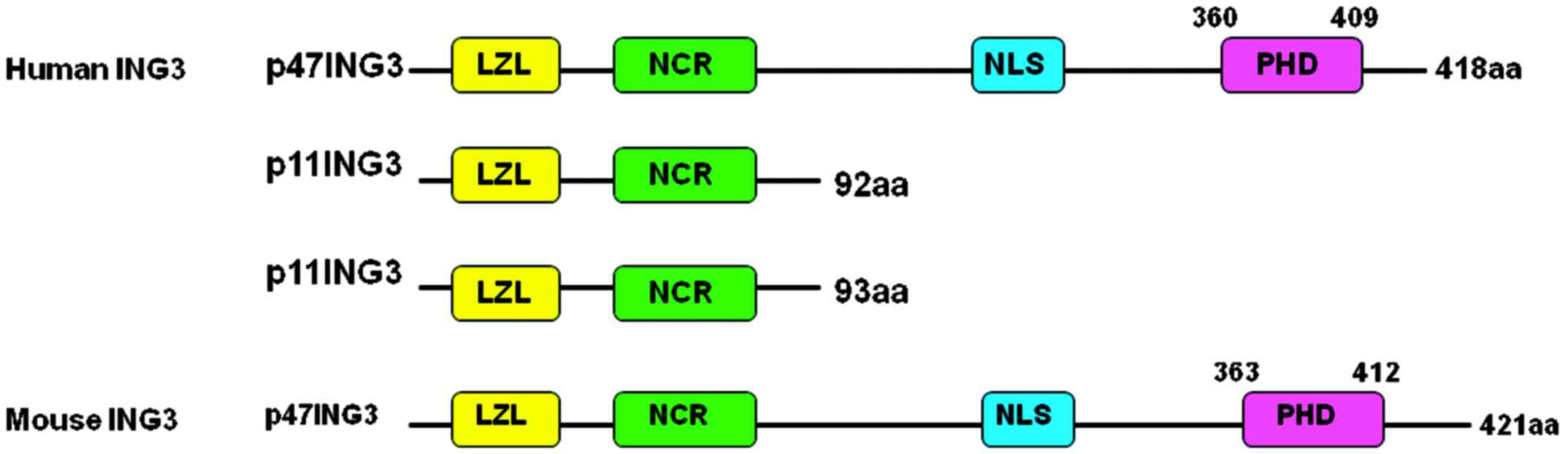
Introduction
The inhibitor of growth (ING) family consists of
five members with various isoforms due to alternative splicing.
Their encoded proteins comprise a highly conserved plant
homeodomain (PHD), a Cys4-His-Cys3 form of
zinc finger that interacts directly with histone H3, and a nuclear
localization sequence (NLS). ING proteins act as receptors and
transducers of stress-activated phosphoinositides, inhibit
angiogenesis, promote cellular senescence or are involved in
various biological processes, including DNA repair, apoptosis, cell
cycle checkpoints, histone methylation and acetylation, and
regulation of transcription by protein-protein or protein-DNA
interaction. They appear to be inactivated in malignancies and
therefore are classified as class II tumor suppressor genes
(1,2).
The human ING3 gene is located at chromosome 7q31.3,
is composed of 12 exons and produces three proteins (Fig. 1), among which p47ING3 controls
p53-mediated transcription, blocks cell cycle control and induces
apoptosis (3). As a significant
chromatin acetylation regulator, ING3 is primarily involved in the
formation of nucleosome NuA4 histone acetyltransferase
multi-subunit complex and is essential for the histone
acetyltransferase activity of Tip60 (2,4). ING3
overexpression decreases the S-phase population of cells and their
colony-forming ability, and induces apoptosis in RKO human colon
carcinoma cells in a p53-mediated manner (5). An additional study has indicated that
ING3 activates p53-transactivated promoters of p21 and
Bcl2-associated X protein (5). In
addition, ING3 has been demonstrated to be capable of enhancing
ultraviolet-induced apoptosis of melanoma cells via a
Fas/caspase-8-dependent signaling pathway, independently of
functional p53 (6). Furthermore, it
was identified that ING3 underwent degradation via its interaction
with subunits of E3 ligase Skp1-Cullin-F-box (SCF) protein complex
in the ubiquitin-proteasome signaling pathway, which provided an
alternative explanation for ING3 downregulation (7). ING3 may also be capable of regulating
asymmetric cell division via the mammalian target of rapamycin
signaling pathway during mouse oocyte maturation (8).
ING3 mRNA is ubiquitously expressed in normal human
tissues, including the testes, skeletal muscle, spleen, heart and
oral mucosa (5). Nuclear ING3
expression is markedly reduced in malignant melanoma compared with
dysplastic nevi, and is significantly associated with a poorer
prognosis for melanoma as an independent factor (9). Gunduz et al (10) demonstrated that loss of heterozygosity
(LOH) resulted in reduced ING3 expression in human head and neck
squamous cell carcinomas (HNSCC). A previous survival analysis
revealed that ING3 downregulation may be considered as an
independent prognostic factor for poor overall survival time in
HNSCC (11). In addition, Borkosky
et al (12) identified that
SSLOH of the ING3 locus was high in solid type tumors of
ameloblastoma. mRNA and protein concentrations of ING3 have been
observed to be downregulated in the majority of hepatocellular
carcinoma (HCC) cases in comparison with matched non-tumor hepatic
tissues, and reduced expression of ING3 protein is correlated with
more aggressive characteristics and adverse prognosis in this tumor
type (13,14). Consistently, ectopic ING3
overexpression in HCC cells was observed to suppress colony
formation, cell proliferation and migration (13,14). These
results suggest that reduced ING3 expression may be associated with
tumorigenesis and the subsequent development of malignancies. Thus,
the present study analyzed the expression profile of ING3 protein
in normal mouse and human tissues, and in human cancer tissues.
Materials and methods
Samples
A total of three male and three female C57BL/6 mice
(8 weeks old; 30–40 g) were maintained under specific pathogen-free
conditions in a temperature-controlled room with a 12-h light/dark
illumination cycle. Standard rodent food and water were supplied
ad libitum. Housing and all procedures were performed
according to guidelines on animal welfare approved by the Committee
for Animal Experiments of Liaoning Medical University. The mice
were sacrificed under sodium pentobarbital anesthesia, and tissue
samples were dissected from the brain, heart, liver, spleen, lung,
kidney, breast, stomach and intestine. All tissues were fixed in
10% neutral formalin, embedded in paraffin and cut into 4-µm
sections. The tissue arrays of human normal tissues (cerebrum,
cerebellum, brain stem, aorta, tongue, thyroid, esophagus, stomach,
intestine, liver, pancreas, lung, trachea, appendix, smooth muscle,
skeletal muscle, heart, testis, bladder and prostate) and cancer
tissues (62 hepatocellular carcinoma, 62 renal clear cell
carcinoma, 62 pancreatic carcinoma, 45 esophageal squamous cell
carcinoma and 31 cervical squamous cell carcinoma cases) were
purchased from Shanghai Outdo Biotech Co., Ltd (Shanghai, China).
Human cervix, endometrium, ovary and breast tissues were sampled
from surgical patients at The First Affiliated Hospital of Liaoning
Medical University (Jinzhou, China). In addition, breast (n=144),
gastric (n=196), colorectal (n=96), ovarian (n=208), endometrial
(n=96) and lung carcinoma (n=192) samples were collected from
patients at the same hospital. Dissected mouse tissues and
collected human normal and cancer tissues, were subjected to tissue
microarray using a tissue microarrayer (AZUMAYA KIN-1; Azumaya
Corporation, Tokyo, Japan). None of the cancer patients had
undergone chemotherapy, radiotherapy or adjuvant treatment prior to
surgery. The patients or their relatives provided written consent
for the use of tumor tissues for clinical research, and the
research protocol was approved by the Ethical and Animal
Experimentation Committees of Liaoning Medical University (Jinzhou,
China).
Immunohistochemistry
Consecutive sections were dewaxed using xylene,
rehydrated in a graded series of alcohol to water, and subjected to
antigen retrieval by irradiation in target retrieval solution (Dako
North America, Inc., Carpinteria, CA, USA) in a microwave oven for
15 min (Oriental Rotor Ltd., Co., Tokyo, Japan). Sections were
subsequently blocked with 5% bovine serum albumin (A8020; Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) for 20
min to prevent non-specific antibody binding. The sections were
incubated with rabbit polyclonal IgG anti-ING3 (#sc-366026; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA; dilution, 1:50) for 15
min, followed by incubation with the secondary anti-rabbit
polyclonal Ig antibody conjugated to horseradish peroxidase (HRP)
(#P0399; HRP; Dako North America, Inc.; ready-to-use) for 60 min.
Following each treatment, the slides were washed using
Tris-buffered saline and Tween 20 (TBST; 3 × 1-min washes). The HRP
was colored with 3,3′-diaminobenzidine. Sections were
counterstained using Mayer's hematoxylin, dehydrated, cleared and
mounted. TBST was utilized as a negative control in place of
primary antibody.
Immunohistochemical evaluation
As indicated in Figs.
2–4, ING3 immunopositivity was
localized to the cytoplasm and/or nucleus. Initially, a strong
expression field was selected under low magnification and all cells
were randomly counted in five different representative fields of
each section, which were assessed blindly by two independent
pathologists. Any inconsistent data was discussed by the
pathologists until a final agreement was reached. The percentages
of counted cells (calculated as the mean percent of positively
stained cells out of the total cells counted) were scored as
follows: 0–10%, negative (−), 11–100%, positive (+).
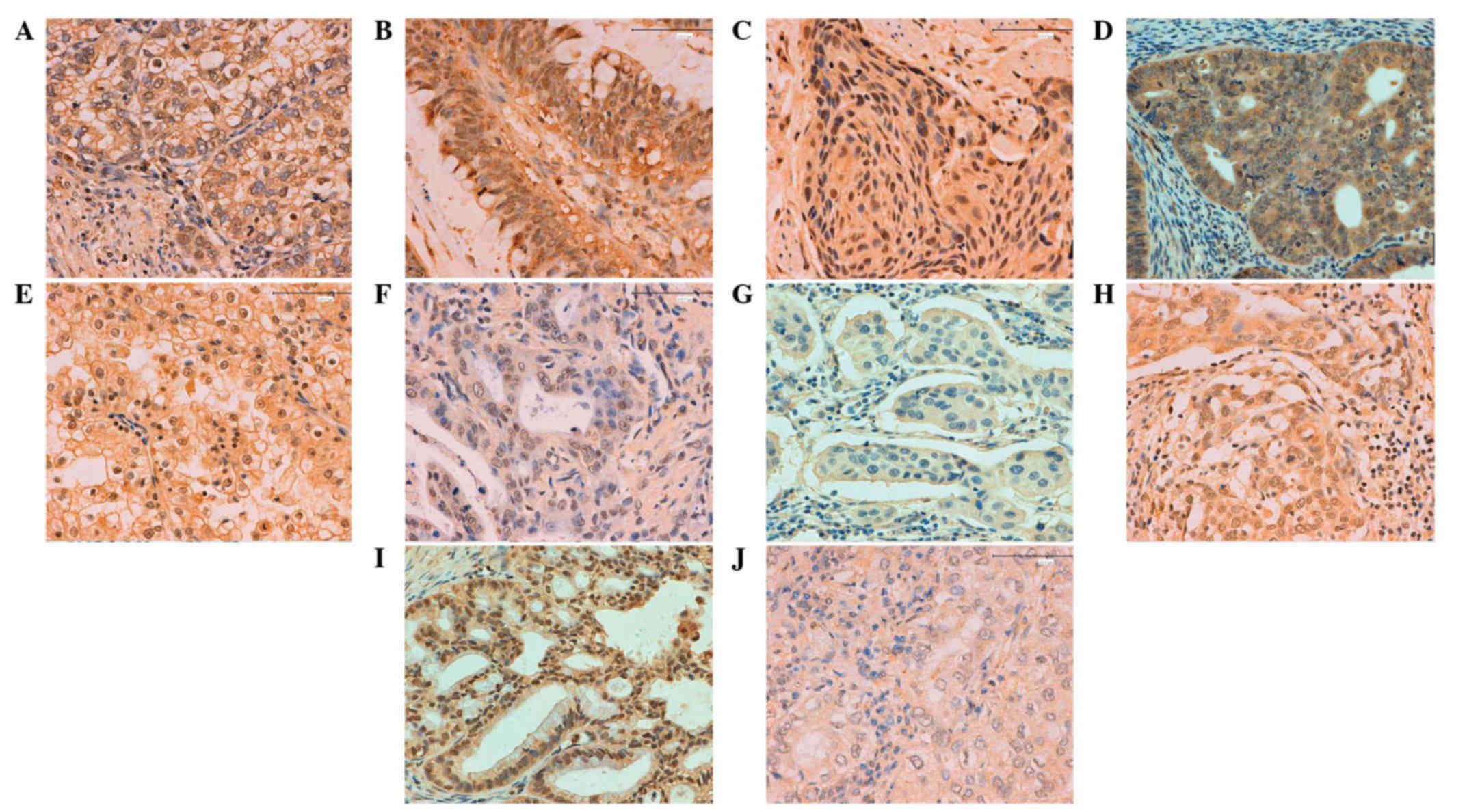 | Figure 4.Inhibitor of growth family, member 3
expression in human cancer detected by immunohistochemistry: (A)
Gastric, (B) colorectal, (C) esophageal, (D) endometrial, (E)
renal, (F) pancreatic, (G) breast, (H) cervical, (I) ovarian and
(J) lung carcinoma. Magnification, ×200. |
Results
ING3 is detectable in a wide range of
cell types in mice
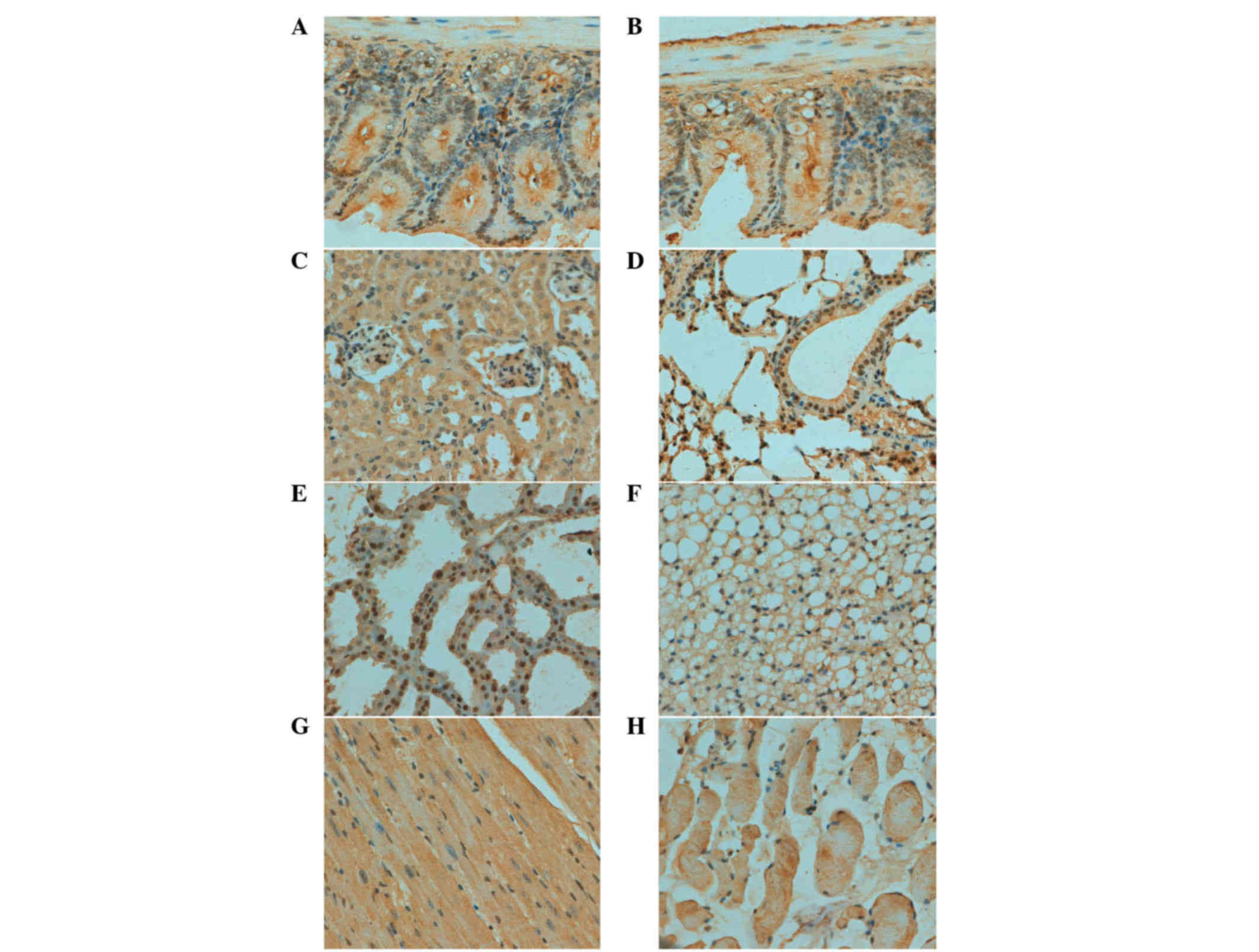
As indicated in Fig.
2, ING3 protein was positively detected in the cytoplasm of
cardiomyocyte, kidney and skeletal muscle cells. A cytoplasmic and
nuclear distribution of ING3 protein was observed in bronchial and
alveolar epithelium, gastric, intestinal and mammary gland cells.
ING3 protein was expressed in the brain, spleen, skin and liver in
a sporadic manner (Table I).
 | Table I.Immunohistochemical examination of
inhibitor of growth family, member 3 protein in mouse normal
tissues. |
Table I.
Immunohistochemical examination of
inhibitor of growth family, member 3 protein in mouse normal
tissues.
| Tissue type | Cell type |
|---|
| Brain | Sporadic |
| Heart | Cardiomyocyte |
| Lung | Bronchial and
alveolar epithelium |
| Kidney | Nephric tubule |
| Stomach | Glandular |
| Intestine | Glandular |
| Spleen | Sporadic |
| Skin | Sporadic |
| Muscle | Striated muscle
cell |
| Fat | Lipocyte |
| Liver | Sporadic |
| Breast | Glandular
epithelium |
ING3 expression is primarily localized
to the cytoplasm in normal human tissues
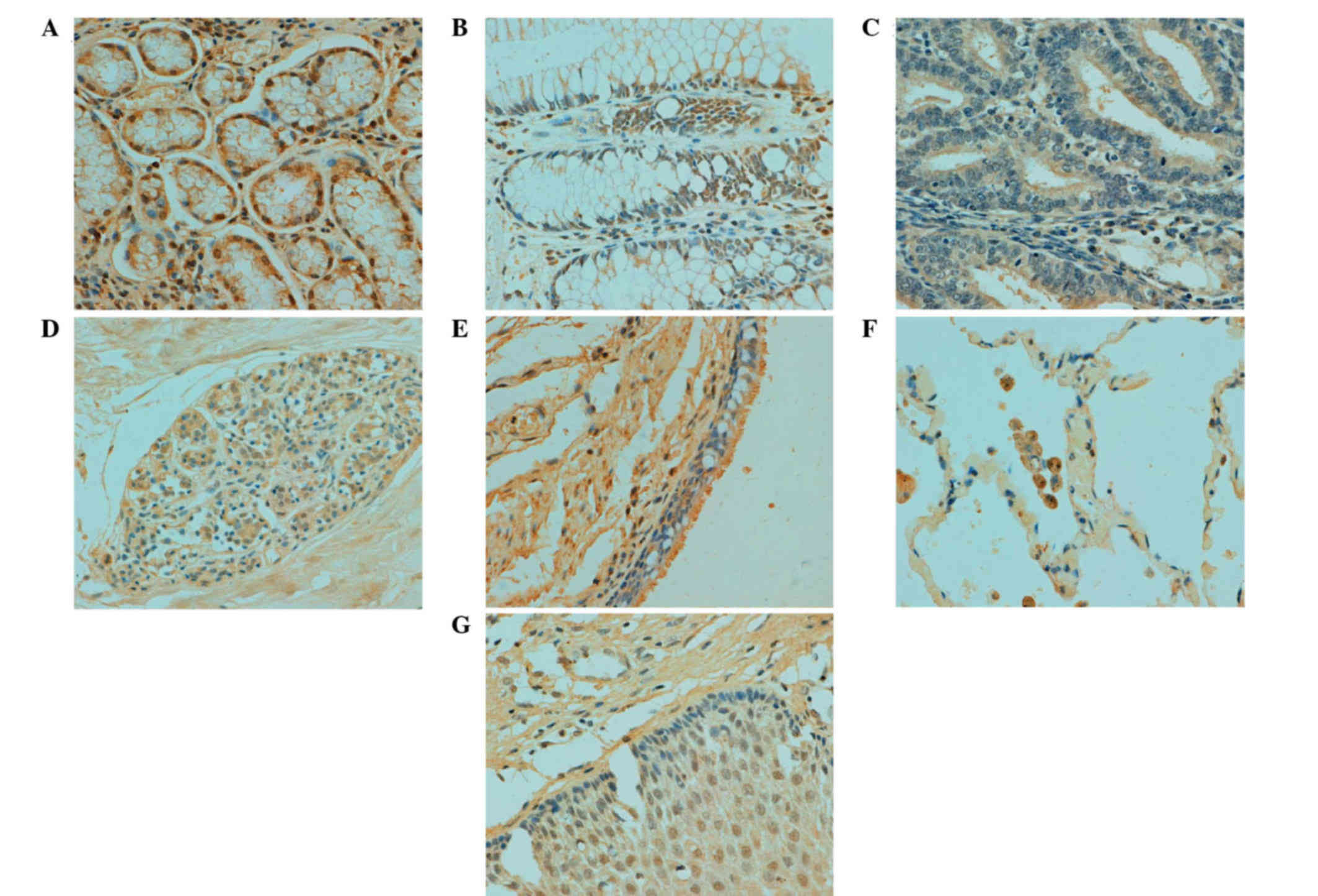
In human tissues, ING3 protein was principally
distributed in the cytoplasm; however, it was observed in both the
cytoplasm and nucleus of tongue, esophagus, stomach, intestine,
lung, skin, appendix, bladder, cervix and breast cells (Table II; Fig.
3). According to the density, ING3 immunoreactivity was
strongly detected in stomach, skin and cervical cells, and was
weakly detected in cerebellum, brain stem, thymus, liver, skeletal
muscle, testis and prostate cells (data not shown).
 | Table II.Immunostaining of ING3 protein in
normal human tissues. |
Table II.
Immunostaining of ING3 protein in
normal human tissues.
|
| ING3 expression |
|---|
|
|
|
|---|
| Tissue type | Nucleus | Cytoplasm |
|---|
| Cerebrum | − | + |
| Cerebellum | − | + |
| Brain stem | − | + |
| Thymus | − | + |
| Hear muscle | − | + |
| Aorta | − | + |
| Tongue | + | + |
| Thyroid | − | + |
| Esophagus | + | + |
| Stomach | + | + |
| Intestine | + | + |
| Liver | − | + |
| Pancreas | − | + |
| Lung | + | + |
| Trachea | − | + |
| Skin | + | + |
| Appendix | + | + |
| Smooth muscle | − | + |
| Skeletal muscle | − | + |
| Heart | − | + |
| Testis | − | + |
| Bladder | + | + |
| Prostate | − | + |
| Cervix | + | + |
| Endometrium | + | + |
| Ovary | − | + |
| Breast | + | + |
ING3 is most frequently expressed in
gynecological types of cancer
In total, ING3-positivity was identified in 424 of
1,194 tested cancer entities (35.5%), with a homogeneous expression
pattern (Fig. 4; Table III). In the majority of cases, ING3
expression was observed to be distributed in the cytoplasm and
nucleus, with the exception of the cytoplasmic distribution in
breast and hepatocellular carcinoma. Among the cancer entities
studied, ING3 was most frequently expressed in cases involving
female cancer types, including ovarian (59.2%; 124/208),
endometrial (47.9%; 46/96), breast (38.9%; 56/144) and cervical
cancers (35.5%; 11/31). ING3-positive cases were more rare in renal
clear cell (17.7%; 11/62), hepatocellular (16.1%; 10/62) and
esophageal carcinomas (17.8%; 8/45).
 | Table III.ING3 expression in various types of
cancer detected by immunohistochemistry. |
Table III.
ING3 expression in various types of
cancer detected by immunohistochemistry.
|
|
|
|
| ING3 expression |
|---|
|
|
|
|
|
|
|---|
| Carcinoma type | Total cases, n | Positive cases,
n | Positive rate,
% | Nucleus | Cytoplasm |
|---|
| Hepatocellular | 62 | 11 | 17.7 | − | + |
| Renal clear
cell | 62 | 10 | 16.1 | + | + |
| Pancreatic | 62 | 23 | 37.1 | + | + |
| Esophageal | 45 | 8 | 17.8 | + | + |
| Cervical | 31 | 11 | 35.5 | + | + |
| Breast | 144 | 56 | 38.9 | − | + |
| Gastric | 196 | 57 | 29.1 | + | + |
| Colorectal | 96 | 28 | 29.2 | + | + |
| Ovarian | 208 | 124 | 59.6 | + | + |
| Endometrial | 96 | 46 | 47.9 | + | + |
| Lung | 192 | 50 | 26.0 | + | + |
Discussion
ING3 protein contains an NLS and a PHD finger motif
at the C-terminus (15). Previously,
Wang et al (9) demonstrated
that nuclear-to-cytoplasmic translocation of ING3 protein led to
reduced nuclear expression in cutaneous melanoma. The degradation
of ING3 by the cytoplasmic SCF (S-phase kinase-associated protein
2)-mediated ubiquitin-proteasome system provided additional
evidence for its cytosolic localization (7). An additional two studies observed a
cytoplasmic expression pattern of ING3 in hepatocytes and HCC
(13,14). In the present study, the expression
level and cellular localization of ING3 protein was characterized
in normal mouse and human tissue, and human cancer tissue. A
positive ING3 signal was observed in the cytoplasm of normal mouse
and human tissue, and in human cancer tissue, and was occasionally
observed in both the cytoplasm and nucleus. Cenzig et al
(16) reported that the mutation or
deletion of the ING5 NLS resulted in its nucleocytoplasmic
translocation. ING1 phosphorylation by 14-3-3 family (17) or Src (18) proteins leads to its cytoplasmic
relocalization for apoptotic induction. Therefore, it was
speculated that chemical modification of ING3 may lead to its
restoration in the cytoplasm, which will require clarification in
future studies.
Amino acid sequence alignment has demonstrated a
high similarity between human p47ING3 and mouse ING3, revealing
that they share 95% identity (1).
Consistently, the present study identified no notable differences
in the patterns of ING3 expression between mouse and human samples.
In human tissue, ING3 protein was strongly detected in stomach,
skin and cervical cells, and was weakly detected in brain, thymus,
liver, skeletal muscle, testis and prostate cells, suggesting a
functional involvement of ING3 in distinct cell types and in the
specific functional state of cells. Therefore, in future studies,
we aim to conditionally ablate the ING3 gene using a
cell-specific promoter and establish an animal model of
ING3-negative tumors. In the relevant literature, ectopic ING3
expression resulted in increased apoptosis via the Fas-mediated
signaling pathway (6) and suppression
of proliferation (5). Therefore, ING3
overexpression in the stomach, skin and cervix may be associated
with regeneration and repair, regardless of whether glandular or
squamous epithelium; this is supported by the observed weaker
expression in organs with low levels of repair and renewal,
including the brain, thymus, skeletal muscle and testis. Notably,
ING2, another member of the ING family, has been reported to be
involved in muscle differentiation via regulating myogenin
transcription (19). As a member of
the ING family, ING3 protein is enriched in heart, skeletal and
smooth muscle cells, which is hypothesized to be associated with
the differentiation of muscles.
ING3 is a candidate tumor suppressor gene, and its
expression is frequently downregulated in tumors (9,14,15). The present study focused on the most
commonly occurring epithelial cancers and demonstrated that female
types of cancer, including breast, ovarian and endometrial,
exhibited higher levels of ING3 expression, indicating that ING3
protein may be involved in estrogen production or may be regulated
by estrogen. It was notable that gastric and colorectal cancers
demonstrated similar levels and patterns of expression of ING3,
which may be due to the similar carcinogenesis and pathological
behaviors of these types of cancer. By contrast, renal clear cell
carcinoma demonstrated the lowest levels of ING3 expression, with a
positive rate of <20%. This knowledge may significantly
facilitate the identification of cancer patients that may
potentially benefit from an ING3-targeting gene therapy. According
to the relevant literature, ING3 protein is involved in the
modulation of p53-mediated transcription, cell cycle control and
apoptosis (1,2). In RKO human colon carcinoma cells, ING3
overexpression reduced colony formation, potentially by reducing
the number of cells in S phase (5).
In combination with these findings, the profiling of ING3
expression may assist with clarification of the role of ING3
expression in disruption of proliferation and apoptosis in various
types of epithelial cancer.
In summary, the present study clarified the
differential expression and/or subcellular location of ING3 in
various tissues, cell types and single cells in normal mouse and
human tissues, and human cancer tissue, suggesting differential
functional involvement. Based on the results of the present study,
it is hypothesized that ING3 may be involved in the repair and
regeneration of organs or tissues, and may have a significant role
in gynecological carcinogenesis.
Acknowledgements
This study was supported by the Liaoning BaiQianWan
Talents Program, a Key Scientific and Technological Project of
Liaoning Province (grant no. 2015408001), Scientific Research Fund
of Liaoning Provincial Education Department (grant no. LJQ2014093)
and the National Natural Science Foundation of China (grant nos.
81172371, 81472544 and 81672700).
References
|
1
|
Ludwig S, Klitzsch A and Baniahmad A: The
ING tumor suppressors in cellular senescence and chromatin. Cell
Biosci. 1:252011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Doyon Y, Cayrou C, Ullah M, Landry AJ,
Côté V, Selleck W, Lane WS, Tan S, Yang XJ and Côté J: ING tumor
suppressor proteins are critical regulators of chromatin
acetylation required for genome expression and perpetuation. Mol
Cell. 21:51–64. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guérillon C, Bigot N and Pedeux R: The ING
tumor suppressor genes: Status in human tumors. Cancer Lett.
345:1–16. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ullah M, Pelletier N, Xiao L, Zhao SP,
Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, et al:
Molecular architecture of quartet MOZ/MORF histone
acetyltransferase complexes. Mol Cell Biol. 28:6828–6843. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagashima M, Shiseki M, Pedeux RM, et al:
A novel PHD-finger motif protein, p47ING3, modulates p53-mediated
transcription, cell cycle control, and apoptosis. Oncogene.
22:343–350. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y and Li G: ING3 promotes UV-induced
apoptosis via Fas/caspase-8 pathway in melanoma cells. J Biol Chem.
281:11887–11893. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen G, Wang Y, Garate M, Zhou J and Li G:
The tumor suppressor ING3 is degraded by SCF(Skp2)-mediated
ubiquitin-proteasome system. Oncogene. 29:1498–1508. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T,
Imai H and Minami N: ING3 is essential for asymmetric cell division
during mouse oocyte maturation. PLoS One. 8:e747492013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Dai DL, Martinka M and Li G:
Prognostic significance of nuclear ING3 expression in human
cutaneous melanoma. Clin Cancer Res. 13:4111–4116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gunduz M, Ouchida M, Fukushima K, Ito S,
Jitsumori Y, Nakashima T, Nagai N, Nishizaki K and Shimizu K:
Allelic loss and reduced expression of the ING3, a candidate tumor
suppressor gene at 7q31, in human head and neck cancers. Oncogene.
21:4462–4470. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gunduz M, Beder LB, Gunduz E, Nagatsuka H,
Fukushima K, Pehlivan D, Cetin E, Yamanaka N, Nishizaki K, Shimizu
K and Nagai N: Downregulation of ING3 mRNA expression predicts poor
prognosis in head and neck cancer. Cancer Sci. 99:531–538. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Borkosky SS, Gunduz M, Beder L, Tsujigiwa
H, Tamamura R, Gunduz E, Katase N, Rodriguez AP, Sasaki A, Nagai N
and Nagatsuka H: Allelic loss of the ING gene family loci is a
frequent event in ameloblastoma. Oncol Res. 18:509–518. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu M, Chen F, Wang Q, Wang K, Pan Q and
Zhang X: Downregulation of inhibitor of growth 3 is correlated with
tumorigenesis and progression of hepatocellular carcinoma. Oncol
Lett. 4:47–52. 2012.PubMed/NCBI
|
|
14
|
Yang HY, Liu HL, Tian LT, Song RP, Song X,
Yin DL, Liang YJ, Qu LD, Jiang HC, Liu JR and Liu LX: Expression
and prognostic value of ING3 in human primary hepatocellular
carcinoma. Exp Biol Med (Maywood). 237:352–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shah S, Smith H, Feng X, Rancourt DE and
Riabowol K: ING function in apoptosis in diverse model systems.
Biochem Cell Biol. 87:117–125. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cengiz B, Gunduz E, Gunduz M, Beder LB,
Tamamura R, Bagci C, Yamanaka N, Shimizu K and Nagatsuka H:
Tumor-specific mutation and downregulation of ING5 detected in oral
squamous cell carcinoma. Int J Cancer. 127:2088–2094. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong W, Russell M, Suzuki K and Riabowol
K: Subcellular targeting of p33ING1b by phosphorylation-dependent
14-3-3 binding regulates p21WAF1 expression. Mol Cell Biol.
26:2947–2954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu L, Thakur S, Leong-Quong RY, Suzuki K,
Pang A, Bjorge JD, Riabowol K and Fujita DJ: Src regulates the
activity of the ING1 tumor suppressor. PLoS One. 8:e609432013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eapen SA, Netherton SJ, Sarker KP, Deng L,
Chan A, Riabowol K and Bonni S: Identification of a novel function
for the chromatin remodeling protein ING2 in muscle
differentiation. PLoS One. 7:e406842012. View Article : Google Scholar : PubMed/NCBI
|