Introduction
Récepteur d'origine nantais (RON) is a receptor
tyrosine kinase and a member of the hepatocyte growth factor
receptor proto-oncogene family (1).
The RON gene encodes the macrophage-stimulating protein (MSP) 1
receptor (1). The activation of RON
by MSP induces tumor progression and invasion, whereas the
inactivation of RON promotes cell apoptosis (2–4). RON is a
heterodimeric protein with a disulfide bond linking a 40-kDa α
chain, containing the extracellular domain for ligand binding, and
a 150-kDa β chain, incorporating the intracellular kinase domain
and a transmembrane domain (5,6). The
binding of MSP to RON activates the intrinsic kinase activity and
induces autophosphorylation of the tyrosine residues in its kinase
domain (7). Alternative splicing of
the RON pre-mRNA produces various proteins, each with distinctive
functions (8). RONΔ165, which is
produced by the exclusion of exon 11, has previously been observed
to accumulate in breast and colon cancer cells, where it acts to
stimulate invasive cell growth and metastasis (9,10).
Previous studies have demonstrated that serine/arginine-rich
splicing factor (SRSF) 1 and heterogeneous nuclear
ribonucleoprotein A1 regulate the epithelial-to-mesenchymal
transition through the regulation of exon 11 alternative splicing
in RON pre-mRNA (10,11). Our previous study also identified
SRSF2 as an important regulator protein for the alternative
splicing of exon 11 (12). Another
isoform, RONΔ160, is produced by the exclusion of exons 5 and 6 and
lacks the first immunoglobulin-plexin-transcription domain in the
extracellular β chain, and therefore induces cellular
transformation, metastasis and protects tumor cells from apoptosis
(13–15). The mechanisms underlying the
alternative splicing of exon 5 and 6 require further
exploration.
Pre-mRNA splicing is required for gene expression in
higher eukaryotes, as introns must be removed from pre-mRNA and
protein-coding exons must be ligated (16). mRNA splicing is a two-step procedure.
Initially, the 5′ splice site is cleaved and two splicing
intermediates are generated, a linear first exon and an
intron-second exon RNA species in a lariat configuration (17); a 2′-5′ phosphodiester bond is formed
between the guanine at the 5′ splice site and the 2′ hydroxyl of an
adenine at a branch point (18).
Secondly, the 3′ splice site is cleaved and the two exons are
ligated together to generate the spliced mRNA (19). Nonsense-mediated mRNA decay (NMD) is a
method of eliminating aberrant mRNA transcripts that contain
premature termination codons (PTCs) (20). If PTCs are included in mRNA, or
partially spliced mRNA, through alternative splicing, then NMD may
occur (21). NMD has a role in the
quantitative posttranslational regulation of gene expression
through specific alternative splicing (22,23);
however, the underlying regulatory mechanisms require further
studies to be fully elucidated.
The results indicated that PTCs at specific
locations affect the splicing and NMD of various isoforms to
alternate degrees. These results may be applicable in treating
certain types of cancer, decreasing cancer-specific or
metastasis-specific RNA isoforms via inducing PTCs using gene
editing.
Materials and methods
Plasmid construction
RON exon 4–7 sequences were amplified from human
genomic DNA using E4Bam.F and E7Xho.R primers (Table I); the products were cloned into a
pcDNA3.1 (+) vector at BamHI and XhoI cleavage sites
to produce a minigene. E5/SC-1, E5/SC-2, E6-7/SC-1 and E6-7/SC-2
constructs were made using overlapping polymerase chain reaction
(PCR); the aforementioned minigene provided the template, and
following primers were used: E5-M1.F/E5-M1.R for the E5/SC-1
minigene, E5-M2.F/E5-M2.R for the E5/SC-2 minigene,
E6Tins.F/E6Tins.R and E7Gdel.F/E7Gdel.R for the E6-7/SC-1 minigene,
and E6Ains.F/E6Ains.R and E7Tdel.F/E7Tdel.R for the E6-7/SC-2
minigene. The PCR cycling settings were as follows: 5 min at 95°C,
followed by 30 cycles of denaturation for 15 sec at 95°C and
annealing for 1 min at 60°C. A 0.5 unit Taq DNA polymerase (Cosmo
Genetech Co., Ltd., Seoul, Korea) and 0.2 mM dNTPs (Cosmo Genetech
Co., Ltd.) were used in the PCR reaction. All of the primer
sequences are listed in Table I.
 | Table I.Primers used for PCR and RT-PCR. |
Table I.
Primers used for PCR and RT-PCR.
| Name | Sequence, 5′-3′ |
|---|
| Construct |
|
|
E4Bam.F |
GTTAGGGATCCGTTTTCCAGGTACCTATCCAAG |
|
E7Xho.R |
ATTACCTCGAGCGTGCTAGCAGACACTCAGTC |
|
E5-M1.F |
CTGACCCTGTGAGGCTCCAACTTC |
|
E5-M1.R |
GAAGTTGGAGCCTCACAGGGTCAG |
|
E5-M2.F |
TCTGGTGCCTTAGGGAACCC |
|
E5-M2.R |
GGGTTCCCTAAGGCACCAGA |
|
E6Tins.F |
GAGGCTTCTCTTTTCATGGTG |
|
E6Tins.R |
CACCATGAAAAGAGAAGCCTC |
|
E7Gdel.F |
TCTTAGGAGCCATGCTGATAG |
|
E7Gdel.R |
TATCAGCATGGCTCCTAAGAG |
|
E6Ains.F |
CCACCGGGCAAAGCACTTCC |
|
E6Ains.R |
GGAAGTGCTTTGCCCGGTGG |
|
E7Tdel.F |
CAACCCCTCTTGGCCCACGG |
|
E7Tdel.R |
CCGTGGGCCAAGAGGGGTTG |
| RT-PCR |
|
|
Exon4.F |
GTTTTCCAGGTACCTATCCAAG |
|
pcDNA.R |
TCCACCACCCTGTTGCTGTA |
Cell culture and transfection
MDA MB 231 cells (ATCC, Manassas, VA, USA) were
grown in RPMI 1640 medium (HyClone; GE Healthcare Life Sciences,
Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS)
(HyClone; GE Healthcare Life Sciences) at 37°C with 5%
CO2. Polyethylenimine (PEI; Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) was used to transfect the minigenes
into the cells.
Reverse transcription (RT)-PCR
A RiboEx reagent (GeneAll Biotechnology Co., Ltd.,
Seoul, Korea) was used to extract the total RNA from the cells. A
total of 40 units ImProm-II™ reverse transcriptase (Promega
Corporation, Madison, WI, USA) was used to reverse transcribe 1 µg
RNA with 100 pmol oligo dT and 0.2 mM each dNTP in 20 µl reaction.
The RT reaction product (1 µl) was amplified by PCR using G-Taq
polymerase (Cosmo Genetech Co., Ltd.) and the following PCR
settings: Denaturing for 5 min at 95°C, followed by 32 cycles of
denaturation for 15 sec at 95°C and annealing for 1 min at 60°C.
The spliced products of the minigenes were detected with Exon4.F
and pcDNA.R primers. The PCR products were loaded onto 1% agarose
gel and run at 100 V for 30 min. following EtBr staining, the DNA
bands were cut and dissolved using a Gel Extraction kit (Cosmo
Genetech Co., Ltd.), then sent for sequencing to confirm the
identity of the bands. All of the primer sequences are listed in
Table I.
Results
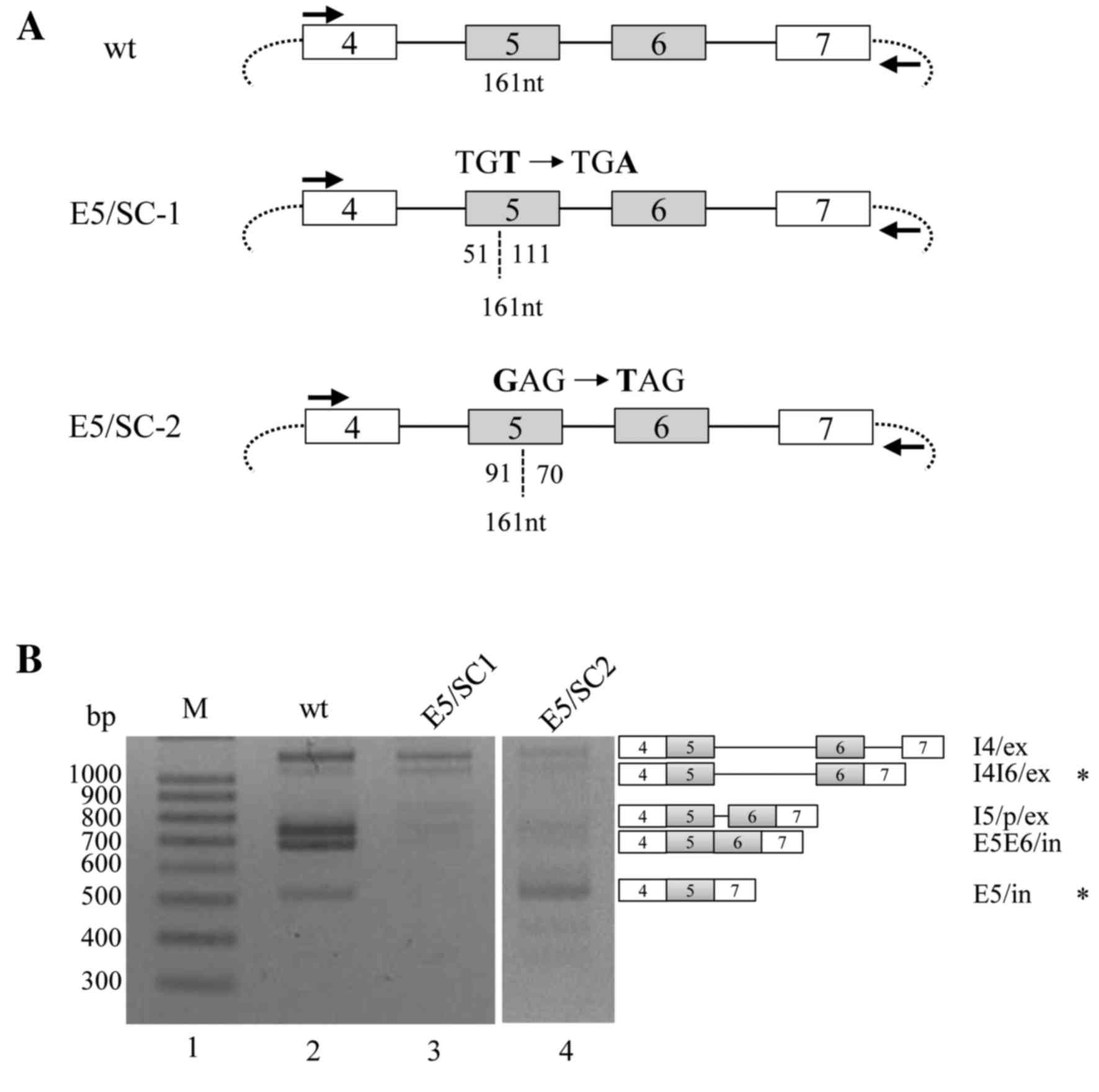
PTCs in exon 5 do not cause
degradation of all spliced and partially spliced isoforms from RON
pre-mRNA
To observe the effects of PTCs on RON pre-mRNA
splicing, a minigene was constructed, in which a TGA stop-codon was
produced through a T-A point mutation, located 51 nucleotides (nt)
downstream from the 3′ splice-site of exon 5 (E5/SC-1; Fig. 1A). RT-PCR analysis of the wild type
(WT) minigene revealed that the following isoforms were produced:
An isoform in which intron 4 is spliced but other introns are not
spliced (I4/ex); an isoform in which intron 4 and intron 6 are
spliced but intron 5 is not spliced (I4I6/ex); an isoform in which
cryptic spliced site in intron 5 activated (I5/p/ex) (24); an isoform in which both exon 5 and 6
are included (E5E6/in); and an isoform in which exon 5 is included
but exon 6 is excluded (E5/in; lane 2; Fig. 1B). The results of the current study
were consistent with the prior hypothesis that PTCs in exons induce
and regulate the NMD signaling pathway. RT-PCR analysis of the
E5/SC-1 minigene revealed that I4/ex was markedly decreased and the
I5/p/ex, E5E6/in and E5/in isoforms were almost absent. However,
the expression of the I4I6/ex isoform from this mutated minigene
was not observed to differ from that of the WT product. The results
of the current study indicate that, although most isoforms were
reduced significantly by PTCs on exon 5, a partially spliced
isoform still survived in the mutant minigene.
Different PTC locations in exon 5 have
various effects on the NMD of the spliced and partially spliced
isoforms of RON pre-mRNA
It was hypothesized that changing the location of
the PTCs in RON exon 5 may affect the alternative splicing of exons
5 and 6 in RON pre-mRNA. A second mutant minigene was constructed,
in which a stop-codon was produced 91 nt downstream from the 3′
splice-site of exon 5, through a G-T point mutation (E5/SC-2;
Fig. 1A). It was observed that the
majority of the exon isoforms, including I4/ex, I4/I6/ex, I5/p/ex
and E5E6/in, were markedly reduced in the E5/SC-2 minigene
(Fig. 1B; lane 4); however, the E5/in
isoform was not reduced in this mutant. Thus, the E5/in isoform was
resistant to degradation by the PTC-mediated NMD signaling pathway.
The results (Fig. 1B) indicate that,
although PTCs on exon 5 induced a marked decrease in the majority
of the spliced or partially spliced mRNA isoforms, specific
isoforms may still survive the PTC-induced NMD. However, the
identities of the surviving isoforms are dependent on the location
of the PTCs in the RON pre-mRNA.
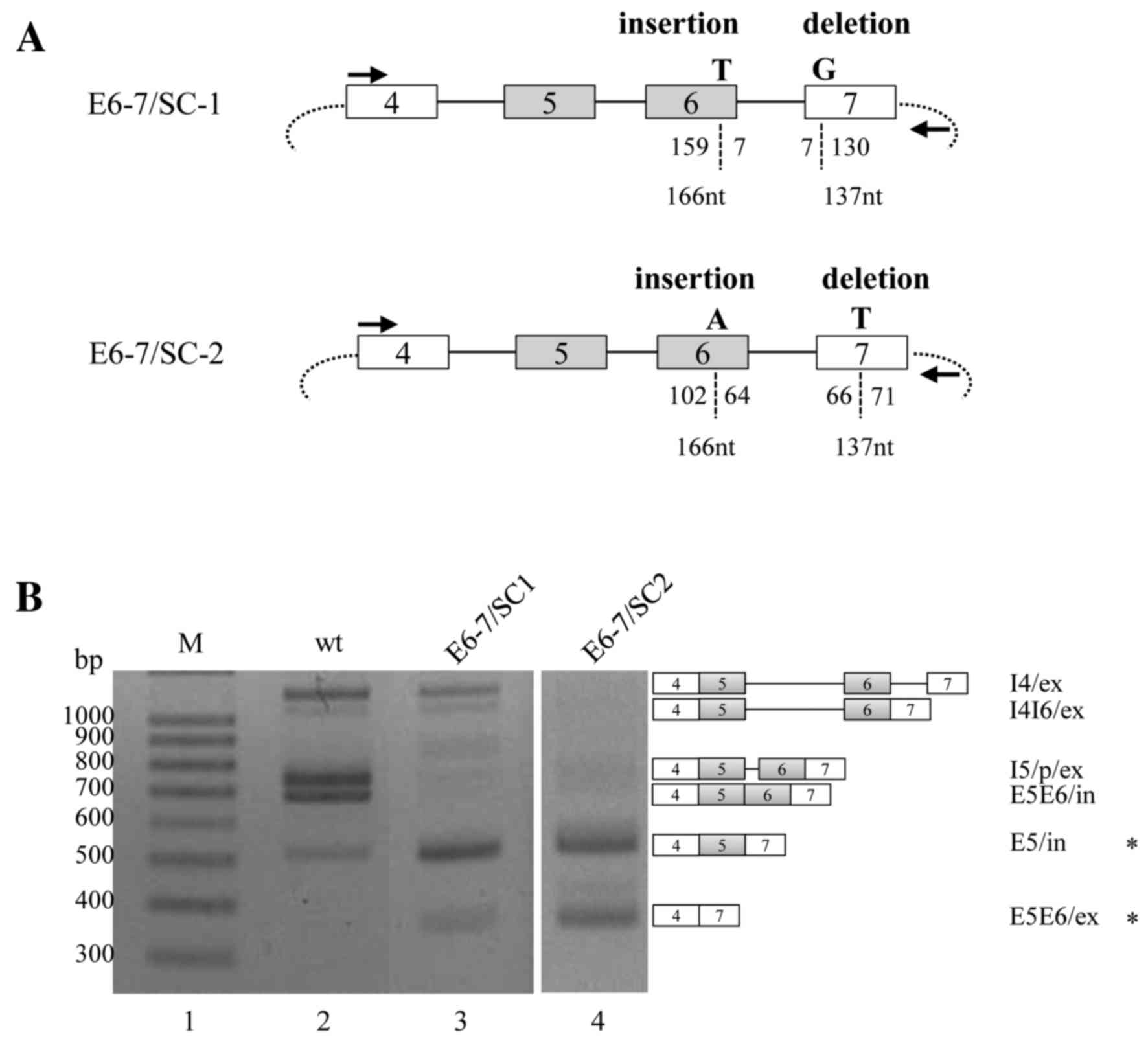
PTCs produced by inserting T or A nts
do not induce NMD of specific isoforms
The current study investigated whether the PTCs
produced by inserting a T residue in exon 6 may have similar
effects on RON pre-mRNA splicing. To ensure that the spliced
product was still capable of encoding the protein, a T residue was
also deleted from exon 7. In the E6-7/SC-1 minigene, the inserted T
was located 7-nt upstream from the 5′ splice site of exon 6, and
the deleted G residue was located 7-nt downstream from the 3′
splice site of exon 7 (Fig. 2A). The
RT-PCR results of the E6-7/SC-1 minigene were consistent with the
PTC-mediated NMD hypothesis, and demonstrated that the majority of
spliced or partially spliced isoforms, including I4/ex, I4I6/ex,
I5/p/ex and E5E6/in, were markedly decreased compared with the WT
minigene. However, the production of the isoform wherein exon 5 was
included but exon 6 was excluded (E5/in) was markedly increased.
Furthermore, the isoform in which both exons 5 and 6 were excluded
(E5E6/ex) was markedly increased; E5E6/ex was not produced from the
WT minigene. Thus, E5/in and E5E6/ex were observed to survive the
NMD signaling pathway.
The present study further investigated whether the
effects of inserted PTCs were dependent on their respective
location. An A residue was inserted 64-nt upstream from the 5′
splice-site of exon 6, and a T residue was deleted 66-nt downstream
from the 3′ splice-site of exon 7 (E6-7/SC-2). The results
demonstrated that, although I4/ex, I4I6/ex, I5/p/ex and E5E6/in
isoform production from the E6-7/SC-2 minigene was markedly
decreased, E5/in isoform was also significantly increased in the
E6-7/SC-1 and E6-7/SC-2 minigenes. In addition, the E5E6/ex isoform
was more markedly increased in the E6-7/SC-2 minigene than in the
E6-7/SC1 minigene (Fig. 2B). In
conclusion, the results demonstrated that PTCs created through both
the insertion and the deletion of certain nts may induce a marked
decrease of the majority of spliced or unspliced isoforms, but also
stimulate the production of specific isoforms, of RON pre-mRNA.
Discussion
It has been demonstrated in a previous study that
mutations in minigenes can affect the splicing of alternative exons
(12). The results of the present
study confirmed that the differential location of PTCs on
alternative exons 5 and 6 may induce NMD of the majority of the
spliced or partially spliced exon isoforms. A minority of RON
alternative-splicing products, exon 6 excluded (E5/in), exon 5 and
6 excluded (E5E6/ex) and intron 5 unspliced isoforms, were observed
to have survived the NMD pathway. Furthermore, the varieties of
these isoforms are dependent on the location of PTCs in the mRNA.
Therefore, the current study provided an insight into the
regulation of NMD in alternative splicing.
The present study investigated the effect of
PTC-mediated NMD on the alternative splicing of the RON
proto-oncogene. The results of the current study were consistent
with those of previous reports, demonstrating that PTCs, created by
the substitution or insertion of nts, induce NMD of the majority of
spliced or unspliced isoforms, as indicated by the significantly
decreased production of these isoforms. However, by contrast with
previous hypotheses (25), specific
isoforms of alternatively spliced RON pre-mRNA were not reduced or
increased as a consequence of the mutations. The isoform identities
varied in the mutant minigenes that harbored PTCs at alternative
locations on exons 5 or 6, and were created using different
approaches. It was observed that the isoform that includes exon 5
but not exon 6 (E5/in) was increased in the majority of
stop-codon-including minigenes; thus the E5/in isoform may have
included an RNA sequence that was resistant to degradation by the
NMD signaling pathway. The results of the current study also
demonstrated that E5E6/ex was increased in the presence of certain
PTCs. The underlying mechanisms that may allow these isoforms to
resist the NMD signaling pathway must be determined through further
studies. It is possible that these isoforms possess specific
sequences that facilitate the recruitment of anti-NMD signaling
pathway proteins, and also include the sequences that promote
alternative splicing.
Although it was previously reported that PTCs cause
degradation of mRNA through NMD, the results of the present study
indicated that PTCs located at different positions may induce the
survival of various spliced or unspliced isoforms. Thus, in
addition to inducing NMD, PTCs have other important roles in the
survival of spliced, unspliced and partially spliced isoforms. The
results suggest that the insertion of PTCs at specific positions by
gene editing may potentially enhance the survival of the isoforms
that encode tumor suppressors or metastasis suppressors. Similarly,
the insertion of PTCs at other specific positions could cause
degradation of the isoforms that produce oncogenes or tumor
metastasis genes.
Acknowledgements
This study was supported by the NRF-2015R1A2A1A1505
4247 grant to Haihong Shen, the NRF-2016R1A2B1007135 grant to
Xuexiu Zheng and Cell Logistics Research Center (grant no.
2016R1A5A1007318) funded by the National Research Foundation (NRF)
of Korea, and an integrative aging research grant at the Gwangju
Institute of Science and Technology (GIST).
References
|
1
|
Ronsin C, Muscatelli F, Mattei MG and
Breathnach R: A novel putative receptor protein tyrosine kinase of
the met family. Oncogene. 8:1195–1202. 1993.PubMed/NCBI
|
|
2
|
Zhao S, Ammanamanchi S, Brattain M, Cao L,
Thangasamy A, Wang J and Freeman JW: Smad4-dependent TGF-beta
signaling suppresses RON receptor tyrosine kinase-dependent
motility and invasion of pancreatic cancer cells. J Biol Chem.
283:11293–11301. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thangasamy A, Rogge J and Ammanamanchi S:
Recepteur d'origine nantais tyrosine kinase is a direct target of
hypoxia-inducible factor-1alpha-mediated invasion of breast
carcinoma cells. J Biol Chem. 284:14001–14010. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Logan-Collins J, Thomas RM, Yu P, Jaquish
D, Mose E, French R, Stuart W, McClaine R, Aronow B, Hoffman RM, et
al: Silencing of RON receptor signaling promotes apoptosis and
gemcitabine sensitivity in pancreatic cancers. Cancer Res.
70:1130–1140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iwama A, Okano K, Sudo T, Matsuda Y and
Suda T: Molecular cloning of a novel receptor tyrosine kinase gene,
STK, derived from enriched hematopoietic stem cells. Blood.
83:3160–3169. 1994.PubMed/NCBI
|
|
6
|
Yoshimura T, Yuhki N, Wang MH, Skeel A and
Leonard EJ: Cloning, sequencing, and expression of human macrophage
stimulating protein (MSP, MST1) confirms MSP as a member of the
family of kringle proteins and locates the MSP gene on chromosome
3. J Biol Chem. 268:15461–15468. 1993.PubMed/NCBI
|
|
7
|
Wang MH, Ronsin C, Gesnel MC, Coupey L,
Skeel A, Leonard EJ and Breathnach R: Identification of the ron
gene product as the receptor for the human macrophage stimulating
protein. Science. 266:117–119. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu Y, Yao HP and Wang MH: Multiple
variants of the RON receptor tyrosine kinase: Biochemical
properties, tumorigenic activities, and potential drug targets.
Cancer Lett. 257:157–164. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Collesi C, Santoro MM, Gaudino G and
Comoglio PM: A splicing variant of the RON transcript induces
constitutive tyrosine kinase activity and an invasive phenotype.
Mol Cell Biol. 16:5518–5526. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonomi S, di Matteo A, Buratti E, Cabianca
DS, Baralle FE, Ghigna C and Biamonti G: HnRNP A1 controls a
splicing regulatory circuit promoting mesenchymal-to-epithelial
transition. Nucleic Acids Res. 41:8665–8679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moon H, Cho S, Loh TJ, Oh HK, Jang HN,
Zhou J, Kwon YS, Liao DJ, Jun Y, Eom S, et al: SRSF2 promotes
splicing and transcription of exon 11 included isoform in Ron
proto-oncogene. Biochim Biophys Acta. 1839:1132–1140. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou YQ, He C, Chen YQ, Wang D and Wang
MH: Altered expression of the RON receptor tyrosine kinase in
primary human colorectal adenocarcinomas: Generation of different
splicing RON variants and their oncogenic potential. Oncogene.
22:186–197. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang MH, Kurtz AL and Chen Y:
Identification of a novel splicing product of the RON receptor
tyrosine kinase in human colorectal carcinoma cells.
Carcinogenesis. 21:1507–1512. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen YQ, Zhou YQ, Angeloni D, Kurtz AL,
Qiang XZ and Wang MH: Overexpression and activation of the RON
receptor tyrosine kinase in a panel of human colorectal carcinoma
cell lines. Exp Cell Res. 261:229–238. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Green MR: Pre-mRNA splicing. Annu Rev
Genet. 20:671–708. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Black DL: Mechanisms of alternative
pre-messenger RNA splicing. Annu Rev Biochem. 72:291–336. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berglund JA, Chua K, Abovich N, Reed R and
Rosbash M: The splicing factor BBP interacts specifically with the
pre-mRNA branchpoint sequence UACUAAC. Cell. 89:781–787. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nelson KK and Green MR: Mammalian U2 snRNP
has a sequence-specific RNA-binding activity. Genes Dev.
3:1562–1571. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Isken O and Maquat LE: The multiple lives
of NMD factors: Balancing roles in gene and genome regulation. Nat
Rev Genet. 9:699–712. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang YF, Imam JS and Wilkinson MF: The
nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem.
76:51–74. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valacca C, Bonomi S, Buratti E, Pedrotti
S, Baralle FE, Sette C, Ghigna C and Biamonti G: Sam68 regulates
EMT through alternative splicing-activated nonsense-mediated mRNA
decay of the SF2/ASF proto-oncogene. J Cell Biol. 191:87–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saltzman AL, Kim YK, Pan Q, Fagnani MM,
Maquat LE and Blencowe BJ: Regulation of multiple core spliceosomal
proteins by alternative splicing-coupled nonsense-mediated mRNA
decay. Mol Cell Biol. 28:4320–4330. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma Q, Zhang K, Yao HP, Zhou YQ, Padhye S
and Wang MH: Inhibition of MSP-RON signaling pathway in cancer
cells by a novel soluble form of RON comprising the entire sema
sequence. Int J Oncol. 36:1551–1561. 2010.PubMed/NCBI
|
|
25
|
Gong Q, Zhang L, Vincent GM, Horne BD and
Zhou Z: Nonsense mutations in hERG cause a decrease in mutant mRNA
transcripts by nonsense-mediated mRNA decay in human long-QT
syndrome. Circulation. 116:17–24. 2007. View Article : Google Scholar : PubMed/NCBI
|