Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide, and is a major cause of morbidity and mortality
(1). Specifically, CRC is the third
most frequent cancer observed in males and the second most frequent
cancer in females (2). CRC represents
~9% of all cancer cases (3).
Diagnosis of and therapy for CRC have advanced significantly over
the last ten years; however, 5-year survival rates remain poor
(4). Therefore, there is a
requirement for the development of novel therapeutic agents with a
high efficacy for the treatment of CRC.
The tumor suppressor protein p53, encoded by the
tumor protein p53 (TP53) gene, prevents the development and
progression of cancer through regulation of a range of cellular
mechanisms, including apoptosis, cell cycle arrest, metabolism and
DNA repair (5,6). In total ~50% of all human cancers have
mutations in TP53 that lead to the production of functionally
inactive p53 (7–9). In addition, these mutations frequently
induce tumor-promoting actions through dominant-negative
inactivation of the remaining wild-type TP53 allele, or apoptosis
resistance, increased tumor aggressiveness and metastatic potential
(6,10,11).
Furthermore, gain-of-function TP53 mutations may produce mutant p53
protein, which exhibits oncogenic effects, including increased cell
proliferation, invasion, metastasis and drug resistance, and
inhibition of apoptosis (12).
A previous study demonstrated that p53 is able to
regulate the expression of specific microRNAs (miRs), which
contributes to tumor suppression by controlling the expression of
the targets of these miRs, which include key effectors of numerous
cellular processes, for example cell cycle progression,
epithelial-mesenchymal transition, stemness, metabolism, cell
survival and angiogenesis (13).
Reciprocally, the activity of p53 has been observed to be under the
control of several miRNAs, including miR-125 and miR-504, which
were identified to negatively regulate p53 expression by directly
targeting the 3′untranslated region (UTR) of TP53 (14,15). In
addition, previous studies have reported that several miRNAs
(miR-29, miR-34a, miR-122, miR-192, miR-194 and miR-215) positively
regulate p53 expression (16–21). miRs are short non-coding RNAs of
between 18 and 24 nucleotides in length, which participate in
numerous biological pathways through regulating the
post-transcriptional expression of genes (22). In total ~1,400 miRs have been
identified in humans thus far, and miRs have been demonstrated to
serve important roles in the pathogenesis of disease (23). For example, miRs have been reported to
regulate the expression of their target mRNAs to affect tumor
growth, invasion and angiogenesis in cancer (24,25).
In the present study, bioinformatic analysis,
performed using TargetScan Software (version 6.2; www.targetscan.org/vert_61), identified that the
3′UTR of p53 mRNA contains a putative miR-600 binding site.
Subsequent ectopic overexpression of miR-600 in vitro using
lentiviral-mediated transduction was observed to decrease cell
proliferation, migration and invasion in mutant p53-expressing
human CRC cell lines, indicating that miR-600 targets p53 mRNA for
degradation in this context. Furthermore, overexpression of miR-600
inhibited the expression of matrix metalloproteinase 9 (MMP-9) and
promoted the expression of E-cadherin and β-catenin proteins.
Similar results were obtained when p53 expression was silenced
using small interfering RNA (siRNA). These results suggest that
targeting the miR-600/p53 network may lead to the identification of
novel therapeutic agents for the treatment of CRC.
Materials and methods
Cell culture
Human CRC cell lines (SW480, SW620 and DLD-1) and
human embryonic kidney cells (HEK293T) were obtained from the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). SW480 and DLD-1 cells were cultured in RPMI-1640 medium,
and SW620 and HEK293T cells were cultured in Dulbecco's modified
Eagle's medium (both Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). All media was supplemented with 10% fetal bovine
serum (FBS) and 100 U/ml penicillin and streptomycin (both
Invitrogen; Thermo Fisher Scientific, Inc.). All cells were
maintained at 37°C in an incubator with 5% CO2.
siRNA transfection
A siRNA was designed to target the human p53 gene
based on the public GenBank sequence (https://www.ncbi.nlm.nih.gov/genbank/) and was
purchased from GenePharma Co., Ltd. (Shanghai, China). The sequence
of p53 siRNA was 5′-GACUCCAGUGGUAAUCUAC-3′. The sequence of
scramble siRNA (control) was 5′-GCAACGGCAUUCCACCUUU-3′. SW620,
SW480 and DLD-1 cells (2×104 cells/well) were seeded
into 6-well plates and were transfected with 50 µM scramble siRNA
[Negative control (NC)] or p53-siRNA using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
Lentiviral vector construction for
miR-600 overexpression
Hsa-miR-600 sequence was obtained from miRBase
database (www.mirbase.org). For the construction
of the lentiviral vector expressing miR-600, miR-600 precursors and
their native context sequences (upstream and downstream flanking
genomic sequences) were amplified from HEK293T cell genomic DNA
using polymerase chain reaction (PCR) with the Takara Ex Taq™ Hot
Start Version kit (Takara Bio, Inc., Otsu, Japan). The PCR reaction
conditions were as follows: 94°C for 5 min, followed by 94°C for 45
sec, 60°C for 45 sec and 72°C for 1 min, for a total of 30 cycles;
a final step at 72°C for 10 min was subsequently performed. The
following primers were used for PCR: Forward-SalI,
5′-gcggtcgacTACTCCTTGATCCATTTCCAT-3′; and reverse-EcoRI,
5′-ggaattcaaaaaGGAACACTTCTTGCATTGTCT-3′ (uppercase letters indicate
the coding primer sequence; lowercase letters refer to the enzyme
loci and the protective bases). The resulting PCR product was
digested with SalI and EcoRI prior to insertion into
a human U6 promoter-containing pBluescript SK (+) plasmid. The
construct obtained was then digested with SalI and
EcoRI, then U6 promoter and subsequent pre-miR-600 genomic
fragments were cloned into the lentiviral plasmid pLUNIG as
previously described (26,27). A lentiviral vector expressing a short
hairpin (sh) RNA targeting firefly luciferase (shLuc target
sequence, 5′-TGCGCTGCTGGTGCCAACCCTATTCT-3′) was used as the
control, as previously described (26,27).
Vesicular stomatitis virus GP pseudotyped lentiviruses were
produced by co-transducing HEK293T cells with lentivirus expression
and packaging plasmids (pMD2.G, pMDL-G/P-RRE and pRSV-REV). A total
of 5×104 CRC cells (SW480, SW620 or DLD-1) were then
transduced with the lentiviruses (lentiviral vector expressing
miR-600 or control) in the presence of 8 µg/ml Polybrene
(Sigma-Aldrich; EMD Millipore, Billerica, MA, USA).
Cell viability assay
Cell viability was measured using the MTT assay.
Briefly, cells were seeded into 96-well plates at a density of
2×103 cells/well and cultured at 37°C for 1, 2, 3 or 4
days. At these time points, 20 µl of MTT (5 mg/ml) solution was
added to each well and cells were incubated for an additional 4 h
at 37°C. The formazan product was dissolved with dimethyl sulfoxide
and plates were incubated for 10 min at room temperature. A
microplate reader was subsequently used to measure the absorbance
at 490 nm. Each condition was determined in quintuplicate, and all
experiments were repeated ≥3 times.
Cell migration and invasion
assays
In vitro cell migration and invasion assays
were performed in 24-well Transwell cell culture chambers with 8 µm
pores (Costar; Corning Incorporated, Cambridge, MA, USA) according
to the manufacturer's instructions. For the migration assay, DLD-1,
SW480 and SW620 cells in starvation medium (without FBS)
(2.5×105/100 µl) were seeded into the Transwell filter
membrane chambers and incubated at 37°C for 16, 24 or 48 h,
respectively. The appropriate culture medium supplemented with 10%
FBS was added to the lower chambers as a chemoattractant. Following
incubation, cells in the lower chambers were fixed using 4%
paraformaldehyde and stained with 0.1% crystal violet solution.
Cells that did not migrate were removed from the upper chamber
surface using a cotton swab, and the number of cells that migrated
to the lower chamber was counted in ≥5 fields (fields were randomly
selected under a light microscope at magnification, ×20). For the
invasion assay, Transwell membranes were pre-coated with 10 µl of
Matrigel (diluted 1:3; BD Biosciences, San Jose, CA, USA) prior to
the process described above.
Western blot analysis
Cells were lysed in a radioimmunoprecipitation assay
lysis buffer (Sigma-Aldrich; EMD Millipore) containing a protease
inhibitor cocktail (Thermo Fisher Scientific, Inc.). The total
concentration of protein obtained was measured using the Pierce BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.). Total protein
(30 µg) was separated by SDS-PAGE using an 8 or 12% gel and
subsequently transferred onto polyvinylidene difluoride membranes
(EMD Millipore). Membranes were blocked in 5% skimmed milk (BD
Biosciences) for 1 h at room temperature and incubated with primary
antibodies overnight at 4°C. Subsequently, the membranes were
incubated with the appropriate horseradish peroxidase-conjugated
secondary antibody for 1 h at room temperature. Antibody binding
was detected using an enhanced chemiluminescence detection system
(GE Healthcare Life Sciences, Pittsburgh, PA, USA). The following
primary antibodies were used: Mouse anti-E-cadherin (diluted
1:3,000; #610181; BD Biosciences) and anti-vimentin (diluted
1:3,000; #5550513; BD Biosciences), rabbit anti-p53 (diluted 1:500;
ab31333; Abcam, Cambridge, UK), rabbit anti-Slug (diluted 1:1,000;
#9585) and anti-β-actin (diluted 1:1,000; #4970, Cell Signaling
Technology, Inc., Danvers, MA, USA), mouse anti-fibronectin
(diluted 1:100; SC18825) and anti-β-catenin (diluted 1:100; SC7199;
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), and rabbit
anti-MMP-9 (diluted 1:1,000; BS1241; Bioworld Technology, Inc., St.
Louis Park, MN, USA). The secondary antibody was horseradish
peroxidase-conjugated goat anti-rabbit (diluted 1:2,000; ab6721;
Abcam) or rabbit anti-mouse (diluted 1:2,000; ab6728; Abcam)
secondary antibody.
Dual-luciferase reporter assay
Putative targets of miR-600 were predicted using
TargetScan Software (version 6.2; www.targetscan.org/vert_61). DNA fragments
corresponding to the 3′UTR of TP53 mRNA containing the putative
miR-600 binding site (wt-P53-3′UTR-sense with SpeI site,
5′-CTAGTTACTGTGAGGGATGTTTGGGAGATGTAAGAAATGTTCTTGA-3′ and
wt-P53-3′UTR-antisense with HindIII site,
5′-AGCTTCAAGAACATTTCTTACATCTCCCAAACATCCCTCACAGTAA-3′), or a mutated
version of this site (mut-P53-3′UTR-sense with SpeI site,
5′-CTAGTTACTGTGAGGGATGTTTGGGAGATAGACGAAATGTTCTTGA-3′ and
mut-P53-3′UTR-antisense with HindIII site,
5′-AGCTTCAAGAACATTTCGTCTATCTCCCAAACATCCCTCACAGTAA-3′), were
chemically synthesized and cloned into the SpeI and
HindIII sites of pMIR-REPORT luciferase vector (Thermo
Fisher Scientific, Inc.). HEK293T cells were seeded into a 24-well
plate at a density of 3×105 cells/well and co-transduced
with pMIR-REPORT-wt-P53-3′UTR or pMIR-REPORT-mut-P53-3′UTR and
miR-600 expression plasmids, and a Renilla plasmid (RLSV40;
Promega Corporation, Madison, WI, USA) as internal normalization.
Cells were cultured at 37°C in an incubator with 5% CO2
for 48 h, and then cells were lysed using Passive Lysis Buffer
(Promega Corporation). Luciferase activity was detected using the
Dual-Luciferase Reporter Assay kit (Promega Corporation) according
to the manufacturer's instructions. Results were obtained from
three independent repeats.
Statistical analysis
One-way analysis of variance (ANOVA) and Student's
t-test were used to statistically compare differences between
groups. Post-hoc tests (Student-Newman-Keuls method) were
used following performance of ANOVA. Statistical analysis was
performed using SPSS software (version 15.0; SPSS, Inc., Chicago,
IL, USA). Results are presented as the mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-600 reduces CRC cell viability,
migration and invasion in vitro
To investigate the effects of miR-600 on CRC cell
growth, migration and invasion, SW480, SW620 and DLD-1 cells were
transduced with lentiviral vectors expressing miR-600 or the
control. The MTT assay was performed to determine cell viability
(Fig. 1). As shown in Fig. 1A, the viability of SW480 cells
transduced with miR-600-containing lentivirus was significantly
suppressed compared with the control group 3 and 4 days following
transduction (P<0.0001). Similar results were observed in SW620
and DLD-1 cells 3 and 4 days post-transduction (P<0.0001 vs. the
control group; Fig. 1A). The impact
of miR-600 overexpression on cell migration and invasion abilities
was measured using Transwell assays in vitro. This
identified that miR-600 overexpression significantly decreased
migration and invasion abilities of SW480 (P<0.0001; Fig. 1B), SW620 (P<0.0001; Fig. 1C) and DLD-1 (migration, P=0.0008 and
invasion, P=0.0001, Fig. 1D) cells
compared with the control group. These results suggest that the
overexpression of miR-600 inhibits proliferation, migration and
invasion in human CRC cells.
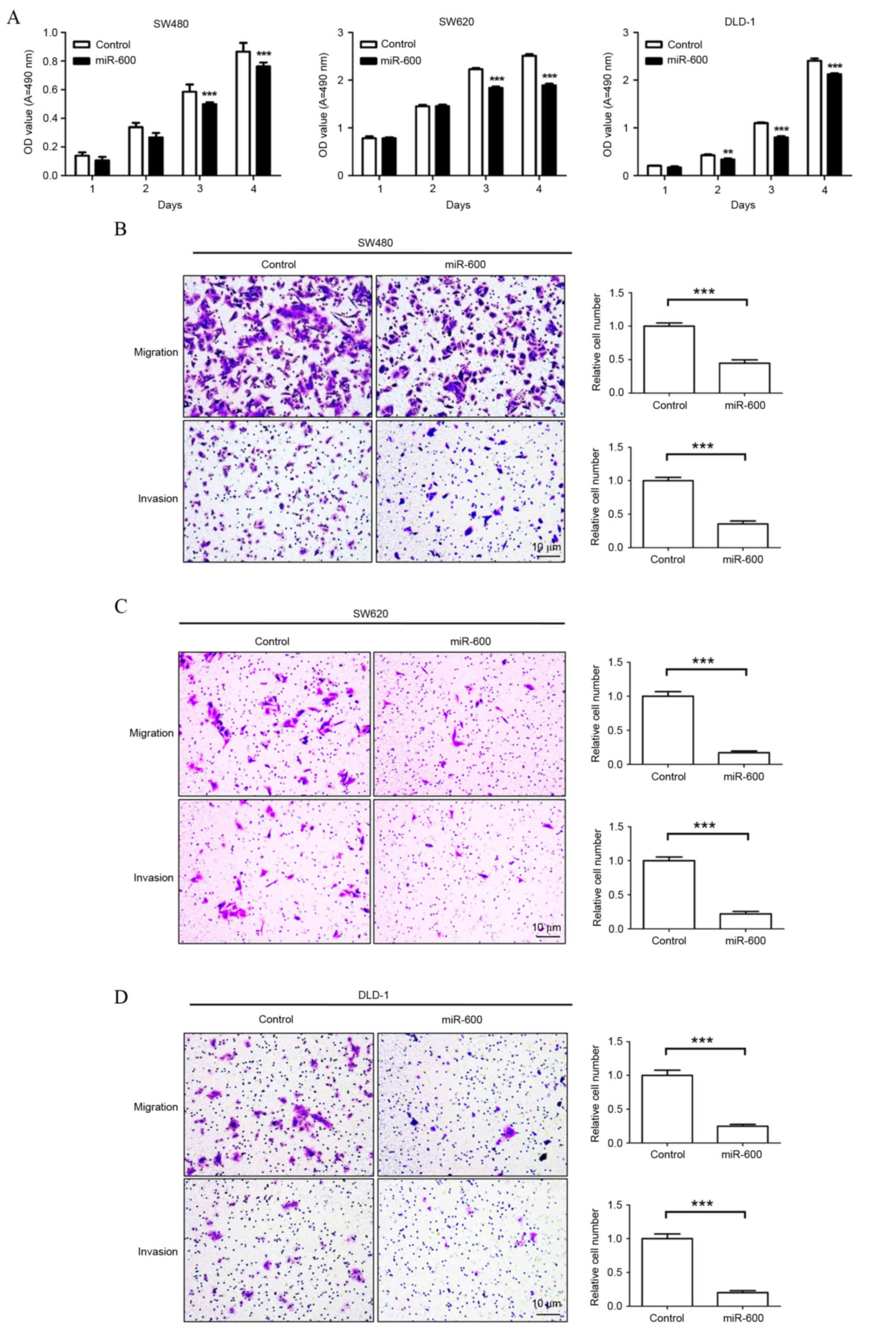 | Figure 1.Ectopic overexpression of miR-600
through lentiviral-mediated transduction inhibits CRC cell
proliferation, migration and invasion in vitro. (A) Effect
of miR-600 overexpression on the viability of CRC cell lines SW480,
SW620 and DLD-1. Effect of miR-600 on the migration and invasion
abilities of (B) SW480, (C) SW620 and (D) DLD-1 cells.
Magnification, ×200; Scale bars, 10 µm. **P<0.01, ***P<0.001
vs. the control group. CRC, colorectal cancer; miR, microRNA; OD,
optical density; A, absorbance. |
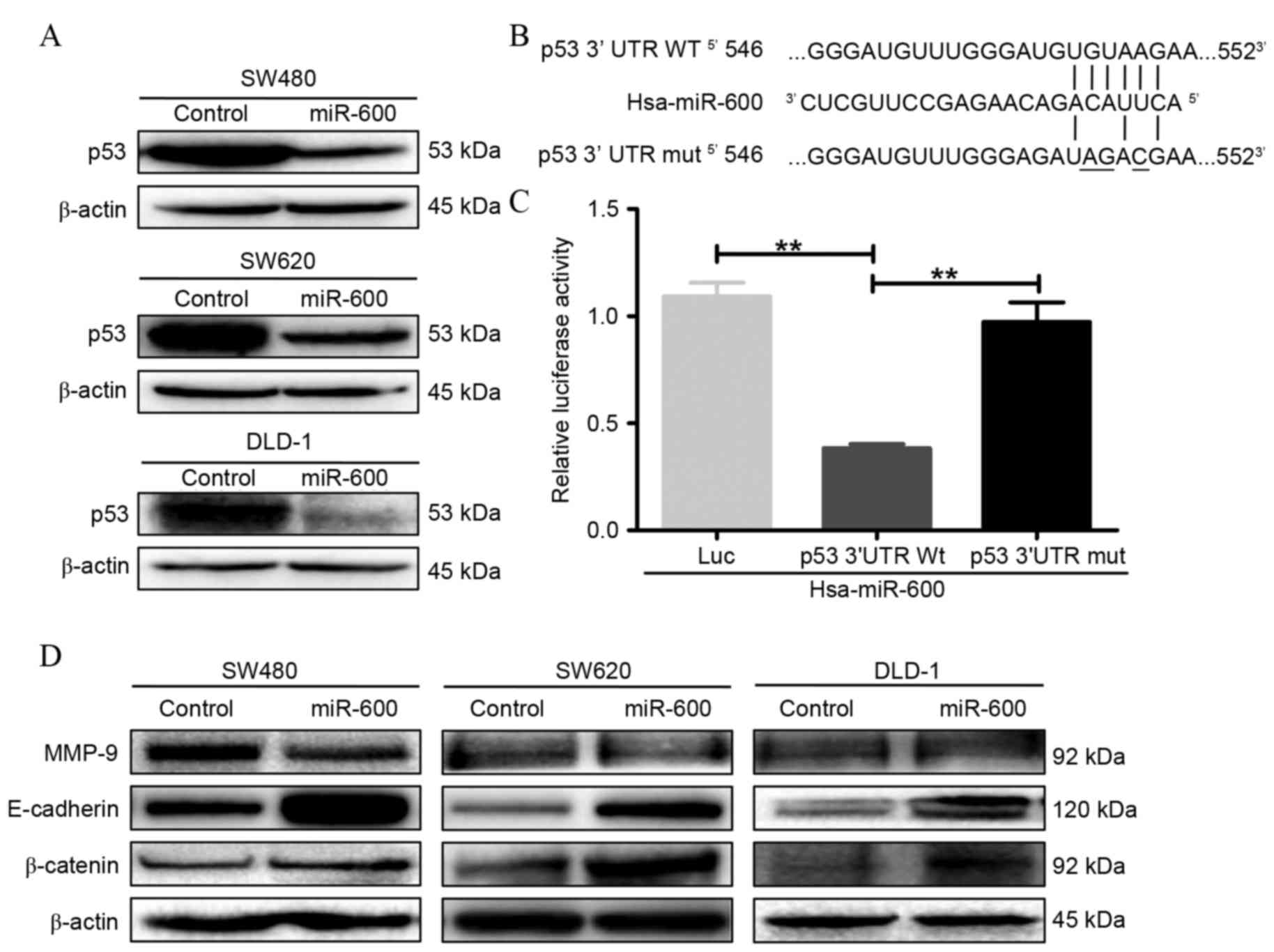
p53 is a direct target of miR-600 in
CRC cells
To identify the molecular target involved in miR-600
regulation of CRC cell proliferation, migration and invasion, the
effects of mutant p53 protein expression were investigated in CRC
cells. As shown in Fig. 2A,
overexpression of miR-600 by lentiviral-mediated transduction
inhibited p53 protein expression in SW480, SW620 and DLD-1 CRC
cells. To further investigate whether p53 is a direct target of
miR-600, bioinformatic analysis was performed using TargetScan
software. This revealed a putative miR-600 binding site in the
3′UTR of p53 mRNA (Fig. 2B).
A luciferase reporter assay was subsequently
performed to determine whether miR-600 directly targeted the 3′UTR
of p53 mRNA in vitro. A fragment of the 3′UTR of p53 with a
wild-type or mutant miR-600 target binding site was cloned into the
pMIR-REPORT luciferase reporter vector. The results of the
dual-luciferase reporter assay demonstrated that miR-600
significantly suppressed luciferase reporter activity at the
wild-type p53 3′UTR site (P=0.0044 vs. the control group), whereas
mutation of the miR-600 binding site blocked this suppressive
effect (P=0.0014 vs. wild-type site; Fig.
2C). These results indicate that miR-600 directly targets the
3′UTR of p53 mRNA to downregulate the expression of p53
protein.
miR-600 modulates
epithelial-mesenchymal transition (EMT)-associated protein
expression by targeting p53 mRNA in mutant p53-expressing human CRC
cell lines
The expression levels of proteins associated with
EMT proteins were detected through western blotting (Fig. 2D). Levels of the epithelial cell
marker proteins E-cadherin and β-catenin, were increased in CRC
cell lines (SW480, SW620 and DLD-1) overexpressing miR-600. In
addition, protein levels of MMP-9, a member of the MMP family,
appeared to be decreased in CRC cells overexpressing miR-600.
siRNA silencing of mutant p53
expression reduces cell viability, migration and invasion in mutant
p53-expressing human CRC cell lines
The effects of p53 mutation on CRC cell viability,
migration and invasion were further investigated in vitro
using the MTT assay, and Transwell migration and invasion assays,
respectively. These assays demonstrated that knockdown of mutant
p53 expression in SW480, SW620 and DLD-1 cells significantly
decreased cell viability compared with the control group at 3 and 4
days post-transfection (P<0.001; Fig.
3A). In addition, the knockdown of mutant p53 expression in
SW480 cells significantly reduced cell migration (91.6%) and
invasion (75.6%) abilities (P<0.0001 vs. the control group;
Fig. 3B). Similarly, migration and
invasion were decreased by 79.3% (P=0.0003) and 84.2% (P=0.0019),
respectively, in SW620 cells (Fig.
3C), and 78.9% (P<0.0001) and 72.2% (P=0.0003),
respectively, in DLD-1 cells (P<0.001; Fig. 3D) compared with the control group.
These results indicate that knockdown of mutant p53 expression in
CRC cells reduces their viability, and inhibits their ability to
migrate and invade.
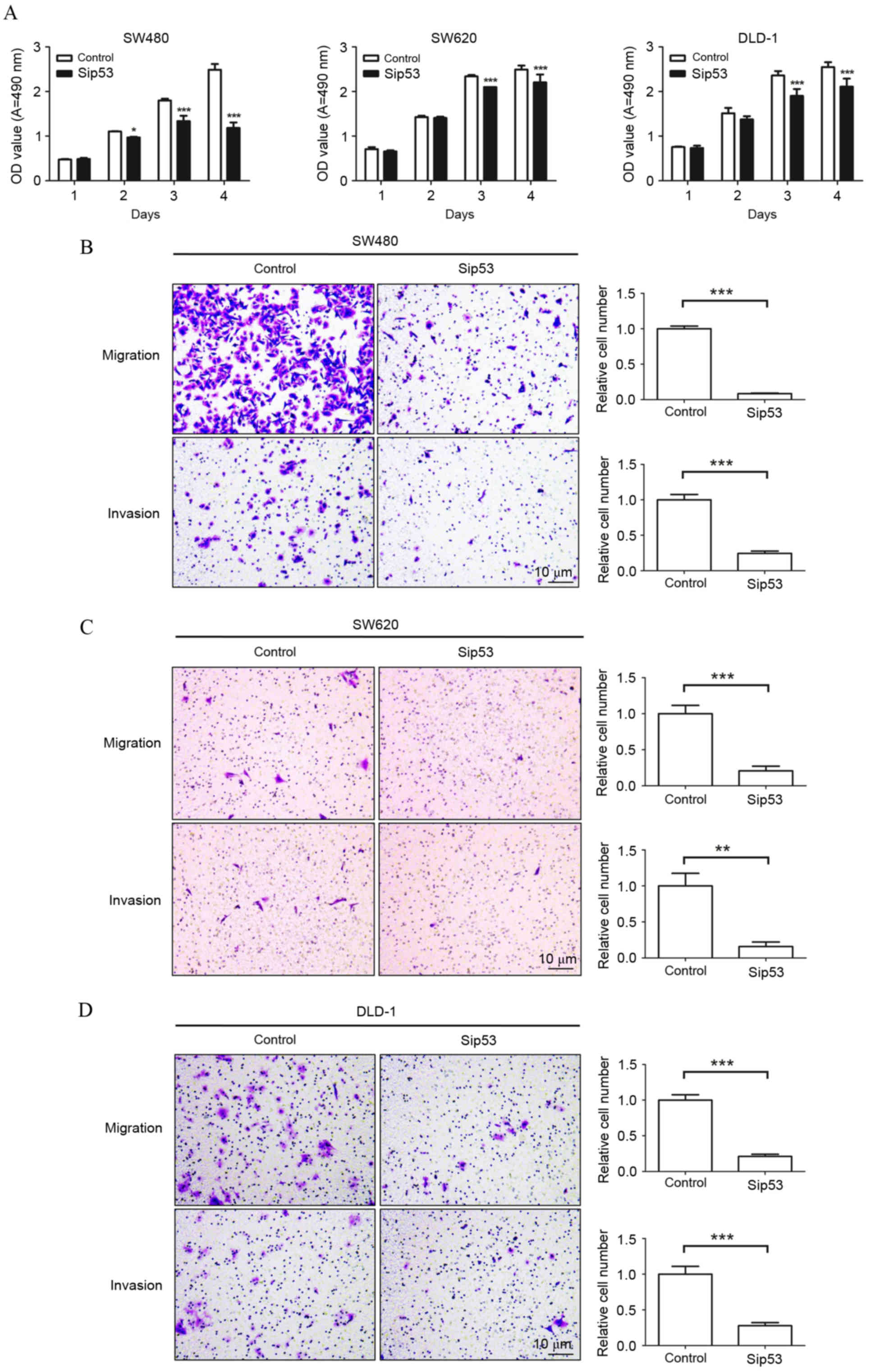 | Figure 3.siRNA silencing of p53 expression
inhibits colorectal cancer cell proliferation, migration and
invasion. (A) The MTT assay was performed to determine the
viability of SW480, SW620 and DLD-1 cells transfected with Sip53. A
total of 50 µM negative control siRNAs or p53 siRNAs were
transfected into SW480, SW620 and DLD-1 cells for 48 h. siRNA
silencing of p53 expression significantly inhibited (B) SW480, (C)
SW620 and (D) DLD-1 cell migration and invasion in a Transwell
assay compared to the control group. Magnification, ×200; Scale
bars, 10 µm. *P<0.05, **P<0.01, ***P<0.001 vs. the control
group. siRNA, small interfering RNA; Sip53, p53-specific siRNA; OD,
optical density; A, absorbance. |
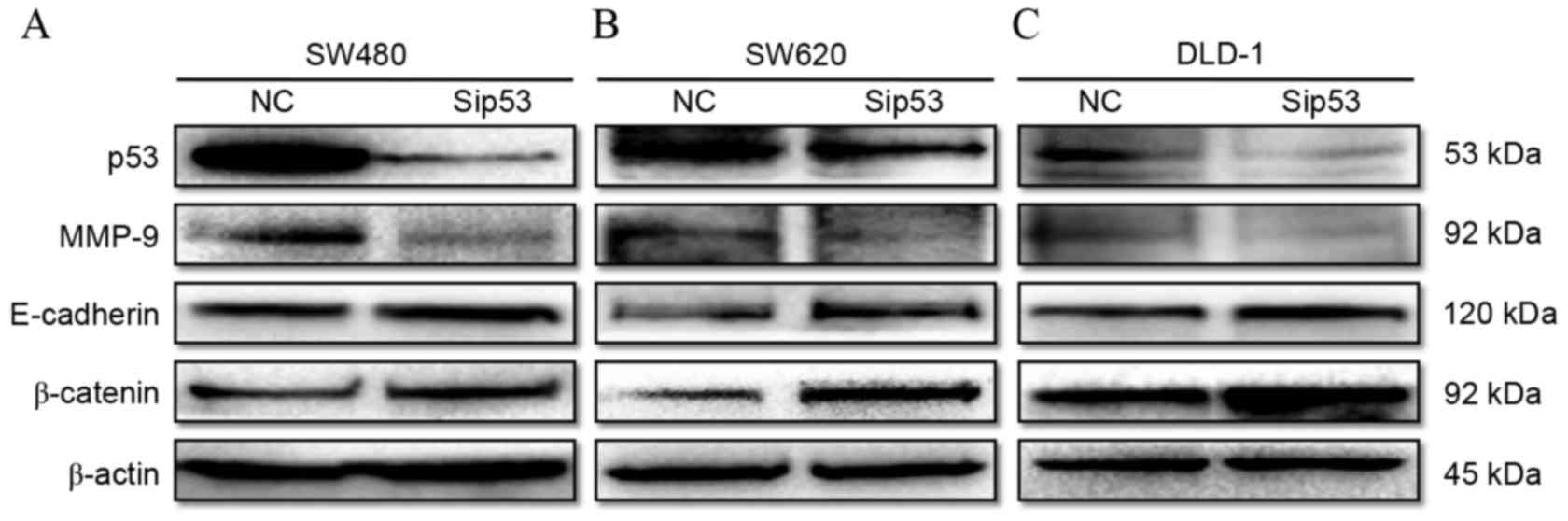
siRNA silencing of mutant p53
expression inhibits the expression of MMP-9 and promotes the
expression of epithelial cell markers in mutant p53-expressing
human CRC cell lines
To evaluate the effect of p53 on the E-cadherin,
β-catenin and MMP-9 protein expression, p53-specific (sip53) and
negative control (NC) siRNAs were transfected into mutant
p53-expressing CRC cell lines (SW480, SW620 and DLD-1). The results
demonstrate that p53 protein expression was markedly decreased 48 h
following siRNA transfection in all cell lines compared with the
cells transfected with the negative control (Fig. 4). Simultaneously, MMP-9 protein
expression was decreased, while E-cadherin and β-catenin protein
expression was increased in SW480 (Fig.
4A), SW620 (Fig. 4B) and DLD-1
(Fig. 4C) cells transfected with
siRNA compared with the negative control cells.
Discussion
Mutation of TP53 is one of the most frequent genetic
alterations in all cancer types, including CRC (28). Although mutation of TP53 may lead to
loss of wild-type p53 activity, some mutated forms of p53 may
result in the gain of oncogenic properties that promote tumor
growth and progression (29). The
ability of mutant p53 to drive enhanced cell growth, invasion and
motility has been confirmed in vitro and in xenograft models
(28–32). Previous studies have demonstrated that
when endogenous expression of mutant p53 was knocked out, the
proliferative capacity and chemoresistance of a number of human
cancer cell lines decreased, while in nude mice tumorigenicity was
also reduced (33,34). In total, ~50% of all human cancer is
caused by p53 mutations (35).
Previous studies have indicated that miRs have
important roles in the pathogenesis of cancer, including the
regulation of tumor proliferation, invasion, metastasis and
angiogenesis through altering the expression levels of target mRNAs
(24,25). It has been reported that ≥1,000 miRs
are found in the human genome and that the expression ~30% of the
human genome is regulated by miRs (36), highlighting the possibility of the
existence of a specific miR that directly regulates p53 protein
expression and its subsequent function. In the present study,
miR-600 was identified as a direct negative regulator of p53
through its binding to the 3′UTR of p53 mRNA. In addition, it was
demonstrated that overexpression of miR-600 repressed the
endogenous level of p53 protein and inhibited cell proliferation,
migration and invasion in mutant p53-expressing human CRC cell
lines (SW480, SW620 and DLD-1) in vitro. Knockdown of
endogenous mutant p53 by siRNA has been reported to reduce cell
growth and chemoresistance in vitro in a number of human
cancer cell lines, and inhibit tumor growth in nude mice (33,34).
Furthermore, silencing of mutant p53 by siRNA was able to induce
cell cycle arrest and apoptosis in human prostate (37) and bladder cancer (38) cells. In the present study, siRNA
silencing of p53 produced a similar phenotype to miR-600
overexpression in SW480, SW620 and DLD-1 cells.
MMP family proteins are associated with
cardiovascular disease and cancer metastasis, and regulated by p53
(39). It has been reported that
wild-type p53 is able to suppress the expression of MMP-9 (40,41).
However, a number of mutant p53 proteins do not possess this
ability. Prior research has revealed that MMP-9 is able to reduce
the expression of E-cadherin (42).
In the current study, overexpression of miR-600 and p53 knockdown
were observed to suppress MMP-9 expression, and promote the
expression of E-cadherin and β-catenin proteins. E-cadherin and
β-catenin are important for epithelial cell-cell adhesion in the
normal epithelium (43). Loss of
E-cadherin is an important event in EMT during tumorigenesis
(44). A previous study identified
that mutant p53 downregulated E-cadherin expression in the CRC cell
line HCT116 (44). Dong et al
(45) reported that mutant
gain-of-function p53 induces EMT through modulation of the
miR-130b-zinc finger E-box binding homeobox 1 axis in endometrial
cancer cells. The current study observed that inhibition of mutant
p53 expression, through overexpression of miR-600 or by siRNA
silencing of mutant p53, induced E-cadherin expression in mutant
p53-expressing human CRC cell lines.
In conclusion, the present study identified that
miR-600 is a direct negative regulator of p53. Overexpression of
miR-600 or siRNA silencing of mutant p53 were observed to inhibit
proliferation, migration and invasion, suppress MMP-9 expression,
and promote the expression of E-cadherin and β-catenin in mutant
p53-expressing human CRC cell lines. The results of the current
study increase the current understanding of the function of miRs in
the upstream regulation of p53. In addition, these results indicate
that targeting mutant p53 through overexpression of miR-600 is a
promising therapeutic strategy for the treatment of CRCs bearing
p53 mutations.
Acknowledgments
The present study was supported by the Zhejiang
Province Natural Science Foundation of China (grant nos.
LY15H160057 and LY17H160056), the National Natural Science
Foundation of China (grant no. 81672385) and Wenzhou Science and
Technology Bureau Program (grant no. Y20140667).
References
|
1
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boyle P and Langman J: ABC of colorectal
cancer: Epidemiology. BMJ. 321:805–808. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lech G, Slotwiński R, Słodkowski M and
Krasnodębski IW: Colorectal cancer tumour markers and biomarkers:
Recent therapeutic advances. World J Gastroenterol. 22:1745–1755.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kruse JP and Gu W: Modes of p53
regulation. Cell. 137:609–622. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oren M and Rotter V: Mutant p53
gain-of-function in cancer. Cold Spring Harb Perspect Biol.
2:a0011072010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kato S, Han SY, Liu W, Otsuka K, Shibata
H, Kanamaru R and Ishioka C: Understanding the function-structure
and function-mutation relationships of p53 tumor suppressor protein
by high-resolution missense mutation analysis. Proc Natl Acad Sci
USA. 100:8424–8429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Milner J and Medcalf E: Cotranslation of
activated mutant p53 with wild type drives the wild-type p53
protein into the mutant conformation. Cell. 65:765–774. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Milner J, Medcalf EA and Cook AC: Tumor
suppressor p53: Analysis of wild-type and mutant p53 complexes. Mol
Cell Biol. 11:12–19. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oren M and Rotter V: Mutant p53
gain-of-function in cancer. Cold Spring Harb Perspect Biol.
2:a0011072010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hermeking H: MicroRNAs in the p53 network:
Micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B,
Korzh V, Lodish HF and Lim B: MicroRNA-125b is a novel negative
regulator of p53. Genes Dev. 23:862–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song
JS, Tang LH, Levine AJ and Feng Z: Negative regulation of tumor
suppressor p53 by microRNA miR-504. Mol Cell. 38:689–699. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng Z, Zhang C, Wu R and Hu W: Tumor
suppressor p53 meets microRNAs. J Mol Cell Biol. 3:44–50. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park SY, Lee JH, Ha M, Nam JW and Kim VN:
miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat
Struct Mol Biol. 16:23–29. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamakuchi M and Lowenstein CJ: MiR-34,
SIRT1 and p53: The feedback loop. Cell Cycle. 8:712–715. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song B, Wang Y, Kudo K, Gavin EJ, Xi Y and
Ju J: miR-192 Regulates dihydrofolate reductase and cellular
proliferation through the p53-microRNA circuit. Clin Cancer Res.
14:8080–8086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pichiorri F, Suh SS, Rocci A, De Luca L,
Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder
H, et al: Downregulation of p53-inducible microRNAs 192, 194, and
215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma
development. Cancer Cell. 18:367–381. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Manfe V, Biskup E, Rosbjerg A, Kamstrup M,
Skov AG, Lerche CM, Lauenborg BT, Odum N and Gniadecki R: miR-122
regulates p53/Akt signalling and the chemotherapy-induced apoptosis
in cutaneous T-cell lymphoma. PloS One. 7:e295412012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stahlhut C and Slack FJ: MicroRNAs and the
cancer phenotype: Profiling, signatures and clinical implications.
Genome Med. 5:1112013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang L, Lai YK, Zhang J, Wang H, Lin MC,
He ML and Kung HF: Targeting S100P inhibits colon cancer growth and
metastasis by Lentivirus-mediated RNA interference and proteomic
analysis. Mol Med. 17:709–716. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Lin MC, Yao H, Wang H, Zhang AQ,
Yu J, Hui CK, Lau GK, He ML, Sung J and Kung HF:
Lentivirus-mediated RNA interference targeting enhancer of zeste
homolog 2 inhibits hepatocellular carcinoma growth through
down-regulation of stathmin. Hepatology. 46:200–208. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li XL, Zhou J, Chen ZR and Chng WJ: P53
mutations in colorectal cancer-molecular pathogenesis and
pharmacological reactivation. World J Gastroenterol. 21:84–93.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou G, Wang J, Zhao M, Xie TX, Tanaka N,
Sano D, Patel AA, Ward AM, Sandulache VC, Jasser SA, et al:
Gain-of-function mutant p53 promotes cell growth and cancer cell
metabolism via inhibition of AMPK activation. Mol Cell. 54:960–974.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lenfert E, Maenz C, Heinlein C, Jannasch
K, Schumacher U, Pantel K, Tolstonog GV, Deppert W and Wegwitz F:
Mutant p53 promotes epithelial-mesenchymal plasticity and enhances
metastasis in mammary carcinomas of WAP-T mice. Int J Cancer.
136:E521–E533. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ji L, Xu J, Liu J, Amjad A, Zhang K, Liu
Q, Zhou L, Xiao J and Li X: Mutant p53 promotes tumor cell
malignancy by both positive and negative regulation of the
transforming growth factor β (TGF-β) pathway. J Biol Chem.
290:11729–11740. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Arjonen A, Kaukonen R, Mattila E, Rouhi P,
Högnäs G, Sihto H, Miller BW, Morton JP, Bucher E, Taimen P, et al:
Mutant p53-associated myosin-X upregulation promotes breast cancer
invasion and metastasis. J Clin Invest. 124:1069–1082. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bossi G, Marampon F, Maor-Aloni R, Zani B,
Rotter V, Oren M, Strano S, Blandino G and Sacchi A: Conditional
RNA interference in vivo to study mutant p53 oncogenic gain of
function on tumor malignancy. Cell Cycle. 7:1870–1879. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bossi G, Lapi E, Strano S, Rinaldo C,
Blandino G and Sacchi A: Mutant p53 gain of function: Reduction of
tumor malignancy of human cancer cell lines through abrogation of
mutant p53 expression. Oncogene. 25:304–309. 2006.PubMed/NCBI
|
|
35
|
Khoo KH, Verma CS and Lane DP: Drugging
the p53 pathway: Understanding the route to clinical efficacy. Nat
Rev Drug Discov. 13:217–236. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu H, Mao Q, Lin Y, Yang K and Xie L: RNA
interference targeting mutant p53 inhibits growth and induces
apoptosis in DU145 human prostate cancer cells. Med Oncol.
28:(Suppl 1). S381–S387. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu HB, Yang K, Xie YQ, Lin YW, Mao QQ and
Xie LP: Silencing of mutant p53 by siRNA induces cell cycle arrest
and apoptosis in human bladder cancer cells. World J Surg Oncol.
11:222013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cohen M, Wuillemin C, Irion O and Bischof
P: Regulation of MMP-9 by p53 in first trimester cytotrophoblastic
cells. Hum Reprod. 23:2273–2281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martelli M, Campana A and Bischof P:
Secretion of matrix metalloproteinases by human endometrial cells
in vitro. J Reprod Fertil. 98:67–76. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu J, Zhan M, Hannay JA, Das P, Bolshakov
SV, Kotilingam D, Yu D, Lazar AF, Pollock RE and Lev D: Wild-type
p53 inhibits nuclear factor-kappaB-induced matrix
metalloproteinase-9 promoter activation: Implications for soft
tissue sarcoma growth and metastasis. Mol Cancer Res. 4:803–810.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nawrocki-Raby B, Gilles C, Polette M,
Martinella-Catusse C, Bonnet N, Puchelle E, Foidart JM, Van Roy F
and Birembaut P: E-cadherin mediates MMP down-regulation in highly
invasive bronchial tumor cells. Am J Pathol. 163:653–661. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chaw SY, Majeed AA, Dalley AJ, Chan A,
Stein S and Farah CS: Epithelial to mesenchymal transition (EMT)
biomarkers-E-cadherin, beta-catenin, APC and Vimentin-in oral
squamous cell carcinogenesis and transformation. Oral Oncol.
48:997–1006. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Roger L, Jullien L, Gire V and Roux P:
Gain of oncogenic function of p53 mutants regulates E-cadherin
expression uncoupled from cell invasion in colon cancer cells. J
Cell Sci. 123:1295–1305. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dong P, Karaayvaz M, Jia N, Kaneuchi M,
Hamada J, Watari H, Sudo S, Ju J and Sakuragi N: Mutant p53
gain-of-function induces epithelial-mesenchymal transition through
modulation of the miR-130b-ZEB1 axis. Oncogene. 32:3286–3295. 2013.
View Article : Google Scholar : PubMed/NCBI
|