Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed cancer and the second leading cause of cancer-associated
mortality in the United States (1).
In China, CRC is the fifth leading cause of cancer-associated
mortality, following lung, liver, gastric and esophageal cancer
(2). The incidence of CRC in China
has increased in recent decades and is predicted to continue rising
due to changes in lifestyle and diet (3,4). Although
novel treatments have been developed, the five-year survival rate
of patients with CRC and distant metastases remains poor at ~13%
(5–7).
CRC develops from the accumulation of genetic and epigenetic
alterations, leading to gene amplifications, the activation of
certain oncogenes or the loss of tumor suppressor genes (6).
Previous studies have demonstrated that the
activation of endoplasmic reticulum (ER) stress in cancer cells may
facilitate their survival and tumor growth; however, certain
studies have revealed that ER stress may inhibit cancer progression
(8,9).
During ER stress, various pathological changes occur to induce ER
calcium depletion and the accumulation of misfolded proteins in the
ER lumen (10). Mammalian cells have
three classes of ER stress sensors, including
protein-kinase-RNA-like-ER kinase (PERK), activating transcription
factor 6 (ATF6) and inositol-requiring enzyme-1α (IRE1α) (10–12). These
sensors are resident ER transmembrane proteins, which regulate the
unfolded protein response (UPR) to manage ER stress (11). The UPR includes the alteration of
protein folding, assembly and degradation programs in order to
reestablish homeostasis and normal ER functioning (11,13).
IRE1α has a dual enzyme activity, as it is a kinase
and a site-specific RNA endonuclease (8,14). IRE1α
is frequently mutated in various types of human cancer (15). One manner in which IRE1α maintains ER
homeostasis is by processing the mRNA encoding X-box binding
protein 1 (XBP1) (16). IRE1α
activates XBP1 protein expression by excising a
26-nucleotide-intron sequence from the un-spliced XBP1 mRNA (XBP1u)
and creating the spliced XBP1 mRNA (XBP1s). The subsequent frame
shift mutation eliminates a stop codon for protein translation
(17,18). XBP1 is a transcription factor that is
involved in tumor growth and survival (9,17). XBP1
expression levels are increased in numerous types of cancer,
including breast cancer (19),
hepatocellular carcinoma (20),
pancreatic cancer (17), and CRC, as
determined in a study of five patients (21); however, XBP1 expression levels may be
decreased in prostate cancer (22).
IRE1α may also induce UPR through the post-transcriptional
modifications of specific ER membrane proteins via regulated
IRE1-dependent decay (RIDD) (8). RIDD
is the process by which IRE1α promotes the degradation of mRNAs
primarily encoding ER-targeted proteins, in order to reduce the
influx of proteins during ER stress (23,24). In
the present study, IRE1α and XBP1 mRNA expression levels in CRC
tissues were analyzed to determine whether they are increased,
compared with the paired control samples.
IRE1β is an analog of IRE1α (25). Whereas IRE1α is expressed
ubiquitously, the expression of IRE1β is restricted to the
epithelium of the gastrointestinal and respiratory tracts (26,27).
Although previous studies have demonstrated that IRE1α and IRE1β
are each able to sense ER stress and protect mice from dextran
sulfate sodium-induced colitis (26,28), they
have differing functions. The two IRE1 proteins have various
substrate specificities; the RNase activity of IRE1α with regard to
XBP1u mRNA is markedly high, compared with IRE1β (27). IRE1α directs cell survival through the
induction of XBP1 mRNA cleavage and the promotion of RIDD (28). IRE1α signaling terminates in the event
of cell apoptosis induced by irremediable ER stress (29–31). IRE1β
is more efficient than IRE1α at degrading 28 s rRNA (16). The cleavage of 28s rRNA may induce
apoptosis, as previously demonstrated in IRE1β-overexpressing HeLa
cells (32). The RNase activity of
IRE1β appears to have broad substrate specificity; it regulates the
stability of the mRNA that encodes certain ER proteins and
maintains ER homeostasis in highly differentiated secretory cells
(16). Thus, IRE1β, but not IRE1α,
degrades the mRNA encoding specific secretory proteins, including
mucin 2 (MUC2) in the intestinal goblet cells (16,33). MUC2
is a macromolecular glycoprotein secreted by goblet cells (34). MUC2 is crucial to host immune system
and protects colon tissues from developing colitis or CRC;
MUC2-deficient mice have been observed to develop spontaneous
colitis and colon cancer (35–38). The
differences between the substrates of IRE1α and IRE1β suggest their
divergent functions in ER stress, and may also reflect their
various roles in the tumorigenesis of CRC (29). Therefore, IRE1β and MUC2 gene
expression profiles in CRC tissue samples were analyzed in the
current study.
In the present study, the expression levels of the
signaling pathways IRE1α-XBP1, IRE1β and MUC2 in colon cancer
tissues were investigated by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR), western blotting and
immunohistochemistry, and their associations with the clinical
features of CRC patients were explored. This study may identify
potential important targets for cancer therapies.
Materials and methods
Patients and tissue samples
The CRC tissue samples were obtained from surgically
resected tumor tissues from patients with colorectal adenocarcinoma
at the First Affiliated Hospital of Henan University of Science and
Technology, between September 1st, 2013 and February 31st, 2014.
The clinicopathological features of the patients recruited for the
current study are listed in Table I.
Tumor tissues and adjacent non-cancerous tissues (serving as
controls) were analyzed. The control non-cancerous tissues were
taken from an area ~5 cm from the lesion. Patients with colorectal
adenocarcinoma were selected, while those patients with mucinous,
signet-ring cell carcinoma, squamous carcinoma, adenosquamous
carcinoma or undifferentiated carcinoma forms of CRC were excluded.
Mucinous CRC was excluded due to its high level of MUC2 expression
compared with that in normal colon tissues (36,39,40).
 | Table I.Clinicopathological characteristics
of the included patients with colorectal carcinoma (n=42). |
Table I.
Clinicopathological characteristics
of the included patients with colorectal carcinoma (n=42).
| Variable | Number of cases
(%) |
|---|
| Gender |
|
|
Male | 18 (42.9) |
|
Female | 24 (57.1) |
| Age (years) |
|
|
<60 | 18 (42.9) |
|
≥60 | 24 (57.1) |
| Tumor size
(cm) |
|
|
<5 | 11 (26.2) |
| ≥5 | 28 (66.7) |
| Data
incomplete | 3 (7.1) |
|
Differentiation |
|
|
WMDC | 11 (26.2) |
|
MDC | 27 (77.1) |
|
PMDC | 4 (9.5) |
| Lymphatic node
metastasis |
|
|
Negative | 15 (35.7) |
|
Positive | 27 (64.3) |
| TNM stage |
|
|
I–II | 24 (57.1) |
|
III–IV | 18 (42.9) |
Two staff pathologists confirmed the diagnosis of
CRC. A section of each tissue sample was fixed in 4%
paraformaldehyde and embedded in paraffin wax for hematoxylin and
eosin staining and immunohistochemistry (IHC). The remaining
section of the tissue sample was stored at −80°C for RNA extraction
and western blot analysis. RNAlater (Qiagen GmbH, Hilden,
Germany; cat. no. 76106) was added immediately following the tissue
sample collection in order to prevent RNA degradation. The tumor
stages were classified according to the 7th edition of the
tumor-node-metastasis (TNM) classification criteria of the American
Joint Committee on Cancer (41).
Informed consent was obtained from all patients and the Clinical
Research Ethics Committee of The First Affiliated Hospital of Henan
University of Science and Technology approved the current
study.
RT-qPCR
Total RNA was extracted using TRIzol®
Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's instructions. A total of 2 µg
total RNA was used for cDNA synthesis using PrimeScript™ RT Master
Mix (Takara Bio, Inc., Otsu, Japan) in a 40 µl reaction mixture (8
µl 5X RT Master Mix; total RNA; diethylpyrocarbonate), as follows:
37°C for 15 min, 85°C for 5 sec and 4°C for 10 min. The primer
sequences for IRE1α, XBP1u, XBP1s, IRE1β, MUC2 and β-actin were
designed using Primer3.0 software (42) and synthesized by Sangon Biotech Co.,
Ltd. (Shanghai, China; Table II).
RT-qPCR was conducted using a CFX96™ Real-Time PCR system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The reaction mixture (25 µl
total volume per well) included 2 µl cDNA, 12.5 µl 2xSYBR Premix Ex
Taq II (Takara Bio, Inc.), 8.5 µl H2O and 2 µl
0.4 µM primers. A two-step method was used due to the 60°C
annealing temperature. The reaction consisted of the following:
95°C for 30 sec, 40 cycles of 95°C for 5 sec and 60°C for 30 sec.
Each tissue sample was assayed in triplicate. The efficiency of the
PCR amplification process was 97–105%. A melting curve analysis was
performed for the PCR products of the target genes in order to
evaluate primer specificity. Relative quantification of the target
gene mRNA expression was conducted using quantification cycle (Cq)
with the formula log102−ΔΔCq (43) and normalized to β-actin. The
difference in mRNA expression was presented as the relative fold
between the groups. A Cq value of >35 was considered to indicate
that a specific gene was not expressed.
 | Table II.Primers sequences for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primers sequences for reverse
transcription-quantitative polymerase chain reaction.
| mRNA | Gene | Primer sequence
(5′-3′) | Amplicon (bp) |
|---|
| NM-001433 | IRE1α | Forward
CTCCGAGCCATGAGAAATAAG | 113 |
|
|
| Reverse
GGGAAGCGAGATGTGAAGTAG |
|
| NM-001079539 | XBP1s | Forward
AAGTGGTAGATTTAGAAGAAGAGAA | 200 |
|
|
| Reverse
ACCTGCTGCGGACTCAG |
|
| NM-005080 | XBP1u | Forward
AGTCCGCAGCACTCAG | 150 |
|
|
| Reverse
GGGTCCAAGTTGTCCAGA |
|
| NM-033266 | IRE1β | Forward
TCCCCTTATAGGACCGGAAC | 147 |
|
|
| Reverse
GTGACTGGCTGGAGAAGGAG |
|
| NM-002457 | MUC2 | Forward
GACACCATCTACCTCACCCG | 103 |
|
|
| Reverse
TGTAGGCATCGCTCTTCTCA |
|
| NM-00110 | β-actin | Forward
AGCACTGTGTTGGCGTACAG | 116 |
|
|
| Reverse
CTCTTCCAGCCTTCCTTCCT |
|
Immunohistological staining for IRE1α,
IRE1β and MUC2
Sections (4 µm) of paraffin-embedded tissue samples
were mounted on poly-L-lysine-coated slides. IHC was performed
using an indirect peroxidase-labelled antibody method as previously
described (44). Briefly, the tissue
sections were dewaxed in xylene, rehydrated with graded alcohol and
antigen retrieval was conducted by microwave-boiling the slides for
10 min in 0.01 mol/l sodium citrate buffer, pH 6.0. Endogenous
peroxidase was blocked by incubation in 3% hydrogen peroxide for 10
min. Following washing in distilled water, non-specific binding was
blocked by 5% bovine serum albumin (Sigma-Aldrich: Merck Millipore,
Darmstadt, Germany) for 20 min at 37°C. The tissue sections were
then incubated overnight at 4°C with rabbit polyclonal antibodies
against IRE1α (Abcam, Cambridge, UK; dilution, 1:200; cat. no.
ab37073) and IRE1β (Abcam; dilution 1:50; cat. no. ab135795), and
an anti-MUC2 mouse monoclonal antibody (Abcam; dilution, 1:500;
cat. no. ab11197). The antigen-antibody complex was then detected
with a biotinylated goat anti-rabbit antibody (Boster Biological
Technology Co., Ltd., Wuhan, China; cat. no. BA1101) and an
anti-mouse antibody (Boster Biological Technology Co., Ltd.; cat.
no. SA1020), subsequently conjugated with streptoavidin-horseradish
peroxidase (HRP) (Boster Biological Technology Co., Ltd.; cat. no.
SA1022) and visualized by reacting with nickel-enhanced
3,3-diaminobenzidine tetrahydrochloride (Solarbio Science and
Technology Co., Ltd.; cat. no. D8230) for color detection. The
tissue sections were then counterstained with hematoxylin. The
negative control sections were obtained by omitting the primary
antibody or by using an unrelated rabbit polyclonal antibody.
The antigen levels in the IHC stained tissue samples
were evaluated in 10 random fields (400x magnification) for each
section. A total of 100 cells/field were categorized as follows: 0,
0–5 cells were stained; 1, 6–25 cells were stained; 2, 26–50 cells
stained; 3, 51–75 cells stained; and 4, 76–100 cells stained. In
addition, the staining intensity was scored as follows: 0, no
staining; 1, weak staining; 2, moderate staining; 3, intense
staining. The intensity score was multiplied by the frequency
score, in order to obtain the final score. A final score of ≥6
indicated high expression levels, whereas a score of <6
indicated low expression levels (45).
Western blotting analysis of
IRE1β
Protein lysates were prepared from collected tissue
samples in radioimmunoprecipitation assay lysis buffer (Solarbio
Science and Technology Co., Ltd.) on ice by homogenization with a
grinder. The supernatant was obtained following centrifugation at
10,800 × g for 15 min at 4°C. A bicinchoninic acid assay
(Solarbio Science and Technology Co., Ltd.,) method was used to
determine the protein concentrations. Protein (30 µg) from each
tissue sample was denatured and resolved by 10% sodium dodecyl
sulfate polyacrylamide gel electrophoresis, and then transferred
onto polyvinylidene difluoride membranes (EMD Millipore, Billerica,
MA, USA). Following blocking for 1 h at 37°C in 5% skim milk, the
membranes were incubated with the anti-IRE1β antibody (Abcam;
dilution 1:200; cat. no. ab135795) for 3 h at 37°C, then washed
four times in 1X TBST. The membranes were subsequently incubated
with HRP-conjugated anti-IgG secondary antibody (Boster Biological
Technology Co., Ltd.; dilution, 1:1,000; cat. no. BA1054) and then
washed four times in 1X TBST. The proteins were visualized using an
enhanced chemiluminescence reagent (Pierce; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. An
anti-β-actin mouse monoclonal antibody (Abcam; dilution, 1:3,000;
cat. no. ab8226) was used to normalize for the protein loading. The
secondary antibody for β-actin was a HRP-conjugated goat anti-mouse
IgG (Boster Biological Technology Co., Ltd.; dilution, 1:1,000;
cat. no. BA1050). ChemiDoc XRS (Bio-Rad Laboratories, Inc.) was
used to capture the images, and the intensity of the images was
quantified using ImageJ software v1.48 (National Institutes of
Health, Bethesda, MD, USA).
Statistical analysis
A Student's t-test and a Mann-Whitney U-test were
used to determined significant differences between the groups. A
Wilcoxon signed-rank test was used for non-parametric data.
Spearman's bivariate analysis was used to determine the correlation
between IRE1β and MUC2 mRNA expression levels. The data are
presented as the mean ± standard deviation. All statistical
analysis was performed using using the SPSS 19.0 statistical
package (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant result.
Results
Clinical characteristics
A total of 42 patients were recruited for the
current study and the clinical features are summarized in Table I. The ages of the patients ranged from
44–82 years (average, 61.3 years). A total of 18 patients were male
and 24 were female. The discrepancy in gender ratio of this study
from the China colorectal report may be due to the gender ratio in
the local area (4,46).
Expression levels of IRE1α, XBP1,
IRE1β and MUC2 mRNA in CRC tissues
XBP1 expression levels are increased in numerous
types of cancer, including CRC (8,17,18). During UPR, IRE1α initiates the
splicing of XBP1u mRNA to XBP1s, generating an XBP1 transcription
factor that regulates a subset of UPR genes to constitute the
IRE1α-XBP1 signaling pathway (8,16). RT-qPCR
was used to analyze the mRNA expression levels of IRE1α, XBP1u and
XBP1s in tissue samples from patients with CRC. The paired colon
tissue samples were analyzed in 31 patients for IRE1α mRNA and 12
patients for XBP1u and XBP1 s mRNAs. It was identified that IRE1α,
XBP1u and XBP1s mRNAs were expressed at similar levels in the CRC
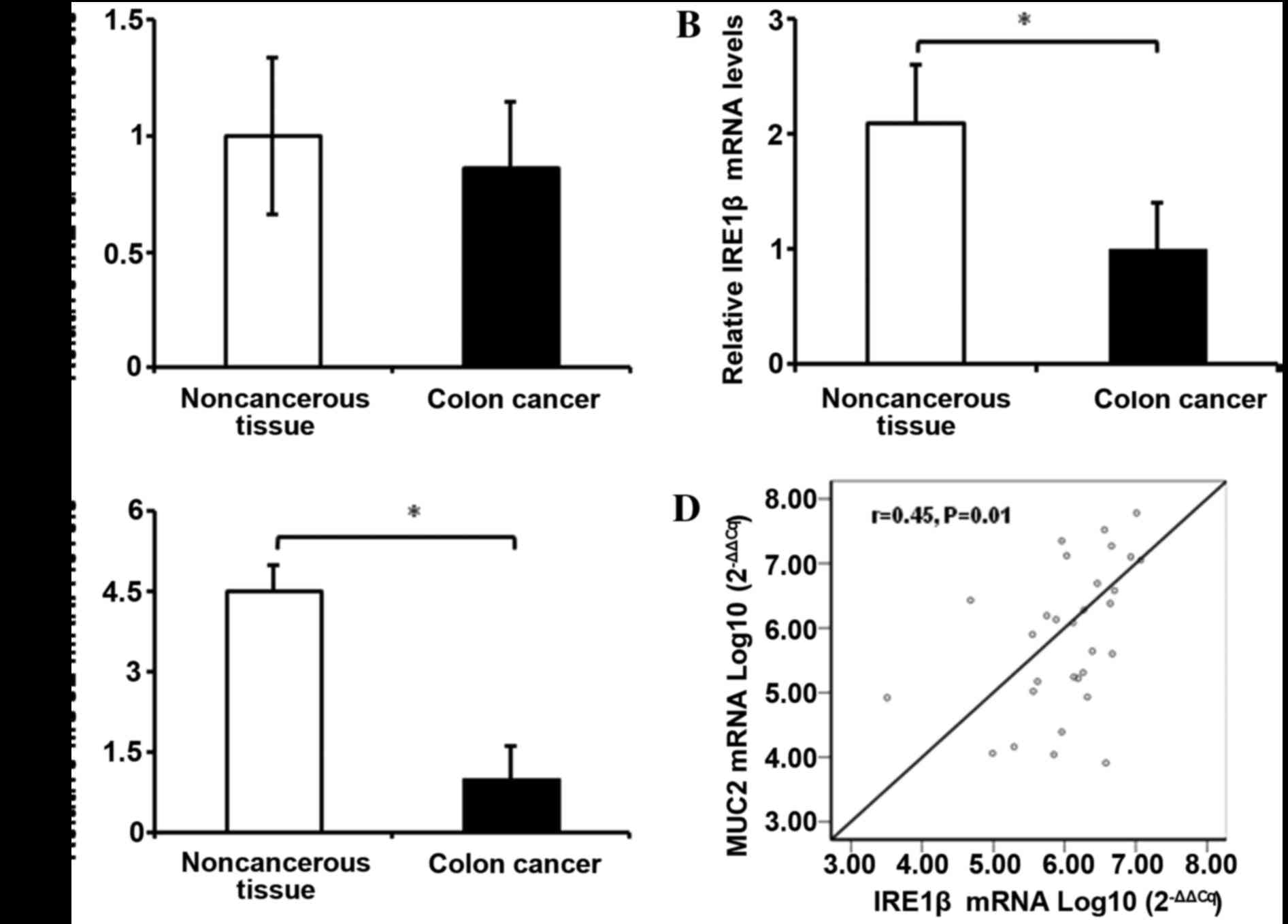
and non-cancerous tissues (Fig. 1A
and Table III).
 | Table III.XBP1u and XBP1s mRNA expression
levels in patients with colorectal cancer. |
Table III.
XBP1u and XBP1s mRNA expression
levels in patients with colorectal cancer.
|
|
|
|
|
| Ratio (C/N) |
|---|
|
|
|
|
|
|
|
|---|
| No. | Patient | Gender | Age (years) |
Differentiation | XBP1u | XBP1s |
|---|
| 1 | C58 | m | 44 | PDC | 2.5 | 0.8 |
| 2 | C57 | f | 65 | MDC | 1.0 | 1.0 |
| 3 | C24 | f | 53 | MDC | 0.8 | 0.8 |
| 4 | C29 | m | 64 | MDC | 1.1 | 1.0 |
| 5 | C31 | m | 61 | MDC | 0.9 | 0.8 |
| 6 | C33 | m | 61 | MDC | 2.0 | 2.5 |
| 7 | C52 | f | 61 | MDC | 1.1 | 0.8 |
| 8 | C53 | f | 60 | MDC | 0.5 | 0.4 |
| 9 | C54 | f | 52 | WMDC | 1.4 | 2.5 |
| 10 | C55 | f | 68 | WDC | 1.1 | 1.3 |
| 11 | C56 | f | 47 | WDC | 5.0 | 5.0 |
| 12 | C59 | f | 47 | WDC | 1.0 | 1.4 |
| Mean ± standard
deviation |
|
|
|
| 1.1±0.5 | 1.0±0.6 |
IRE1β, an analog of IRE1α, is specifically expressed
in the epithelium of the gastrointestinal and respiratory tracts;
MUC2 expression in the intestine is regulated by IRE1β, but not by
IRE1α (27,47). IRE1β and MUC2 mRNA expression levels
were evaluated in 35 patients with CRC. The mRNA expression levels
of IRE1β and MUC2 in the cancerous tissues were 2.1 and 4.5-fold
lower, respectively, compared with in the adjacent non-cancerous
colon mucosa (Fig. 1B and 1C). It was
identified that MUC2 mRNA expression levels were positively
correlated with IRE1β mRNA expression levels (r=0.45; P=0.01;
Fig. 1D).
mRNA expression levels of IRE1β, but
not IRE1α or MUC2, were associated with lower clinical stages,
metastasis and poor differentiation in CRC
To evaluate the clinical significance of changes in
the mRNA expression levels of IRE1β, IRE1α and MUC2 in CRC tissues,
the association between the mRNA expression levels of these genes
and the clinicopathological features of the patients with CRC, were
analyzed. It was identified that IRE1β mRNA expression levels were
significantly associated with tumor differentiation (P=0.049),
lymph node metastasis (P=0.043) and TNM stage (P=0.018) (Table IV), but not with gender (P=0.709),
age (P=0.558) and T-stage classification (P=0.384) (48,49) (data
not presented). Although the mRNA expression levels of IRE1β in the
tumor tissues were low compared with the adjacent normal tissues,
in poor-moderately differentiated CRC tissues the IRE1β mRNA
expression levels were high as compared with in well-differentiated
CRC tissues. Furthermore, those patients with lymphatic metastasis
or stage III–IV CRC, had high IRE1β mRNA expression levels, as
compared with those patients without lymphatic metastasis or stage
I–II CRC. IRE1α and MUC2 mRNA expression levels were not observed
to be significantly associated with patient clinicopathological
features (Table IV).
 | Table IV.IRE1α, IRE1β and MUC2 mRNA expression
levels in patients with CRC. |
Table IV.
IRE1α, IRE1β and MUC2 mRNA expression
levels in patients with CRC.
|
| IRE1α mRNA | IRE1β mRNA | MUC2 mRNA |
|---|
|
|
|
|
|
|---|
| Variables | n | Ratio of C/N | P-value | n | Ratio of C/N | P-value | n | Ratio of C/N | P-value |
|---|
| Total patients | 31 |
|
| 35 |
|
| 35 |
|
|
| Clinical stage |
|
| 0.328 |
|
| 0.018a |
|
| 0.355 |
|
I+II | 17 | 1.0±0.1 |
| 21 | 3.7±0.6 |
| 20 | 1.0±0.4 |
|
|
III+IV | 14 | 1.3±0.2 |
| 14 | 1.0±0.3 |
| 15 | 2.4±1.2 |
|
| LN metastasis |
|
| 0.135 |
|
| 0.043 |
|
| 0.692 |
|
Yes | 12 | 1.5±0.1 |
| 11 | 3.3±0.5 |
| 13 | 1.5±0.7 |
|
| No | 19 | 1.0±0.1 |
| 24 | 1.0±0.3 |
| 22 | 1.0±0.4 |
|
|
Differentiation |
|
| 0.605 |
|
| 0.049a |
|
| 0.519 |
|
Well | 9 | 1.1±0.1 |
| 8 | 1.0±0.3 |
| 10 | 1.0±0.5 |
|
|
Moderate or Poor | 22 | 1.0±0.1 |
| 27 | 4.9±0.9 |
| 25 | 2.0±0.9 |
|
Immunohistochemistry of IRE1α, IRE1β
and MUC2 in CRC tissues
ER stress is emerging as an important factor in
tumor pathogenesis (8,9). However, to the best of our knowledge,
there are no previous reports on the role of IRE1α in the
tumorigenesis of CRC. Although it was demonstrated in the current
study that the mRNA expression levels of IRE1α are similar in CRC
and adjacent normal tissues, as IRE1α regulates ER stress at the
protein level, the IRE1α protein expression levels were also
evaluated using IHC (Fig. 2A and B).
IRE1α was expressed in the plasma membrane of non-cancerous colon
epithelium and IRE1α was stained at apical surface of cancerous
epithelial cells in the colon. In submucosa, certain unidentified
cells also had positive nuclei staining for IRE1α. Again, there was
no significant difference in IRE1α protein expression levels
between CRC and non-cancerous tissue (Fig. 2C).
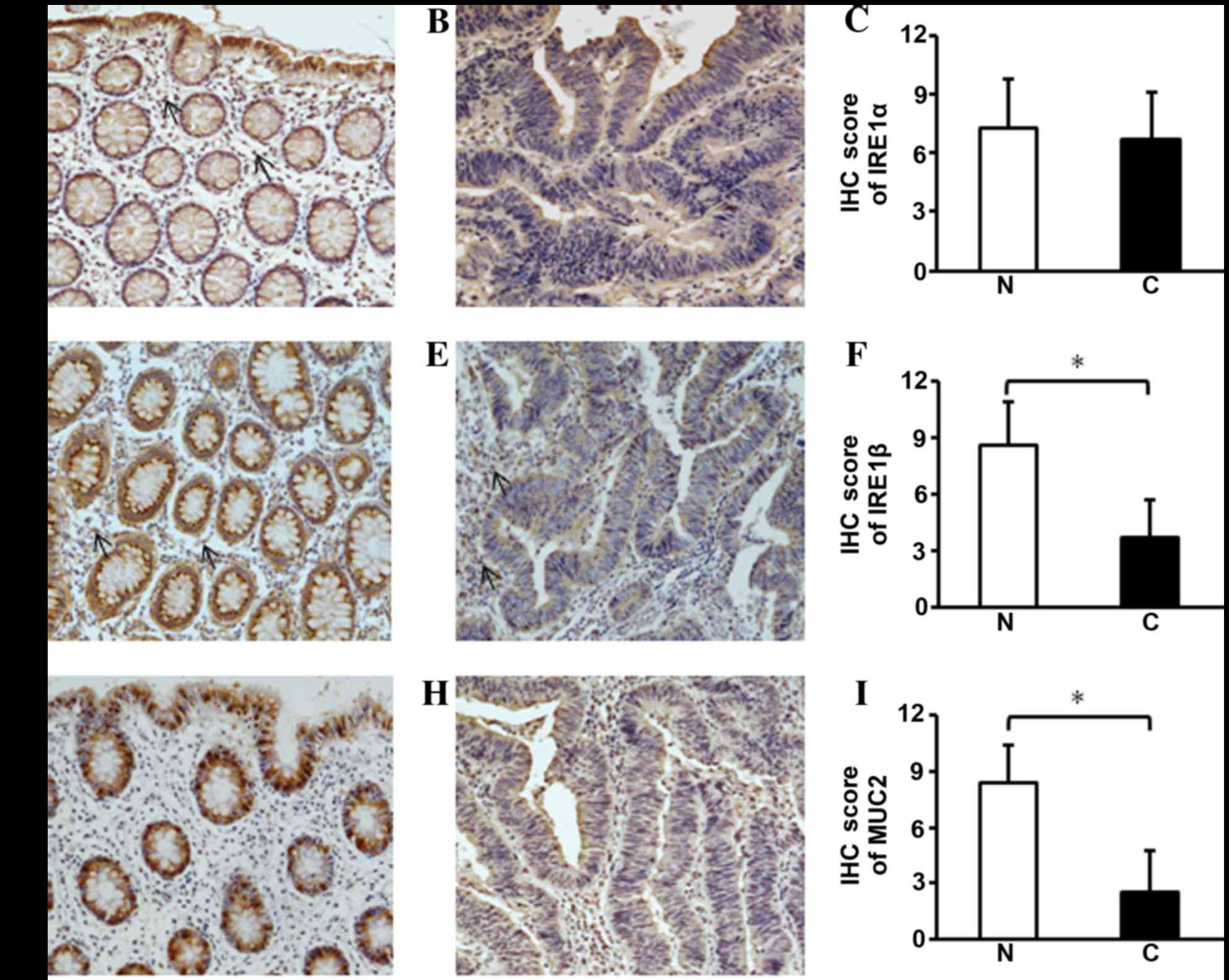 | Figure 2.Immunohistochemistry of IRE1α, IRE1β
and MUC2 in CRC tissues. (A) IRE1α was expressed in the cytoplasm
of epithelial cells in the crypts surface of non-cancerous tissues.
In the submucosa unidentified cells were observed to have weakly
positive staining (arrow). (B and C) In CRC tissues there was a
similar level of the IRE1α protein; (D) IRE1β was observed in the
cytoplasm above the nuclei of the non-cancerous colonic epithelial
cells. (E and F) In the CRC tissues, the cytoplasmic staining for
IRE1β was faint, suggesting the expression of IRE1β was decreased;
in the submucosa of the non-cancerous tissues and cancerous tissues
there were also unidentified cells with weakly positive staining
(arrow). (G) The staining for MUC2 was intensely positive in the
goblet cells of the non-cancerous colonic epithelium; (H and I) in
the CRC tissues MUC2 staining was barely visible. Representative
immunostaining for (A and B) IRE1α, (D and E) IRE1β and (G and H)
MUC2 and the semi-quantification of (C) IRE1α, (F) IRE1β and (I)
MUC2 for each tissue group. The data are presented as the mean ±
standard deviation (n=35 for all three groups). *P<0.05, vs. the
non-cancerous tissues. N, noncancerous tissues; C, cancerous
tissues; IRE1, inositol-requiring enzyme 1; MUC2, mucin 2; CRC,
colorectal cancer; IHC, immunohistochemistry. |
Aberrant mucin accumulation in the goblet cells of
mouse colons was observed when the IRE1β gene was deleted (33), and IRE1β was revealed to be required
for airway mucin excretion (27). In
the current study, it was also identified that the IRE1β protein
was expressed in colon epithelial cells, including in goblet cells.
However, by contrast with a prior animal study (33), the results did not demonstrate a
predominant IRE1β-positive staining in human colon goblet cells.
Above the nuclei of the epithelial cells, a strong positive
staining was observed in non-cancerous tissue samples (Fig. 2D). In the cytoplasm of CRC tissues,
IRE1β positive staining was comparatively low (Fig. 2E and F); in normal and cancerous
tissues, there were unidentified cells that were weakly stained in
the submucosa. Similar to the mRNA expression of IRE1β, the
downregulation of IRE1β was significantly associated with tumor
differentiation (P=0.047), lymph node metastasis (P=0.009) and TNM
stage (P=0.001) (Table V). No
significant association was identified between IRE1β expression
levels and other clinicopathological factors, including gender
(P=0.998), age (P=0.115) and tumor size (P=0.742) (data not
presented).
 | Table V.Association between the
immunohistochemical staining of IRE1β and the clinical
characteristics of patients with colorectal carcinoma (n=35). |
Table V.
Association between the
immunohistochemical staining of IRE1β and the clinical
characteristics of patients with colorectal carcinoma (n=35).
|
|
| IRE1β |
|---|
|
|
|
|
|---|
| Variable | Patients, n | Low, n | High, n | P-value |
|---|
| Clinical stage |
|
|
| 0.001 |
|
I+II | 21 | 17 | 4 |
|
|
III+IV | 14 | 2 | 12 |
|
| Lymph node
metastasis |
|
|
| 0.009 |
|
Yes | 11 | 2 | 9 |
|
|
No1 | 24 | 17 | 7 |
|
| Pathologic
differentiation |
|
|
| 0.047 |
|
Well | 8 | 7 | 1 |
|
|
Moderate or Poor | 27 | 12 | 15 |
|
MUC2 protein is present in the goblet cells of the
colon epithelium (34,50). It has been reported that the IHC
staining of MUC2 in goblet cells exhibits whole-cell filled or
peri-nuclear staining patterns (51,52). In
the present study, the MUC2 staining pattern was half-filled in the
goblet cells, and MUC2 expression levels were significantly
decreased in these CRC tissues (P<0.001; Fig. 2G-I). Similar to MUC2 mRNA expression
levels, the MUC2 protein expression levels, as quantified by IHC
staining, were not significantly associated with the
clinicopathological factors of patients with CRC (data not
presented).
Western blot analysis of IRE1β
expression in CRC tissues
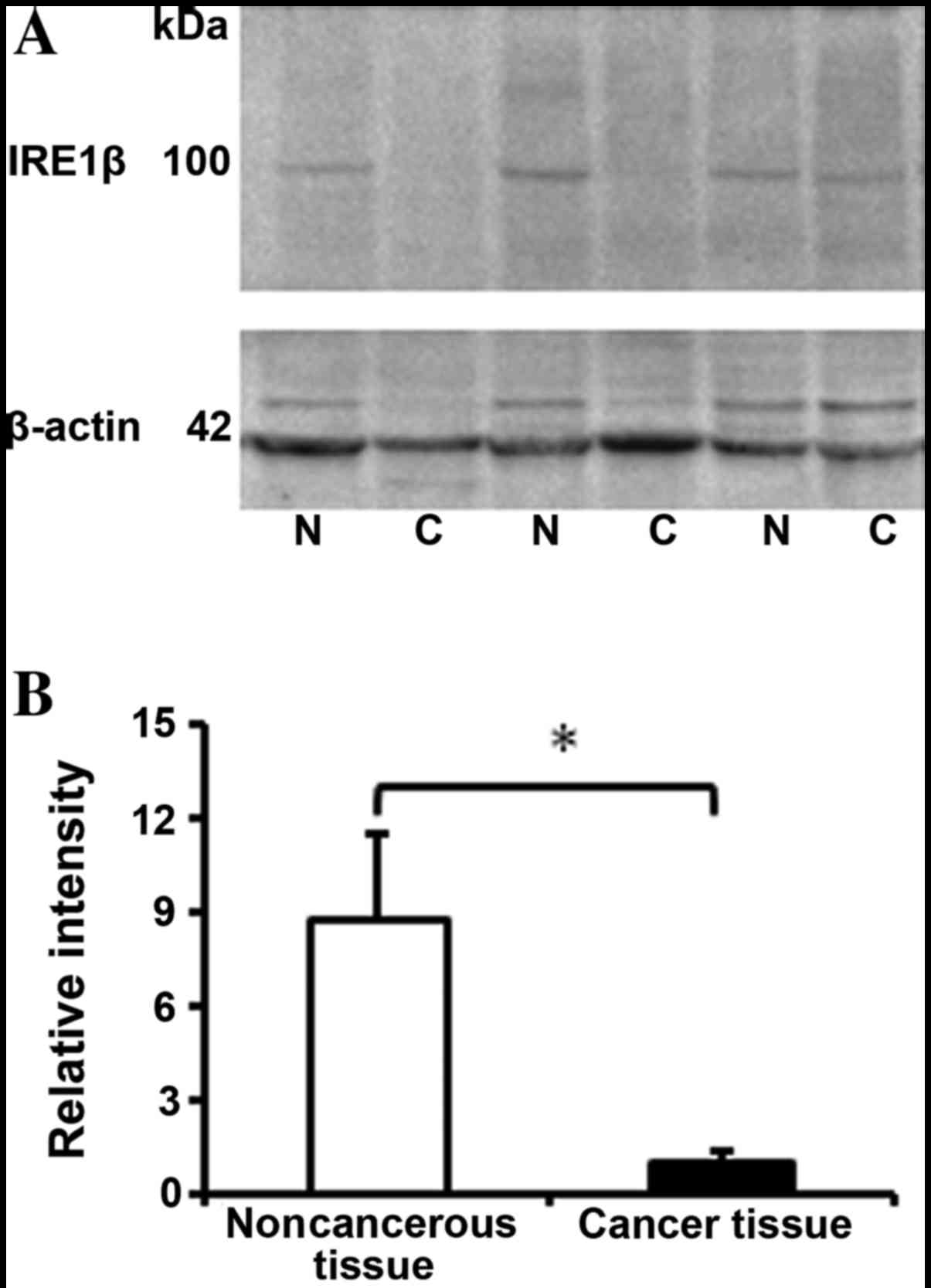
The protein expression levels of IRE1β in 13 paired
CRC and adjacent normal tissue samples were evaluated using western
blotting. Similar to the expression levels of IRE1β in the mRNA and
IHC results, a significant difference was identified in the IRE1β
protein expression levels, which were 8-fold lower in cancerous
tissues, compared with the adjacent normal control tissues
(Fig. 3A and B; P<0.001), which
indicated that the expression level was decreased in CRC tissues at
the transcriptional and translational levels.
Discussion
ER stress affects tumorigenesis and elevation of the
XBP1 transcription factor has previously been reported in numerous
types of cancer, including CRC (9,19,21). As XBP1 mRNA is processed by IRE1α, the
IRE1α, XBP1u and XBP1s mRNA expression levels, and IRE1α protein
expression levels, were analysed in cancerous and adjacent normal
colon tissue samples; however, no significant difference was
observed between the two tissues. Although Fujimoto et al
(21) identified that XBP1 gene and
protein expression levels were increased in 4/5 CRC tissues, the
sample size in their study was small. In the current study, XBP1u
and XBP1s gene expression levels were analyzed in 12 cases of CRC.
Additionally, the IRE1α mRNA and protein expression levels were
evaluated and no significant changes were observed in CRC tissues,
compared with the adjacent non-cancerous tissues. Therefore, the
results suggest that the IRE1α-XBP1 signaling pathway does not have
an important role in the progression of CRC.
Colon epithelial cells also express IRE1β, an analog
of the ubiquitous IRE1α, which has differing functions to IRE1β
with regard to cell survival and apoptosis (29–32,47). IRE1β
is inefficient at cleaving XBP1 mRNA and directly interacts with
unfolded proteins in the ER by association with glucose-regulated
protein 78, which is crucial regulator of ER stress (26). Therefore, the IRE1β expression levels
in patients with CRC were also analyzed. The present study
demonstrated that the mRNA and protein expression levels of IRE1β
were significantly decreased in CRC tissues. It is possible that
the decreased expression levels reflect the transition of normal
epithelial cells to cancerous cells. The IHC results revealed
positive IRE1β staining in all epithelial cells, suggesting that
IRE1β affects not only goblet cells, but also epithelial cells. A
previous study reported that IRE1β regulates lipid absorption by
mediating the transcription of microsomal triacylglycerol transfer
protein expression in the colon epithelium (53). However, the association between the
change in lipid absorption and the tumorigenesis of CRC requires
further study for elucidation. It was hypothesized that decreased
IRE1β expression levels may be associated with tumorigenesis, as
IRE1β is a protective factor for colitis and is involved in cell
apoptosis (26,32). Intestinal inflammation and cell
apoptosis are putatively associated with occurrence of CRC
(54). Analysis of the mRNA
expression levels of IRE1β in tumor tissues revealed that higher
IRE1β mRNA expression levels were significantly associated with
poor tumor differentiation, lymph node metastasis and later TNM
stage. The results suggest that IRE1β may be involved in the
development of CRC. The results of the present study are concordant
with the in vitro study conducted by Dai et al
(55), who identified that the mRNA
expression levels of IRE1β were high in undifferentiated Caco-2
cells, and were correspondingly decreased following the
differentiation of these cells (55).
Two previous studies have reported that IRE1β is an
important regulator of MUC2 secretion (28,33). In
animal model studies, IRE1β−/− mice were more
susceptible to dextran sodium sulfate (DSS) -induced colitis,
compared with wild type mice (26).
The loss of intestinal mucin also increases the sensitivity of mice
to DSS-induced colitis (56). IRE1β
is essential for UPR in goblet cells, and MUC2 is its target
protein (27,33). In the current study, MUC2 expression
levels were revealed to be decreased in colorectal adenocarcinomas.
The results are concordant with previous studies, in which
nonmucinous CRC tissues were negative for MUC2 expression (57,58). This
may be due to a decreased number of goblet cells in non-mucinous
CRC tissues, as the expression of MUC2 was positively correlated
with the mRNA expression levels of IRE1β. However, it has
previously been revealed that the methylation of the MUC2 promoter
and the loss of functional tumor protein 53 may decrease MUC2
expression levels in CRC tissues (59,60).
Correlation analysis in the present study indicated that the
expression levels of MUC2 were positively associated with the mRNA
expression levels of IRE1β. IRE1β and MUC2 act as protective
factors to maintain the intestinal physiological homeostasis, and
the suppression of the two proteins may be associated with the
tumorigenesis of CRC.
In conclusion, the results of the current study
revealed that the decreased expression levels of IRE1β in CRC
tissues were associated with clinical features of patients with
CRC. IRE1β gene expression levels were positively correlated with
those of MUC2, indicating that IRE1β and MUC2 may be involved in
the tumorigenesis of CRC. The association of IRE1β and MUC2, as
well as the significance of IRE1β in CRC, require further studies
in order to identify novel therapeutic targets for this type of
cancer.
Acknowledgements
The authors would like to thank Dr Yonggan Fan, Dr
Xianli Liu and Dr Wenchao Zhao for their help with patient
recruitment and pathological consultation in the present study.
This study was supported by a grant (grant no. 81370487 to Q.Gao)
from The National Natural Science Foundation of China.
Glossary
Abbreviations
Abbreviations:
|
ATF6
|
activating transcription factor 6
|
|
CRC
|
colorectal cancer
|
|
ER
|
endoplasmic reticulum
|
|
IRE1
|
inositol-requiring enzyme 1
|
|
MUC
|
mucin
|
|
PERK
|
protein-kinase-RNA-like-ER kinase
|
|
RIDD
|
regulated IRE1-dependent decay
|
|
UPR
|
unfolded protein response
|
|
XBP1
|
X-box binding protein 1
|
References
|
1
|
Fleming M, Ravula S, Tatishchev SF and
Wang HL: Colorectal carcinoma: Pathologic aspects. J Gastrointest
Oncol. 3:153–173. 2012.PubMed/NCBI
|
|
2
|
Zhao P, Dai M, Chen W and Li N: Cancer
trends in China. Jpn J Clin Oncol. 40:281–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li L and Ma BB: Colorectal cancer in
Chinese patients: Current and emerging treatment options. Onco
Targets Ther. 7:1817–1828. 2014.PubMed/NCBI
|
|
4
|
Dai Z, Zheng RS, Zou XN, Zhang SW, Zeng
HM, Li N and Chen WQ: Analysis and prediction of colorectal cancer
incidence trend in China. Zhonghua Yu Fang Yi Xue Za Zhi.
46:598–603. 2012.(In Chinese). PubMed/NCBI
|
|
5
|
Stintzing S: Management of colorectal
cancer. F1000Prime Rep. 6:1082014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim ER and Kim YH: Clinical application of
genetics in management of colorectal cancer. Intest Res.
12:184–193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
SEER Stat Fact Sheets: Colon and Rectum
Cancer). National Cancer Institute; Bethesda, MD: https://seer.cancer.gov/statfacts/html/colorect.htmlSeptember
1–2015
|
|
8
|
Manié SN, Lebeau J and Chevet E: Cellular
mechanisms of endoplasmic reticulum stress signaling in health and
disease. 3. Orchestrating the unfolded protein response in
oncogenesis: An update. Am J Physiol Cell Physiol. 307:C901–C907.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dicks N, Gutierrez K, Michalak M,
Bordignon V and Agellon LB: Endoplasmic reticulum stress, genome
damage, and cancer. Front Oncol. 5:112015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parmar VM and Schröder M: Sensing
endoplasmic reticulum stress. Adv Exp Med Biol. 738:153–168. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gardner BM, Pincus D, Gotthardt K,
Gallagher CM and Walter P: Endoplasmic reticulum stress sensing in
the unfolded protein response. Cold Spring Harb Perspect Biol.
5:a0131692013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao Q, Esworthy RS, Kim BW, Synold TW,
Smith DD and Chu FF: Atherogenic diets exacerbate colitis in mice
deficient in glutathione peroxidase. Inflamm Bowel Dis.
16:2043–2054. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McMillan DR, Gething MJ and Sambrook J:
The cellular response to unfolded proteins: Intercompartmental
signaling. Curr Opin Biotechnol. 5:540–545. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Papandreou I, Denko NC, Olson M, Van
Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F,
Niwa M and Koong AC: Identification of an Ire1alpha endonuclease
specific inhibitor with cytotoxic activity against human multiple
myeloma. Blood. 117:1311–1314. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Greenman C, Stephens P, Smith R, Dalgliesh
GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C,
et al: Patterns of somatic mutation in human cancer genomes.
Nature. 446:153–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakamura D, Tsuru A, Ikegami K, Imagawa Y,
Fujimoto N and Kohno K: Mammalian ER stress sensor IRE1β
specifically down-regulates the synthesis of secretory pathway
proteins. FEBS Lett. 585:133–138. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koong AC, Chauhan V and Romero-Ramirez L:
Targeting XBP-1 as a novel anti-cancer strategy. Cancer Biol Ther.
5:756–759. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoshida H, Oku M, Suzuki M and Mori K:
pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded
protein response activator pXBP1(S) in mammalian ER stress
response. J Cell Biol. 172:565–575. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fujimoto T, Onda M, Nagai H, Nagahata T,
Ogawa K and Emi M: Upregulation and overexpression of human X-box
binding protein 1 (hXBP-1) gene in primary breast cancers. Breast
Cancer. 10:301–306. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shuda M, Kondoh N, Imazeki N, Tanaka K,
Okada T, Mori K, Hada A, Arai M, Wakatsuki T, Matsubara O, et al:
Activation of the ATF6, XBP1 and grp78 genes in human
hepatocellular carcinoma: A possible involvement of the ER stress
pathway in hepatocarcinogenesis. J Hepatol. 38:605–614. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fujimoto T, Yoshimatsu K, Watanabe K,
Yokomizo H, Otani T, Matsumoto A, Osawa G, Onda M and Ogawa K:
Overexpression of human X-box binding protein 1 (XBP-1) in
colorectal adenomas and adenocarcinomas. Anticancer Res.
27:127–131. 2007.PubMed/NCBI
|
|
22
|
Takahashi S, Suzuki S, Inaguma S, Ikeda Y,
Cho YM, Nishiyama N, Fujita T, Inoue T, Hioki T, Sugimura Y, et al:
Down-regulation of human X-box binding protein 1 (hXBP-1)
expression correlates with tumor progression in human prostate
cancers. Prostate. 50:154–161. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Y and Brandizzi F: IRE1: ER stress
sensor and cell fate executor. Trends Cell Biol. 23:547–555. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Coelho DS and Domingos PM: Physiological
roles of regulated Ire1 dependent decay. Front Genet. 5:762014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Welihinda AA, Tirasophon W and Kaufman RJ:
The cellular response to protein misfolding in the endoplasmic
reticulum. Gene Expr. 7:293–300. 1999.PubMed/NCBI
|
|
26
|
Bertolotti A, Wang X, Novoa I, Jungreis R,
Schlessinger K, Cho JH, West AB and Ron D: Increased sensitivity to
dextran sodium sulfate colitis in IRE1beta-deficient mice. J Clin
Invest. 107:585–593. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martino MB, Jones L, Brighton B, Ehre C,
Abdulah L, Davis CW, Ron D, O'Neal WK and Ribeiro CM: The ER stress
transducer IRE1β is required for airway epithelial mucin
production. Mucosal Immunol. 6:639–654. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang HS, Chen Y, Fan L, Xi QL, Wu GH, Li
XX, Yuan TL, He SQ, Yu Y, Shao ML, et al: The Endoplasmic Reticulum
Stress Sensor IRE1α in Intestinal Epithelial Cells Is Essential for
Protecting against Colitis. J Biol Chem. 290:15327–15336. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin JH, Li H, Yasumura D, Cohen HR, Zhang
C, Panning B, Shokat KM, Lavail MM and Walter P: IRE1 signaling
affects cell fate during the unfolded protein response. Science.
318:944–949. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hetz C: The unfolded protein response:
Controlling cell fate decisions under ER stress and beyond. Nat Rev
Mol Cell Biol. 13:89–1026. 2012.PubMed/NCBI
|
|
31
|
Hetz C and Glimcher LH: Fine-tuning of the
unfolded protein response: Assembling the IRE1alpha interactome.
Mol Cell. 35:551–561. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iwawaki T, Hosoda A, Okuda T, Kamigori Y,
Nomura-Furuwatari C, Kimata Y, Tsuru A and Kohno K: Translational
control by the ER transmembrane kinase/ribonuclease IRE1 under ER
stress. Nat Cell Biol. 3:158–164. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsuru A, Fujimoto N, Takahashi S, Saito M,
Nakamura D, Iwano M, Iwawaki T, Kadokura H, Ron D and Kohno K:
Negative feedback by IRE1β optimizes mucin production in goblet
cells. Proc Natl Acad Sci USA. 110:2864–2869. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Johansson ME and Hansson GC: Mucus and the
goblet cell. Dig Dis. 31:305–309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Johansson ME, Phillipson M, Petersson J,
Velcich A, Holm L and Hansson GC: The inner of the two Muc2
mucin-dependent mucus layers in colon is devoid of bacteria. Proc
Natl Acad Sci USA. 105:15064–15069. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kawashima H: Roles of the gel-forming MUC2
mucin and its O-glycosylation in the protection against colitis and
colorectal cancer. Biol Pharm Bull. 35:1637–1641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Van der Sluis M, De Koning BA, De Bruijn
AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J,
Van Seuningen I, Renes IB and Einerhand AW: Muc2-deficient mice
spontaneously develop colitis, indicating that MUC2 is critical for
colonic protection. Gastroenterology. 131:117–129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Velcich A, Yang W, Heyer J, Fragale A,
Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K and
Augenlicht L: Colorectal cancer in mice genetically deficient in
the mucin Muc2. Science. 295:1726–1729. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Imai Y, Yamagishi H, Fukuda K, Ono Y,
Inoue T and Ueda Y: Differential mucin phenotypes and their
significance in a variation of colorectal carcinoma. World J
Gastroenterol. 19:3957–3968. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Debunne H and Ceelen W: Mucinous
differentiation in colorectal cancer: Molecular, histological and
clinical aspects. Acta Chir Belg. 113:385–390. 2013.PubMed/NCBI
|
|
41
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Untergasser A, Cutcutache I, Koressaar T,
Ye J, Faircloth BC, Remm M and Rozen SG: Primer3-new capabilities
and interfaces. Nucleic Acids Res. 40:e1152012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu FX, Su YL, Zhang H, Kong JY, Yu H and
Qian BY: Prognostic implications for high expression of MiR-25 in
lung adenocarcinomas of female non-smokers. Asian Pac J Cancer
Prev. 15:1197–1203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao Q, Meijer MJ, Kubben FJ, Sier CF,
Kruidenier L, van Duijn W, van den Berg M, van Hogezand RA, Lamers
CB and Verspaget HW: Expression of matrix metalloproteinases-2 and
−9 in intestinal tissue of patients with inflammatory bowel
diseases. Dig Liver Dis. 37:584–592. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu C, Huang Z, Jiang H and Shi F: The
sirtuin 3 expression profile is associated with pathological and
clinical outcomes in colon cancer patients. Biomed Res Int.
2014:8712632014.PubMed/NCBI
|
|
46
|
Liu S, Zheng R, Zhang M, Zhang S, Sun X
and Chen W: Incidence and mortality of colorectal cancer in China,
2011. Chin J Cancer Res. 27:22–28. 2015.PubMed/NCBI
|
|
47
|
Imagawa Y, Hosoda A, Sasaka S, Tsuru A and
Kohno K: RNase domains determine the functional difference between
IRE1alpha and IRE1beta. FEBS Lett. 582:656–660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Piemonte M: [TNM - classification of
malignant tumors (VI edition - 2002). Innovations in the
classification of head and neck neoplasms]. Acta Otorhinolaryngol
Ital. 23:132–135. 2003.(In Italian). PubMed/NCBI
|
|
49
|
Wei Q, Huang X, Fu B, Liu J, Zhong L, Yang
Q and Zhao T: IMP3 expression in biopsy specimens of colorectal
cancer predicts lymph node metastasis and TNM stage. Int J Clin Exp
Pathol. 8:11024–3213. 2015.PubMed/NCBI
|
|
50
|
Kesari MV, Gaopande VL, Joshi AR,
Babanagare SV, Gogate BP and Khadilkar AV: Immunohistochemical
study of MUC1, MUC2 and MUC5AC in colorectal carcinoma and review
of literature. Indian J Gastroenterol. 34:63–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Walsh MD, Clendenning M, Williamson E,
Pearson SA, Walters RJ, Nagler B, Packenas D, Win AK, Hopper JL,
Jenkins MA, et al: Expression of MUC2, MUC5AC, MUC5B, and MUC6
mucins in colorectal cancers and their association with the CpG
island methylator phenotype. Mod Pathol. 26:1642–1656. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Okudaira K, Kakar S, Cun L, Choi E, Wu
Decamillis R, Miura S, Sleisenger MH, Kim YS and Deng G: MUC2 gene
promoter methylation in mucinous and non-mucinous colorectal cancer
tissues. Int J Oncol. 36:765–775. 2010.PubMed/NCBI
|
|
53
|
Iqbal J, Dai K, Seimon T, Jungreis R,
Oyadomari M, Kuriakose G, Ron D, Tabas I and Hussain MM: IRE1beta
inhibits chylomicron production by selectively degrading MTP mRNA.
Cell Metab. 7:445–455. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Terzić J, Grivennikov S, Karin E and Karin
M: Inflammation and colon cancer. Gastroenterology.
138:2101–2114.e5. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dai K, Khatun I and Hussain MM: NR2F1 and
IRE1beta suppress microsomal triglyceride transfer protein
expression and lipoprotein assembly in undifferentiated intestinal
epithelial cells. Arterioscler Thromb Vasc Biol. 30:568–574. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Heazlewood CK, Cook MC, Eri R, Price GR,
Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ,
et al: Aberrant mucin assembly in mice causes endoplasmic reticulum
stress and spontaneous inflammation resembling ulcerative colitis.
PLoS Med. 5:e542008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Manne U, Weiss HL and Grizzle WE: Racial
differences in the prognostic usefulness of MUC1 and MUC2 in
colorectal adenocarcinomas. Clin Cancer Res. 6:4017–4025.
2000.PubMed/NCBI
|
|
58
|
Matsuda K, Masaki T, Watanabe T, Kitayama
J, Nagawa H, Muto T and Ajioka Y: Clinical significance of MUC1 and
MUC2 mucin and p53 protein expression in colorectal carcinoma. Jpn
J Clin Oncol. 30:89–94. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gratchev A, Siedow A, Bumke-Vogt C, Hummel
M, Foss HD, Hanski ML, Kobalz U, Mann B, Lammert H, Mansmann U, et
al: Regulation of the intestinal mucin MUC2 gene expression in
vivo: Evidence for the role of promoter methylation. Cancer Lett.
168:71–80. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ookawa K, Kudo T, Aizawa S, Saito H and
Tsuchida S: Transcriptional activation of the MUC2 gene by p53. J
Biol Chem. 277:48270–48275. 2002. View Article : Google Scholar : PubMed/NCBI
|