Drug Targets. 12:316–328. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kent OA, McCall MN, Cornish TC and
Halushka MK: Lessons from miR-143/145: the importance of cell-type
localization of miRNAs. Nucleic Acids Res. 42:7528–7538. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Segditsas S and Tomlinson I: Colorectal
cancer and genetic alterations in the Wnt pathway. Oncogene.
25:7531–7537. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Zhang P, Wang F, Zhang H, Yang Y,
Shi C, Xia Y, Peng J, Liu W, Yang Z and Qin H: Elevated oncofoetal
miR-17-5p expression regulates colorectal cancer progression by
repressing its target gene P130. Nat Commun. 3:12912012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Strillacci A, Valerii MC, Sansone P,
Caggiano C, Sgromo A, Vittori L, Fiorentino M, Poggioli G, Rizzello
F, Campieri M and Spisni E: Loss of miR-101 expression promotes
Wnt/β-catenin signalling pathway activation and malignancy in colon
cancer cells. J Pathol. 229:379–389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vermeulen L, De Sousa E Melo F, van der
Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M,
Merz C, Rodermond H, et al: Wnt activity defines colon cancer stem
cells and is regulated by the microenvironment. Nat Cell Biol.
12:468–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Le Grand F, Jones AE, Seale V, Scimè A and
Rudnicki MA: Wnt7a activates the planar cell polarity pathway to
drive the symmetric expansion of satellite stem cells. Cell Stem
Cell. 4:535–547. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hwang WL, Jiang JK, Yang SH, Huang TS, Lan
HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW and Yang MH:
MicroRNA-146a directs the symmetric division of Snail-dominant
colorectal cancer stem cells. Nat Cell Biol. 16:268–280. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Carstens JL, Lovisa S and Kalluri R:
Microenvironment-dependent cues trigger miRNA-regulated feedback
loop to facilitate the EMT/MET switch. J Clin Invest.
124:1458–1460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang JX, Mai SJ, Huang XX, Wang FW, Liao
YJ, Lin MC, Kung HF, Zeng YX and Xie D: MiR-29c mediates
epithelial-to-mesenchymal transition in human colorectal carcinoma
metastasis via PTP4A and GNA13 regulation of β-catenin signaling.
Ann Oncol. 25:2196–2204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen X, Guo X, Zhang H, Xiang Y, Chen J,
Yin Y, Cai X, Wang K, Wang G, Ba Y, et al: Role of miR-143
targeting KRAS in colorectal tumorigenesis. Oncogene. 28:1385–1392.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pagliuca A, Valvo C, Fabrizi E, di Martino
S, Biffoni M, Runci D, Forte S, De Maria R and Ricci-Vitiani L:
Analysis of the combined action of miR-143 and miR-145 on oncogenic
pathways in colorectal cancer cells reveals a coordinate program of
gene repression. Oncogene. 32:4806–4813. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tsang WP and Kwok TT: The miR-18a*
microRNA functions as a potential tumor suppressor by targeting on
K-Ras. Carcinogenesis. 30:953–959. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY,
Cui YM, Qi L, Wu P, Jiao HL, Xie YJ, et al: MicroRNA-30b functions
as a tumour suppressor in human colorectal cancer by targeting
KRAS, PIK3CD and BCL2. J Pathol. 232:415–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sebio A, Paré L, Pàez D, Salazar J,
González A, Sala N, del Río E, Martín-Richard M, Tobeña M, Barnadas
A and Baiget M: The LCS6 polymorphism in the binding site of let-7
microRNA to the KRAS 3′-untranslated region: its role in the
efficacy of anti-EGFR-based therapy in metastatic colorectal cancer
patients. Pharmacogenet Genomics. 23:142–147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ota T, Doi K, Fujimoto T, Tanaka Y, Ogawa
M, Matsuzaki H, Kuroki M, Miyamoto S, Shirasawa S and Tsunoda T:
KRAS up-regulates the expression of miR-181a, miR-200c and miR-210
in a three-dimensional-specific manner in DLD-1 colorectal cancer
cells. Anticancer Res. 32:2271–2275. 2012.PubMed/NCBI
|
|
42
|
Ciardiello F and Tortora G: EGFR
antagonists in cancer treatment. N Engl J Med. 358:1160–1174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lièvre A, Bachet JB, Boige V, Cayre A, Le
Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al: KRAS
mutations as an independent prognostic factor in patients with
advanced colorectal cancer treated with cetuximab. J Clin Oncol.
26:374–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Manceau G, Imbeaud S, Thiébaut R, Liébaert
F, Fontaine K, Rousseau F, Génin B, Le Corre D, Didelot A, Vincent
M, et al: Hsa-miR-31-3p expression is linked to progression-free
survival in patients with KRAS wild-type metastatic colorectal
cancer treated with anti-EGFR therapy. Clin Cancer Res.
20:3338–3347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Saridaki Z, Weidhaas JB, Lenz HJ,
Laurent-Puig P, Jacobs B, De Schutter J, De Roock W, Salzman DW,
Zhang W, Yang D, et al: A let-7 microRNA-binding site polymorphism
in KRAS predicts improved outcome in patients with metastatic
colorectal cancer treated with salvage cetuximab/panitumumab
monotherapy. Clin Cancer Res. 20:4499–4510. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pichler M, Winter E, Ress AL, Bauernhofer
T, Gerger A, Kiesslich T, Lax S, Samonigg H and Hoefler G: MiR-181a
is associated with poor clinical outcome in patients with
colorectal cancer treated with EGFR inhibitor. J Clin Pathol.
67:198–203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pichler M, Winter E, Stotz M, Eberhard K,
Samonigg H, Lax S and Hoefler G: Down-regulation of
KRAS-interacting miRNA-143 predicts poor prognosis but not response
to EGFR-targeted agents in colorectal cancer. Br J Cancer.
106:1826–1832. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fabbri M and Calin GA: Epigenetics and
miRNAs in human cancer. Adv Genet. 70:87–99. 2010.PubMed/NCBI
|
|
49
|
Toyota M, Suzuki H, Sasaki Y, Maruyama R,
Imai K, Shinomura Y and Tokino T: Epigenetic silencing of
microRNA-34b/c and B-cell translocation gene 4 is associated with
CpG island methylation in colorectal cancer. Cancer Res.
68:4123–4132. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vinci S, Gelmini S, Mancini I, Malentacchi
F, Pazzagli M, Beltrami C, Pinzani P and Orlando C: Genetic and
epigenetic factors in regulation of microRNA in colorectal cancers.
Methods. 59:138–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lujambio A, Ropero S, Ballestar E, Fraga
MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A,
Sanchez-Cespedes M, Git A, et al: Genetic unmasking of an
epigenetically silenced microRNA in human cancer cells. Cancer Res.
67:1424–1429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, Prosper F and Garcia-Foncillas J:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen WS, Leung CM, Pan HW, Hu LY, Li SC,
Ho MR and Tsai KW: Silencing of miR-1-1 and miR-133a-2 cluster
expression by DNA hypermethylation in colorectal cancer. Oncol Rep.
28:1069–1076. 2012.PubMed/NCBI
|
|
54
|
Suzuki H, Takatsuka S, Akashi H, Yamamoto
E, Nojima M, Maruyama R, Kai M, Yamano HO, Sasaki Y, Tokino T, et
al: Genome-wide profiling of chromatin signatures reveals
epigenetic regulation of MicroRNA genes in colorectal cancer.
Cancer Res. 71:5646–5658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Deng G, Kakar S and Kim YS: MicroRNA-124a
and microRNA-34b/c are frequently methylated in all histological
types of colorectal cancer and polyps and in the adjacent normal
mucosa. Oncol Lett. 2:175–180. 2011.PubMed/NCBI
|
|
56
|
Ueda Y, Ando T, Nanjo S, Ushijima T and
Sugiyama T: DNA methylation of microRNA-124a is a potential risk
marker of colitis-associated cancer in patients with ulcerative
colitis. Dig Dis Sci. 59:2444–2451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Grady WM, Parkin RK, Mitchell PS, Lee JH,
Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz
AM, et al: Epigenetic silencing of the intronic microRNA
hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene.
27:3880–3888. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Balaguer F, Link A, Lozano JJ, Cuatrecasas
M, Nagasaka T, Boland CR and Goel A: Epigenetic silencing of
miR-137 is an early event in colorectal carcinogenesis. Cancer Res.
70:6609–6618. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yan H, Choi AJ, Lee BH and Ting AH:
Identification and functional analysis of epigenetically silenced
microRNAs in colorectal cancer cells. PLoS One. 6:e206282011.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Humphreys KJ, Cobiac L, Le Leu RK, Van der
Hoek MB and Michael MZ: Histone deacetylase inhibition in
colorectal cancer cells reveals competing roles for members of the
oncogenic miR-17–92 cluster. Mol Carcinog. 52:459–474. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Siemens H, Neumann J, Jackstadt R,
Mansmann U, Horst D, Kirchner T and Hermeking H: Detection of
miR-34a promoter methylation in combination with elevated
expression of c-Met and β-catenin predicts distant metastasis of
colon cancer. Clin Cancer Res. 19:710–720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Takahashi Y, Iwaya T, Sawada G, Kurashige
J, Matsumura T, Uchi R, Ueo H, Takano Y, Eguchi H, Sudo T, et al:
Up-regulation of NEK2 by microRNA-128 methylation is associated
with poor prognosis in colorectal cancer. Ann Surg Oncol.
21:205–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun J, Song Y, Wang Z, Wang G, Gao P, Chen
X, Gao Z and Xu H: Clinical significance of promoter region
hypermethylation of microRNA-148a in gastrointestinal cancers. Onco
Targets Ther. 7:853–863. 2014.PubMed/NCBI
|
|
64
|
Cho WC: Epigenetic alteration of microRNAs
in feces of colorectal cancer and its clinical significance. Expert
Rev Mol Diagn. 11:691–694. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang Y, Wang X, Xu B, Wang B, Wang Z,
Liang Y, Zhou J, Hu J and Jiang B: Epigenetic silencing of miR-126
contributes to tumor invasion and angiogenesis in colorectal
cancer. Oncol Rep. 30:1976–1984. 2013.PubMed/NCBI
|
|
66
|
Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z,
Chen Z, Qiu F, Xu J and Huang J: miRNA-27b targets vascular
endothelial growth factor C to inhibit tumor progression and
angiogenesis in colorectal cancer. PLoS One. 8:e606872013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang F, Ma YL, Zhang P, Shen TY, Shi CZ,
Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y and Qin HL: SP1
mediates the link between methylation of the tumour suppressor
miR-149 and outcome in colorectal cancer. J Pathol. 229:12–24.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Menigatti M, Staiano T, Manser CN,
Bauerfeind P, Komljenovic A, Robinson M, Jiricny J, Buffoli F and
Marra G: Epigenetic silencing of monoallelically methylated miRNA
loci in precancerous colorectal lesions. Oncogenesis. 2:e562013.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Meng X, Wu J, Pan C, Wang H, Ying X, Zhou
Y, Yu H, Zuo Y, Pan Z, Liu RY and Huang W: Genetic and epigenetic
down-regulation of microRNA-212 promotes colorectal tumor
metastasis via dysregulation of MnSOD. Gastroenterology.
145:426–436.e1-e6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tang JT, Wang JL, Du W, Hong J, Zhao SL,
Wang YC, Xiong H, Chen HM and Fang JY: MicroRNA 345, a
methylation-sensitive microRNA is involved in cell proliferation
and invasion in human colorectal cancer. Carcinogenesis.
32:1207–1215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tanaka T, Arai M, Wu S, Kanda T, Miyauchi
H, Imazeki F, Matsubara H and Yokosuka O: Epigenetic silencing of
microRNA-373 plays an important role in regulating cell
proliferation in colon cancer. Oncol Rep. 26:1329–1335.
2011.PubMed/NCBI
|
|
73
|
Hinoue T, Weisenberger DJ, Lange CP, Shen
H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk
CM, et al: Genome-scale analysis of aberrant DNA methylation in
colorectal cancer. Genome Res. 22:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Weber B, Stresemann C, Brueckner B and
Lyko F: Methylation of human microRNA genes in normal and
neoplastic cells. Cell Cycle. 6:1001–1005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Christman JK: 5-Azacytidine and
5-aza-2′-deoxycytidine as inhibitors of DNA methylation:
mechanistic studies and their implications for cancer therapy.
Oncogene. 21:5483–5495. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ceppi P, Mudduluru G, Kumarswamy R, Rapa
I, Scagliotti GV, Papotti M and Allgayer H: Loss of miR-200c
expression induces an aggressive, invasive and chemoresistant
phenotype in non-small cell lung cancer. Mol Cancer Res.
8:1207–1216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Vrba L, Jensen TJ, Garbe JC, Heimark RL,
Cress AE, Dickinson S, Stampfer MR and Futscher BW: Role for DNA
methylation in the regulation of miR-200c and miR-141 expression in
normal and cancer cells. PLoS One. 5:e86972010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt
L, Ramanathan R, Hansen TB, Villadsen SB, Gao S, Ostenfeld MS,
Borre M, et al: Coordinated epigenetic repression of the miR-200
family and miR-205 in invasive bladder cancer. Int J Cancer.
128:1327–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Davalos V, Moutinho C, Villanueva A, Boque
R, Silva P, Carneiro F and Esteller M: Dynamic epigenetic
regulation of the microRNA-200 family mediates epithelial and
mesenchymal transitions in human tumorigenesis. Oncogene.
31:2062–2074. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hartnett L and Egan LJ: Inflammation, DNA
methylation and colitis-associated cancer. Carcinogenesis.
33:723–731. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Valeri N, Braconi C, Gasparini P, Murgia
C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat
F, et al: MicroRNA-135b promotes cancer progression by acting as a
downstream effector of oncogenic pathways in colon cancer. Cancer
Cell. 25:469–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu CW, Ng SC, Dong Y, Tian L, Ng SS, Leung
WW, Law WT, Yau TO, Chan FK, Sung JJ and Yu J: Identification of
microRNA-135b in stool as a potential noninvasive biomarker for
colorectal cancer and adenoma. Clin Cancer Res. 20:2994–3002. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kanaan Z, Rai SN, Eichenberger MR, Barnes
C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE,
et al: Differential microRNA expression tracks neoplastic
progression in inflammatory bowel disease-associated colorectal
cancer. Hum Mutat. 33:551–560. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Koukos G, Polytarchou C, Kaplan JL,
Morley-Fletcher A, Gras-Miralles B, Kokkotou E, Baril-Dore M,
Pothoulakis C, Winter HS and Iliopoulos D: MicroRNA-124 regulates
STAT3 expression and is down-regulated in colon tissues of
pediatric patients with ulcerative colitis. Gastroenterology.
145:842–852 e2. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang F, Zhang P, Ma Y, Yang J, Moyer MP,
Shi C, Peng J and Qin H: NIRF is frequently upregulated in
colorectal cancer and its oncogenicity can be suppressed by let-7a
microRNA. Cancer Lett. 314:223–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Brueckner B, Stresemann C, Kuner R, Mund
C, Musch T, Meister M, Sültmann H and Lyko F: The human let-7a-3
locus contains an epigenetically regulated microRNA gene with
oncogenic function. Cancer Res. 67:1419–1423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hu S, Dong TS, Dalal SR, Wu F, Bissonnette
M, Kwon JH and Chang EB: The microbe-derived short chain fatty acid
butyrate targets miRNA-dependent p21 gene expression in human colon
cancer. PLoS One. 6:e162212011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Shin S, Lee EM, Cha HJ, Bae S, Jung JH,
Lee SM, Yoon Y, Lee H, Kim S, Kim H, et al: MicroRNAs that respond
to histone deacetylase inhibitor SAHA and p53 in HCT116 human colon
carcinoma cells. Int J Oncol. 35:1343–1352. 2009.PubMed/NCBI
|
|
91
|
Falkenberg KJ and Johnstone RW: Histone
deacetylases and their inhibitors in cancer, neurological diseases
and immune disorders. Nat Rev Drug Discov. 13:673–691. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Rada-Iglesias A, Enroth S, Ameur A, Koch
CM, Clelland GK, Respuela-Alonso P, Wilcox S, Dovey OM, Ellis PD,
Langford CF, et al: Butyrate mediates decrease of histone
acetylation centered on transcription start sites and
down-regulation of associated genes. Genome Res. 17:708–719. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Jeltsch A: Beyond Watson and Crick: DNA
methylation and molecular enzymology of DNA methyltransferases.
Chembiochem. 3:274–293. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Borralho PM, Kren BT, Castro RE, da Silva
IB, Steer CJ and Rodrigues CM: MicroRNA-143 reduces viability and
increases sensitivity to 5-fluorouracil in HCT116 human colorectal
cancer cells. FEBS J. 276:6689–6700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang Y, Wang Z, Chen M, Peng L, Wang X,
Ma Q, Ma F and Jiang B: MicroRNA-143 targets MACC1 to inhibit cell
invasion and migration in colorectal cancer. Mol Cancer. 11:232012.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gregersen LH, Jacobsen A, Frankel LB, Wen
J, Krogh A and Lund AH: MicroRNA-143 down-regulates Hexokinase 2 in
colon cancer cells. BMC Cancer. 12:2322012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Qian X, Yu J, Yin Y, He J, Wang L, Li Q,
Zhang LQ, Li CY, Shi ZM, Xu Q, et al: MicroRNA-143 inhibits tumor
growth and angiogenesis and sensitizes chemosensitivity to
oxaliplatin in colorectal cancers. Cell Cycle. 12:1385–1394. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li
JJ, Röcken C, Ebert MP, Kwok TT and Sung JJ: MicroRNA-143 targets
DNA methyltransferases 3A in colorectal cancer. Br J Cancer.
101:699–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu
R, Pan Z, Kang T and Huang W: MicroRNA-342 inhibits colorectal
cancer cell proliferation and invasion by directly targeting DNA
methyltransferase 1. Carcinogenesis. 32:1033–1042. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yeung F, Hoberg JE, Ramsey CS, Keller MD,
Jones DR, Frye RA and Mayo MW: Modulation of NF-kappaB-dependent
transcription and cell survival by the SIRT1 deacetylase. EMBO J.
23:2369–2380. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Huffman DM, Grizzle WE, Bamman MM, Kim JS,
Eltoum IA, Elgavish A and Nagy TR: SIRT1 is significantly elevated
in mouse and human prostate cancer. Cancer Res. 67:6612–6618. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kriegl L, Vieth M, Kirchner T and Menssen
A: Up-regulation of c-MYC and SIRT1 expression correlates with
malignant transformation in the serrated route to colorectal
cancer. Oncotarget. 3:1182–1193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Kaller M, Liffers ST, Oeljeklaus S,
Kuhlmann K, Röh S, Hoffmann R, Warscheid B and Hermeking H:
Genome-wide characterization of miR-34a induced changes in protein
and mRNA expression by a combined pulsed SILAC and microarray
analysis. Mol Cell Proteomics. 10:M111.010462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Padi SK, Zhang Q, Rustum YM, Morrison C
and Guo B: MicroRNA-627 mediates the epigenetic mechanisms of
vitamin D to suppress proliferation of human colorectal cancer
cells and growth of xenograft tumors in mice. Gastroenterology.
145:437–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Dou L, Zheng D, Li J, Li Y, Gao L, Wang L
and Yu L: Methylation-mediated repression of microRNA-143 enhances
MLL-AF4 oncogene expression. Oncogene. 31:507–517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing
C and Liu Z: Altered expression of MiR-148a and MiR-152 in
gastrointestinal cancers and its clinical significance. J
Gastrointest Surg. 14:1170–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Pavicic W, Perkiö E, Kaur S and Peltomäki
P: Altered methylation at microRNA-associated CpG islands in
hereditary and sporadic carcinomas: a methylation-specific
multiplex ligation-dependent probe amplification (MS-MLPA)-based
approach. Mol Med. 17:726–735. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Takahashi M, Cuatrecasas M, Balaguer F,
Hur K, Toiyama Y, Castells A, Boland CR and Goel A: The clinical
significance of MiR-148a as a predictive biomarker in patients with
advanced colorectal cancer. PLoS One. 7:e466842012. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhu A, Xia J, Zuo J, Jin S, Zhou H, Yao L,
Huang H and Han Z: MicroRNA-148a is silenced by hypermethylation
and interacts with DNA methyltransferase 1 in gastric cancer. Med
Oncol. 29:2701–2709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y,
Qi YT, Xu Q, Li W, Lu B, et al: A regulatory circuit of
miR-148a/152 and DNMT1 in modulating cell transformation and tumor
angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 5:3–13.
2013. View Article : Google Scholar : PubMed/NCBI
|