Introduction
Previous studies have established that c-Myc serves
important roles in cell cycle regulation, cell growth,
differentiation, transformation, apoptosis and genomic instability
(1,2).
The c-Myc protein also fulfills essential functions in damage
repair processes; in particular, this protein is associated not
only with the promoters of the double-stranded break repair genes
Nijmegen breakage syndrome 1, Ku70, Rad51, breast cancer 2, early
onset, Rad50, Rad54L and DNA-dependent protein kinase catalytic
subunit (DNA-PKcs), but also with the expression of various
mismatch repair genes (3–5). The inhibition of proper DNA-damage
repair by c-Myc can lead to genomic instability, causing tumor
cells to become sensitive to specific drugs and/or radiation
(6). Our prior studies (7,8) revealed
the existence of a novel signaling pathway for maintaining the
stability of the c-Myc protein: DNA-PKcs/protein kinase B
(Akt)/glycogen synthase kinase 3 (GSK3)/c-Myc. Our recent study
determined that c-Myc participates in the repair process of
ionizing radiation (IR)-induced DNA double-strand breaks (9). It has been established that IR can cause
a variety of DNA damage and abnormal checkpoints of cell cycle;
each is able to induce mitotic catastrophe (10). In 2012, Kessler et al (11) reported that a SUMOylation-dependent
transcriptional sub-program is required for the carcinogenic
effects of the c-Myc gene. The inactivation of SUMO-activating
enzyme (SAE) 2 leads to the excessive activation of the c-Myc gene,
which induces mitotic catastrophe and cell death (11). These types of SUMOylation-dependent
Myc switcher genes are essential for mitotic spindle function
(11). The results of this study
(11) provide further evidence for
the involvement of c-Myc in mitosis. However, whether c-Myc is
directly involved in the process of ionizing radiation-induced
mitotic catastrophe remains unclear. Therefore, the current study
aimed to investigate this issue.
Materials and methods
Materials and reagents
Human cervical epithelial cells (HeLa-NC) purchased
from the Institute of Biochemistry and Cell Biology, Shanghai
Institute for Biological Sciences, Chinese Academy of Sciences
(Shanghai, China) and c-Myc silenced cells (HeLa-630 cells stably
expressing myc-shRNA-630 were generated in our previous study)
(9), and were maintained in the
laboratory of the School of Radiation Medicine and Protection
(Suzhou, China) (9). The HeLa-NC and
HeLa-630 cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (Biological Industries,
Beit Haemek, Israel) at 37°C in a humidified incubator with 5%
CO2. Protease inhibitor tablets were purchased from
Roche Diagnostics GmbH (Mannheim, Germany). Microarray detection
was performed with assistance from Shanghai Kang-Chen Biotechnology
(Shanghai, China).
mRNA microarray. RNA extraction
TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was used to extract total RNA
according to the manufacturer's instructions, and the resulting RNA
solutions were stored at −70°C. The ratio of OD260/OD280 was
utilized to determine the purity, and the product of OD260
multiplied by 40 µg/ml was the concentration of RNA samples.
Denaturing agarose gel electrophoresis was used to detect RNA
integrity (12).
The synthesis of cDNA and the
fluorescent labeling of samples
Once double-stranded cDNA was synthesized, the DNA
was labeled using a NimbleGen One-Color labeling kit (Roche
NimbleGen, Inc., Madison, WI, USA). The samples were then labeled
with Cy3-random nonamers (supplied in the kit). Based on the
concentrations (determined using a NanoDrop ND-1000; Thermo Fisher
Scientific, Inc.) of the labeled products, the volume of the
Cy3-labeled sample in each tube was calculated (1 µg cDNA in 40 µl
diluted Cy3-random nonamers). Equal quantities of samples were used
for subsequent stages of the hybridization experiments.
Genome-wide microarray
hybridization
NimbleGen microarray (Roche NimbleGen, Inc.) was
detected by Shanghai Kang-Chen Biotechnology. A total of six
microarrays were used, including three biological replicates for
HeLa-NC and HeLa-630 cells. The NimbleGen Hybridization System 4
(Roche NimbleGen, Inc.) was used for hybridization. The
hybridization reaction was performed using a hybridization kit
(Roche NimbleGen, Inc.) in accordance with NimbleGen's
instructions. Once the reaction was complete, the array was
subjected to elution using a Wash Buffer kit (Roche NimbleGen,
Inc.) and spun dry by centrifugation (200 × g, 25°C, 5 min).
Image acquisition and data
analysis
Microarray results were analyzed using a GenePix
4000 B single-channel scanner (Axon Instruments, Inc., Union City,
CA, USA). NimbleScan v.2.5 (Roche NimbleGen, Inc.) was used to read
the values of the raw microarray signals (532 nm); these signal
values were then corrected and normalized according to the
NimbleGen's instructions. GeneSpring v11.0 software (Agilent
Technologies, Inc., Santa Clara, CA, USA) was used for statistical
analysis, clustering and pathway analysis and visualizations. A
threshold value of 2 was established; thus, increases and decreases
≥2-fold were regarded as cases of upregulation and downregulation,
respectively.
Quantification of the proportion of
multinucleated cells by flow cytometry
HeLa-NC and HeLa-630 cells were seeded into 60-mm
culture dishes with 5×105 cells in each dish. The dishes were then
exposed to 4 Gy of 60Coγ radiation. Cells were collected 48 h
post-IR. The collected cells were washed with PBS, fixed in
pre-cooled 70% ethanol at −20°C for 24 h, and centrifuged at 700 ×
g, at 4°C for 10 min. The fixative was discarded, cells were washed
twice in PBS and 10 mg/ml RNase (Sangon Biotech Co., Ltd.,
Shanghai, China) was added to the cell samples to a final
concentration of 50 µg/ml, which were then incubated at 37°C for 30
min. Propidium iodide was then added to the cell samples to a final
concentration of 50 µg/ml, and cells were analyzed by flow
cytometry (CyFlow Green; PARTEC GmbH, Münster, Germany). These
experiments were repeated three times.
Immunofluorescence analysis of
multinucleated cells and mitotic catastrophe
HeLa-NC and HeLa-630 cells (104 cells) were seeded
onto cover slips (circular; diameter, 12 mm; Thermo Fisher
Scientific, Inc.). These cells were cultured overnight in a 37°C
incubator and then exposed to 4 Gy of 60Coγ radiation. Cells were
collected at 48 h post-IR. The collected cells were washed twice in
PBS, fixed with 4% paraformaldehyde at room temperature for 30 min,
washed three times with PBS for 10 min per wash, permeabilized in a
0.3% Triton X-100 solution on ice for 15 min and then blocked in a
solution of PBS and 1% fetal bovine serum albumin (Biological
Industries) at room temperature for 1 h. Each cell sample was then
incubated overnight at 4°C with a 1:200 dilution of a primary
anti-α-tubulin (cat no. 2125; Cell Signaling Technology, Inc.,
Danvers, MA, USA) or an anti-γ-tubulin antibody (cat no. ab16504;
Abcam, Cambridge, UK). Following this incubation, samples were
washed three times in PBS for 10 min per wash and then incubated
with Alexa Fluor® 488 Phalloidin (green; cat no. 8878)
or Alexa Fluor® 647 Phalloidin (red; cat no. 8940)
labeled secondary antibody (Cell Signaling Technology, Inc.) at
room temperature for 1 h (incubation began in the dark). Finally,
each sample was washed three times with PBS for 10 min per wash,
stained with DAPI (2 mg/ml; Cell Signaling Technology, Inc.) for 10
min at room temperature and imaged using confocal laser scanning
microscopy (FV1000; FV10-ASW software; Olympus Corporation, Tokyo,
Japan).
Statistical analysis
Experimental data are expressed as the mean ±
standard deviation. The SPSSv.10.0 software package (SPSS, Inc.,
Chicago, IL, USA) was used to process and analyze the data.
Differences between groups were determined using unpaired
two-tailed Student's t-tests. P<0.05 was considered to indicate
a statistically significant difference.
Results
An increase in multinucleated cells
within the HeLa-630 cell population
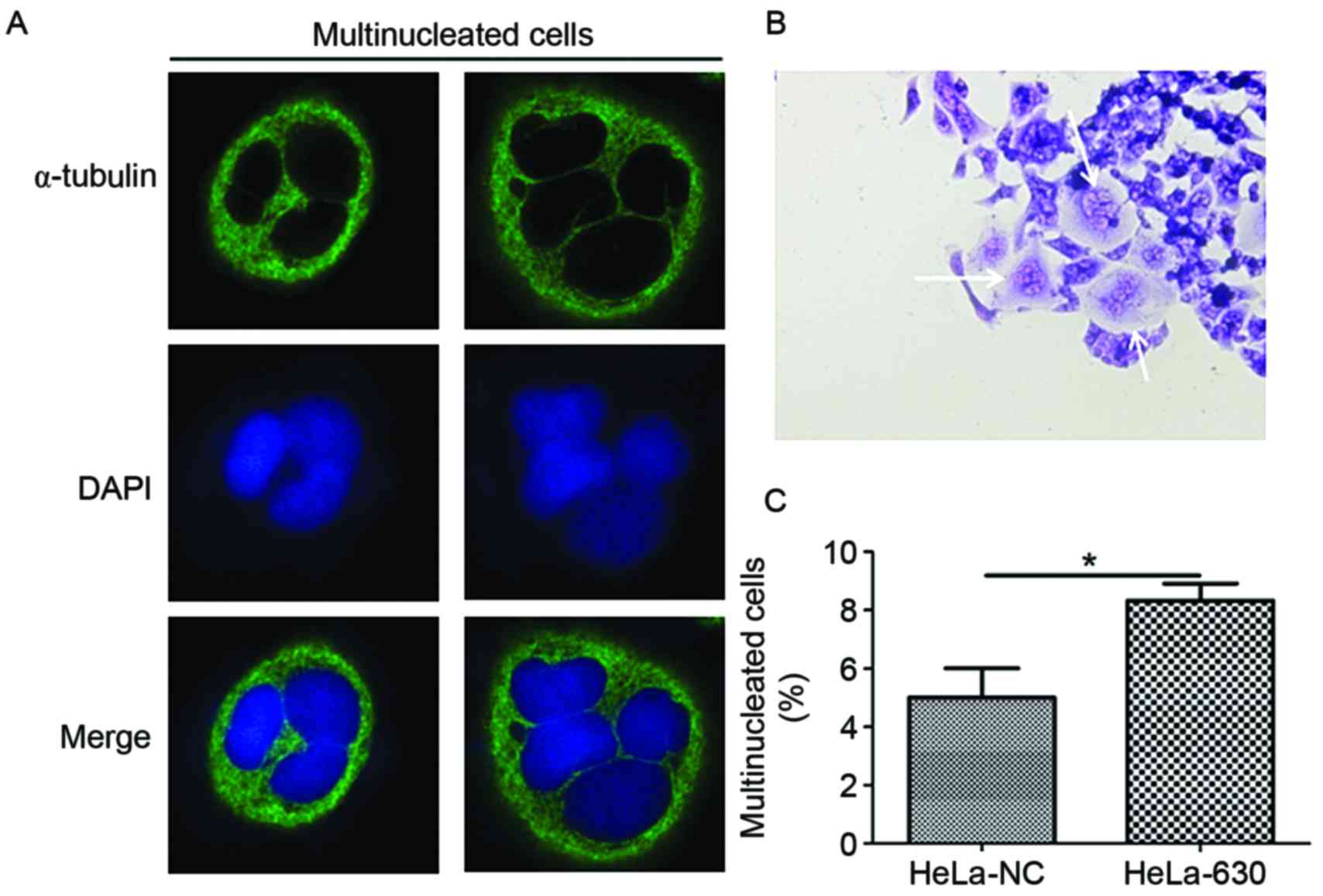
Light microscopy observation revealed that
multinucleation frequently occurred in HeLa-630 cells during the
G418 selection and growth process (Fig.
1A). Therefore, immunofluorescence and flow cytometry were used
to further examine the HeLa-630 cells. Immunofluorescence results
demonstrated that multinucleated cells, including cells with 3–4
nuclei, were identified among HeLa-630 and HeLa-NC cells (Fig. 1B). The flow cytometry results
indicated that there were a greater number of multinucleated cells
among the HeLa-630 cell population compared with the HeLa-NC cell
population (Fig. 1C).
Differentially expressed genes in
HeLa-630 cells
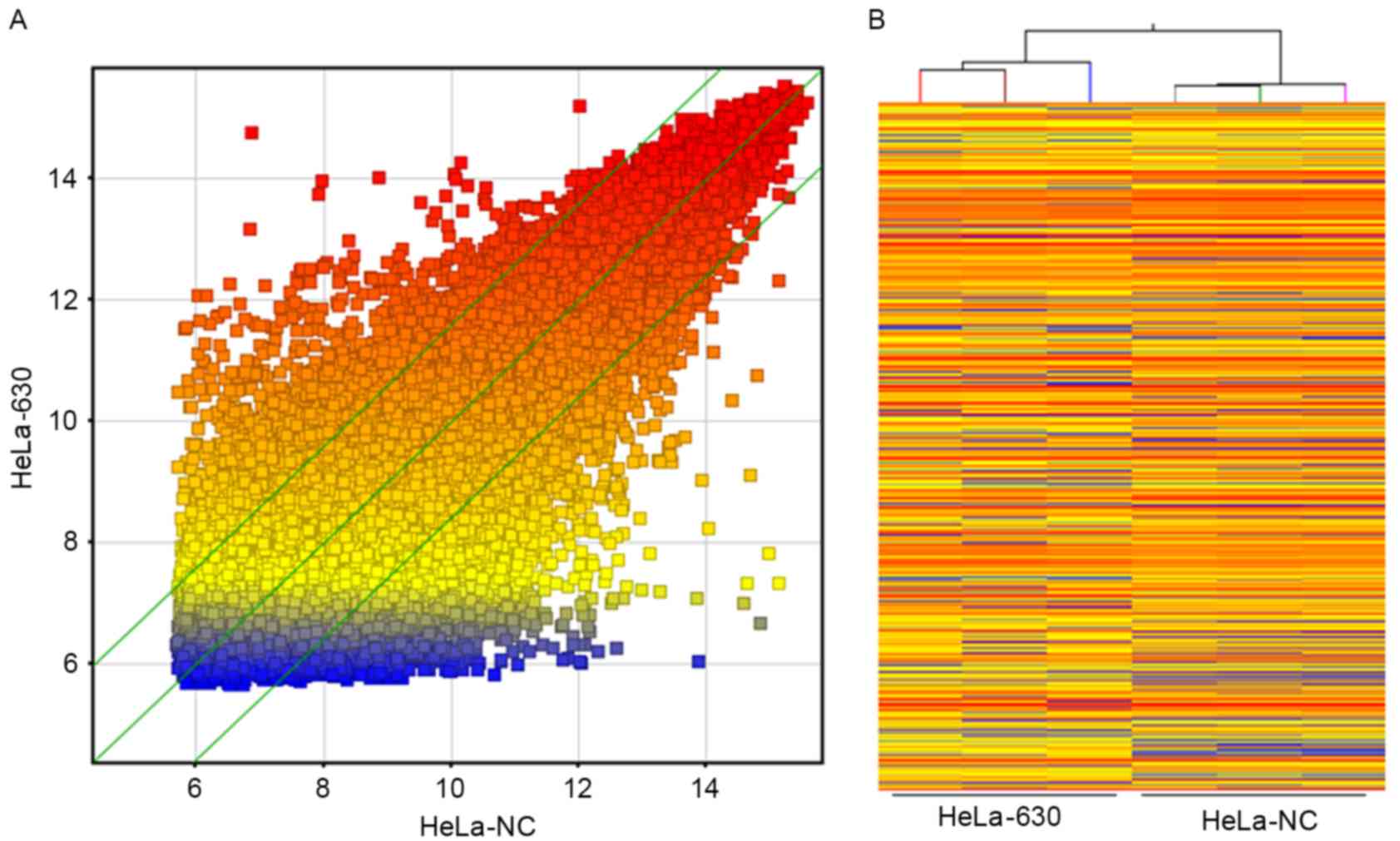
Whole-genome expression profiles for three dishes of
HeLa-NC cells and three dishes of HeLa-630 cells revealed that
≤2745 genes exhibited differential expression in these two cell
lines, based on the statistical significance parameters of
P<0.05 and a fold change of 2; of these genes, 1,098 were
upregulated in HeLa-630 cells and 1,647 were downregulated in
HeLa-630 cells. These results are presented as a cluster analysis
and a scatter plot in Fig. 2.
GeneSpring v.11.0 software was used to perform GO
analysis and signaling pathway analysis of differentially expressed
genes from the whole-genome expression profile microarray results,
in order to investigate the biological significance of these genes.
In the analysis results, changes in gene expression levels are
presented in terms of the three GO domains, as follows: Biological
process (BP); molecular function (MF); cellular component (CC). The
first 10 terms in each category for downregulated and upregulated
genes are arranged from highest to lowest p-value in Tables I and II, respectively.
 | Table I.GO analysis of the biological
functions of downregulated mRNAs. |
Table I.
GO analysis of the biological
functions of downregulated mRNAs.
| GO.ID | Term | Count | Pop.Hits |
|---|
| BP |
|
GO:0007020 | Microtubule
nucleation | 6 | 10 |
|
GO:0010718 | Positive regulation
of epithelial to mesenchymal transition | 10 | 18 |
|
GO:0010770 | Positive regulation
of cell morphogenesis involved in differentiation | 10 | 19 |
|
GO:0048820 | Hair follicle
maturation | 6 | 12 |
|
GO:0030206 | Chondroitin sulfate
biosynthetic process | 5 | 10 |
|
GO:0060325 | Face
morphogenesis | 5 | 10 |
|
GO:0045197 | Establishment or
maintenance of epithelial cell apical/basal polarity | 5 | 11 |
|
GO:0070723 | Response to
cholesterol | 5 | 11 |
|
GO:0010717 | Regulation of
epithelial to mesenchymal transition | 12 | 27 |
|
GO:0051298 | Centrosome
duplication | 7 | 16 |
| CC |
|
GO:0000242 | Pericentriolar
material | 5 | 12 |
|
GO:0009925 | Basal plasma
membrane | 7 | 22 |
|
GO:0045178 | Basal part of
cell | 8 | 26 |
|
GO:0005868 | Cytoplasmic dynein
complex | 3 | 10 |
|
GO:0005952 | cAMP-dependent
protein kinase complex | 3 | 10 |
|
GO:0012507 | ER to Golgi
transport vesicle membrane | 5 | 17 |
|
GO:0043073 | Germ cell
nucleus | 5 | 17 |
|
GO:0005901 | caveolae | 16 | 55 |
|
GO:0030663 | COPI coated vesicle
membrane | 4 | 14 |
|
GO:0005801 | cis-Golgi
network | 6 | 22 |
| MF |
|
GO:0005160 | Transforming growth
factor-β receptor binding | 9 | 19 |
|
GO:0070411 | I-SMAD binding | 5 | 11 |
|
GO:0004115 | 3′,5′-cyclic-AMP
phosphodiesterase activity | 7 | 16 |
|
GO:0005072 | Transforming growth
factor-β receptor, cytoplasmic mediator activity | 4 | 10 |
|
GO:0004675 | Transmembrane
receptor protein serine/threonine kinase activity | 7 | 19 |
|
GO:0051010 | Microtubule
plus-end binding | 4 | 11 |
|
GO:0051879 | Hsp90 protein
binding | 4 | 11 |
|
GO:0005024 | Transforming growth
factor-β receptor activity | 6 | 17 |
|
GO:0004697 | Protein kinase C
activity | 5 | 15 |
|
GO:0045295 | γ-catenin
binding | 4 | 12 |
 | Table II.GO analysis of the biological
functions of upregulated mRNAs. |
Table II.
GO analysis of the biological
functions of upregulated mRNAs.
| GO.ID | Term | Count | Pop.Hits |
|---|
| BP |
|
GO:0007512 | Adult heart
development | 5 | 12 |
|
GO:0032098 | Regulation of
appetite | 5 | 12 |
|
GO:0042749 | Regulation of
circadian sleep/wake cycle | 6 | 15 |
|
GO:0045187 | Regulation of
circadian sleep/wake cycle, sleep | 6 | 15 |
|
GO:0022410 | Circadian
sleep/wake cycle process | 6 | 16 |
|
GO:0050802 | Circadian
sleep/wake cycle, sleep | 6 | 16 |
|
GO:0001977 | Renal system
process involved in regulation of blood volume | 4 | 11 |
|
GO:0050482 | Arachidonic acid
secretion | 4 | 11 |
|
GO:0042745 | Circadian
sleep/wake cycle | 6 | 17 |
|
GO:0050879 | Multicellular
organismal movement | 8 | 23 |
| CC |
|
GO:0005865 | Striated muscle
thin filament | 5 | 14 |
|
GO:0000786 | Nucleosome | 18 | 66 |
|
GO:0001772 | Immunological
synapse | 3 | 12 |
|
GO:0034362 | Low-density
lipoprotein particle | 3 | 12 |
|
GO:0034364 | High-density
lipoprotein particle | 5 | 24 |
|
GO:0034361 | Very-low-density
lipoprotein particle | 4 | 20 |
|
GO:0034385 | Triglyceride-rich
lipoprotein particle | 4 | 20 |
|
GO:0042734 | Presynaptic
membrane | 7 | 39 |
|
GO:0032993 | Protein-DNA
complex | 18 | 102 |
|
GO:0032994 | Protein-lipid
complex | 6 | 35 |
| MF |
|
GO:0051183 | Vitamin transporter
activity | 5 | 15 |
|
GO:0015250 | Water channel
activity | 3 | 10 |
|
GO:0015026 | Co-receptor
activity | 6 | 21 |
|
GO:0030547 | Receptor inhibitor
activity | 4 | 14 |
|
GO:0005372 | Water transmembrane
transporter activity | 3 | 11 |
|
GO:0005184 | Neuropeptide
hormone activity | 6 | 23 |
|
GO:0005313 | L-glutamate
transmembrane transporter activity | 3 | 12 |
|
GO:0015172 | Acidic amino acid
transmembrane transporter activity | 3 | 12 |
|
GO:0016918 | Retinal
binding | 3 | 12 |
|
GO:0048019 | Receptor antagonist
activity | 3 | 13 |
GO analysis for downregulated genes in HeLa-630
cells revealed that the primary terms in the BP domain included
references to microtubule nucleation, regulation of
epithelial-mesenchymal transition (EMT), morphological regulation
during cell differentiation, follicle maturation, chondroitin
sulfate synthesis, the establishment or maintenance of basal
epithelial cell polarity, response to cholesterol, centrosome
duplication and various other processes. Major terms in the GO CC
domain included references to pericentriolar material, the
extracellular matrix, the basal region of the cell, the cytoplasmic
dynein complex, the cAMP-dependent protein kinase complex, ER-Golgi
transport vesicle membranes and the Golgi network, among numerous
other components. Major terms in the GO MF domain included
references to transforming growth factor-β (TGF-β) receptor
binding, cytoplasmic mediator activity, transmembrane receptor
serine/threonine kinase activity, microtubule binding, heat shock
protein 90 binding, protein kinase C activity and other
functions.
GO analysis for the upregulated genes in HeLa-630
cells revealed that the main terms in the BP domain included
references to adult heart development, appetite regulation,
circadian sleep/wake cycle regulation, renal system processes
involved in blood volume regulation, arachidonic acid secretion,
multicellular organismal movement and other processes. The main
terms in the GO CC domain included references to striated muscle
thin filaments, nucleosomes, immunological synapses, low-density
lipoprotein, high-density lipoprotein, very-low-density
lipoprotein, the presynaptic membrane, protein-DNA complexes and
protein-lipid complexes, among various other components. Major
terms in the GO MF domain included references to vitamin transport
activity, water channel activity, co-receptor activity, receptor
inhibitor activity, water transmembrane transporter activity,
neuropeptide hormone activity, L-glutamate transmembrane
transporter activity, acidic amino acid transmembrane transporter
activity, retinal binding, receptor antagonist activity and
numerous other functions.
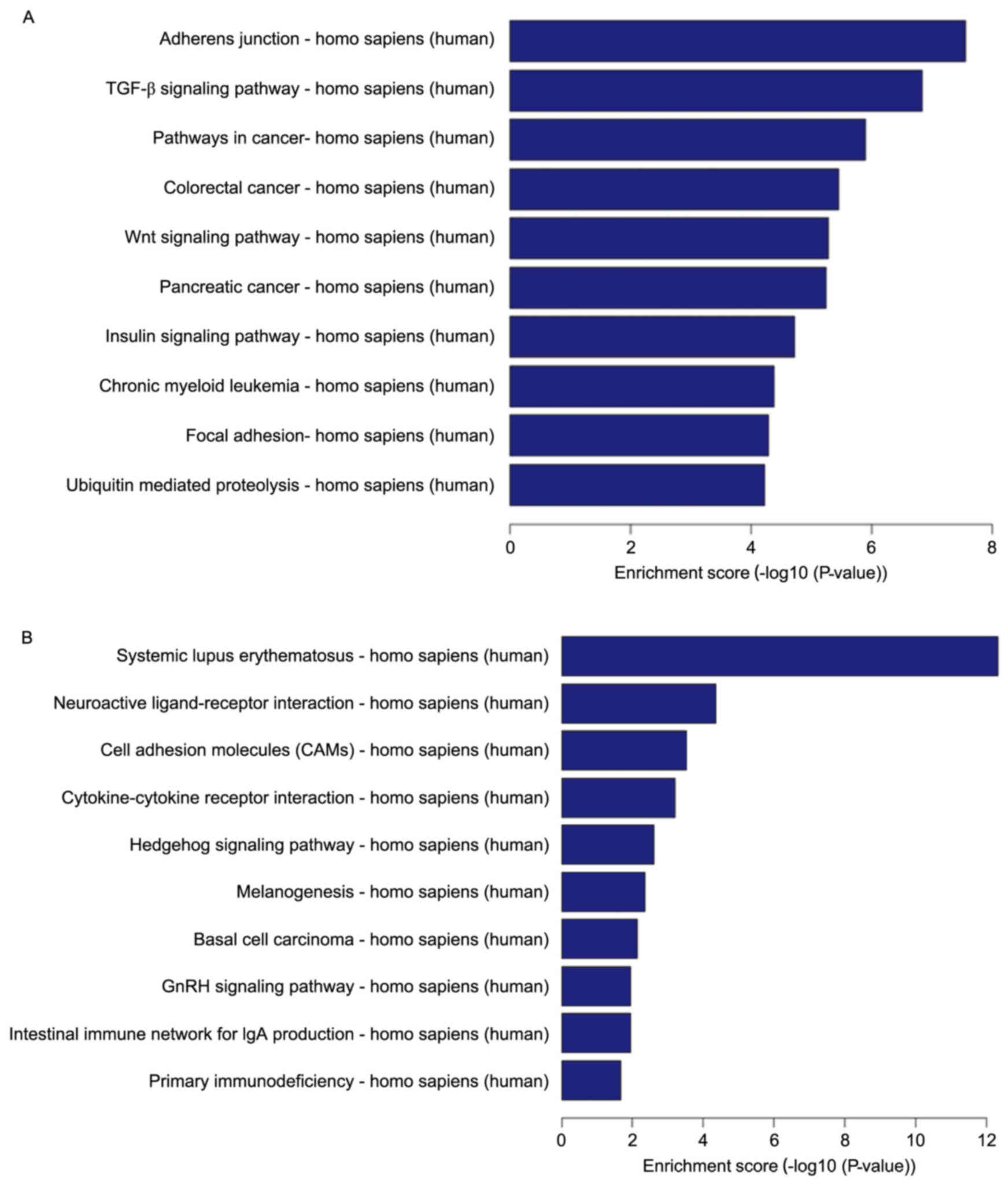
The pathway analysis for the 10 results with the
highest p-values of among the downregulated differentially
expressed genes and the pathway analysis of upregulated genes are
presented in Fig. 3. Downregulated
differentially expressed genes were mainly principally involved in
adherens junctions, the TGF-β signaling pathway, pathways in
cancer, the Wnt signaling pathway, the insulin signaling pathway,
focal adhesions and ubiquitin-mediated proteolysis, among other
diverse activities functions (Fig.
3A). Upregulated differentially expressed genes were mainly
primarily involved in systematic lupus erythematosus, neuroactive
ligand-receptor interactions, cell adhesion, cytokine-cytokine
receptor interactions, the Hedgehog signaling pathway,
melanogenesis, basal cell carcinoma, the gonadotropin-releasing
hormone (GnRH) signaling pathway, the intestinal immune network for
IgA production, and primary immunodeficiency, among various other
activities (Fig. 3B).
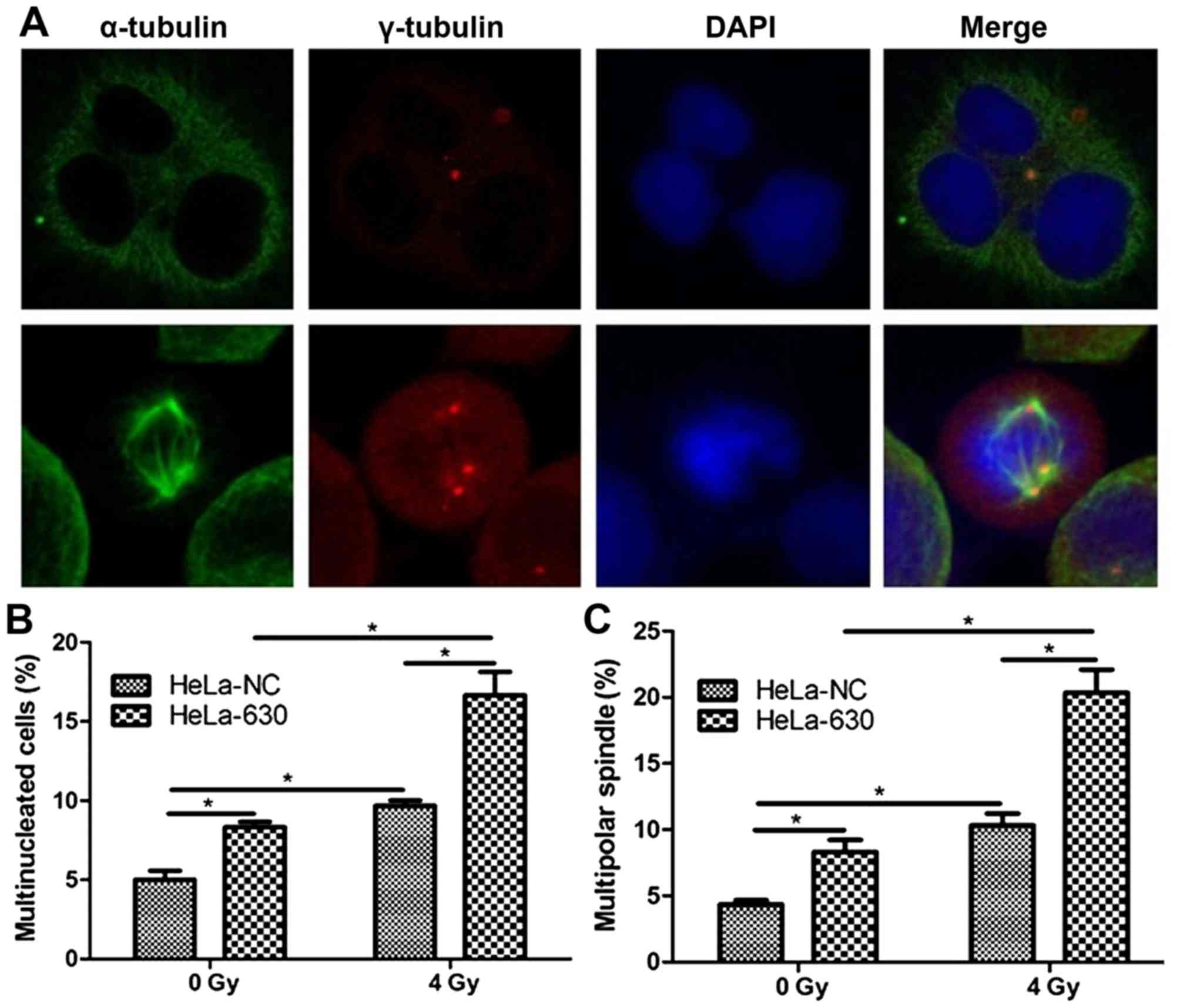
Mitotic catastrophe in HeLa-630 cells
following irradiation
As indicated in Fig.
4, following the exposure of HeLa-630 cells to 4 Gy of 60Coγ
radiation, various abnormalities were observed, including centriole
abnormalities and multipolar spindles. Flow cytometry revealed
that, at 48 h post radiation, the proportion of multinucleated
cells was significantly greater in the HeLa-630 cell group, as
compared with in the HeLa-NC cell group (P<0.05).
Discussion
C-Myc was initially discovered as a proto-oncogene
with activity in numerous types of human malignancies; however,
c-Myc is also a transcription factor and a certain level of c-Myc
protein expression is required for the growth and proliferation of
normal cells (13,14). However, the overexpression of c-Myc in
transgenic mice induces malignant T-cell lymphoma and acute myeloid
leukemia (15); in these mice, the
inhibition of c-Myc expression produces significant anti-tumor
effects. In c-Myc knockout mice, normal embryonic development
proceeds for 9.5–10.5 days, but will subsequently cease (16); this finding indicates that, in
mammals, c-Myc serves a vital role in normal embryonic development
and in overall growth and developmental processes. In addition,
c-Myc promotes the self-renewal and differentiation of stem and
progenitor cells in mice and contributes to maintaining embryonic
stem cell characteristics (17,18).
Takahashi et al (19) used a
retroviral vector to co-transfect the four reprogramming factors
octamer-binding transcription factor 4 (Oct4), SRY-box 2 (Sox2),
c-Myc and Krüppel-like factor 4 (Klf4) into mouse/human fibroblasts
and produced induced pluripotent stem cells (iPSCs). Furthermore,
this generation of iPSCs does not require egg cells or embryos;
instead, only the introduction of the four transcriptionally active
genes Oct4, SOX2, c-Myc and Klf4 into the cells is required
(20). These findings suggest that
c-Myc has essential functions in cell proliferation, growth and
transformation. Recent studies have demonstrated that c-Myc, its
target genes and its microRNAs form a complex regulatory network
that serves a vital role in the maintenance of cancer stem cells,
EMT, metastasis and angiogenesis (21). The exploration of c-Myc functions may
provide novel perspectives regarding the development of cancer
treatments that target c-Myc.
In the laboratory of the School of Radiation
Medicine and Protection, RNA interference techniques were utilized
to establish a cell model in which c-Myc expression was inhibited
(22). In the current study, an
increased number of multinucleated cells were observed in the
HeLa-630 cell group. There are two major causes for the formation
of multinucleated cells (e.g., polyploidy). First, cell fusion can
occur, wherein the cytoplasm of independent cells mixes, resulting
in the generation of syncytia (23).
Second, multinucleated cells may be produced when nuclear division
occurs in mononuclear cells, but cellular division does not occur
due to failed cytokinesis (24). In
this second case, polyploid cells formed by the failure of
cytokinesis cannot effectively undergo mitosis, thus, these cells
must eventually die. This fate is regarded as a form of
proliferative cell death (25).
Therefore, the proportion of multinucleated cells was elevated in
the HeLa-630 cell population due to a reduction in c-Myc protein
expression. The results of GO analysis of the downregulated genes
in these cells revealed that changes in the three BP, MF and CC
domains suggested that c-Myc is involved in the regulation of cell
growth and morphology. Following c-Myc silencing, abnormalities in
microtubule nucleation, centrosome duplication and pericentriolar
material occur; these phenomena may help to explain why the
proportion of multinucleated cells is elevated in HeLa-630 cells.
In our prior studies (7,8), a novel signaling pathway was proposed
for maintaining the stability of the c-Myc protein:
DNA-PKcs/Akt/GSK3/c-Myc. Previous findings have identified DNA-PKcs
as an upstream regulator of c-Myc protein stability (26). Inhibiting the expression or activity
of DNA-PKcs leads to various cellular abnormalities during mitosis,
includingthe presence of extraneous centrosomes, abnormal spindle
structure and microtubule damage (27). Furthermore, due to cytokinesis
failure, the inhibition of DNA-PKcs induces various phenotypic
changes, including an increase in the proportion of multinucleated
cells (28). An elevated proportion
of multinucleated cells due to DNA-PKcs inactivation may occur not
only as a result of damage from ionizing radiation, but also under
natural growth conditions (29). From
the results of the current study, it may be hypothesized that the
c-Myc protein serves an important role in the maintenance of
karyotype stability by DNA-PKcs.
The results of GO analysis of the upregulated genes
in HeLa-630 cells revealed that changes in the three BP, MF and CC
domains suggested that the c-Myc protein is also involved in the
regulation of DNA synthesis and metabolism, protein metabolism and
the regulation of ion concentrations, among various other
functions. It has previously been reported that c-Myc is involved
in the regulation of DNA synthesis and metabolism via promoting the
transcription of the carbamoyl-phosphate synthetase 2/aspartate
transcarbamylase/dihydroorotasegene (30). This metabolism-regulating function may
be involved in the formation of multinucleated cells.
Downregulated differentially expressed genes were
primarily involved in the adherens junctions and the TGF-β
signaling pathway. An increasing body of evidence indicates that
alterations in the levels of cadherin-dependent adherens junctions
regulate the stability of tight junction complexes (31). Reduced expression of the c-Myc gene
repressed E-cadherin promoter activity, and subsequently decreased
E-cadherin mRNA and protein levels (32). Blocking TGF-β upregulates E-cadherin
and reduces migration and invasion of hepatocellular carcinoma
cells (33). Therefore, c-Myc serves
a role in cancer via E-cadherin and the TGF-β pathway. Based on the
comprehensive findings of the aforementioned investigations and the
results of the current study, it is evident that c-Myc is involved
in mitotic processes. With regard to the significance of this
finding in radiotherapy, a series of previous reports have
demonstrated that the overexpression of c-Myc contributes to cancer
radioresistance (34,35). One study suggested that c-Myc
downregulation sensitizes melanoma cells to radiotherapy by
inhibiting MLH1 and MSH2 mismatch repair proteins in a
p53-independent manner (4). Another
study revealed that the sensitization of non-Hodgkin's lymphoma
cells to ionizing radiation by rituximab is achieved through the
inhibition of c-Myc expression (36).
In the present study, it was observed that irradiation causes a
greater amount of mitotic catastrophe in HeLa-630 cells than
HeLa-NC cells. It maybe concluded that the radiosensitization of
certain tumor cell types may be achieved by inducing mitotic
catastrophe through the inhibition of c-Myc expression. In
conclusion, the results of the current study demonstrated that
inhibition of c-Myc expression accounts for an increased number of
multinucleated cells in HeLa cells, and that the radiosensitization
of inactivating c-Myc oncogene may be associated with an increased
degree of mitotic catastrophe.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81172706) and by the
Priority Academic Program Development of Jiangsu Higher Education
Institutions.
References
|
1
|
Cowling VH and Cole MD: Mechanism of
transcriptional activation by the Myc oncoproteins. Semin Cancer
Biol. 16:242–252. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Adhikary S and Eilers M: Transcriptional
regulation and transformation by Myc proteins. Nat Rev Mol Cell
Biol. 6:635–645. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiang YC, Teng SC, Su YN, Hsieh FJ and Wu
KJ: c-Myc directly regulates the transcription of the NBS1 gene
involved in DNA double-strand break repair. J Biol Chem.
278:19286–19291. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bucci B, D'Agnano I, Amendola D, Citti A,
Raza GH, Miceli R, De Paula U, Marchese R, Albini S, Felsani A, et
al: Myc down-regulation sensitizes melanoma cells to radiotherapy
by inhibiting MLH1 and MSH2 mismatch repair proteins. Clin Cancer
Res. 11:2756–2767. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bindra RS and Glazer PM: Co-repression of
mismatch repair gene expression by hypoxia in cancer cells: Role of
the Myc/Max network. Cancer Lett. 252:93–103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bristow RG, Ozcelik H, Jalali F, Chan N
and Vesprini D: Homologous recombination and prostate cancer: A
model for novel DNA repair targets and therapies. Radiother Oncol.
83:220–230. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
An J, Huang YC, Xu QZ, Zhou LJ, Shang ZF,
Huang B, Wang Y, Liu XD, Wu DC and Zhou PK: DNA-PKcs plays a
dominant role in the regulation of H2AX phosphorylation in response
to DNA damage and cell cycle progression. BMC Mol Biol. 11:182010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
An J, Xu QZ, Sui JL, Bai B and Zhou PK:
Downregulation of c-Myc protein by siRNA-mediated silencing of
DNA-PKcs in HeLa cells. Int J Cancer. 117:531–537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cui F, Fan R, Chen Q, He Y, Song M, Shang
Z, Zhang S, Zhu W, Cao J, Guan H and Zhou PK: The involvement of
c-Myc in the DNA double-strand break repair via regulating
radiation-induced phosphorylation of ATM and DNA-PKcs activity. Mol
Cell Biochem. 406:43–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eriksson D, Löfroth PO, Johansson L,
Riklund KA and Stigbrand T: Cell cycle disturbances and mitotic
catastrophes in HeLa Hep2 cells following 2.5 to 10 Gy of ionizing
radiation. Clin Cancer Res. 13:5501s–5508s. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kessler JD, Kahle KT, Sun T, Meerbrey KL,
Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et
al: A SUMOylation-dependent transcriptional subprogram is required
for Myc-driven tumorigenesis. Science. 335:348–353. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Masek T, Vopalensky V, Suchomelova P and
Pospisek M: Denaturing RNA electrophoresis in TAE agarose gels.
Anal Biochem. 336:46–50. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dang CV: c-Myc target genes involved in
cell growth, apoptosis, and metabolism. Mol Cell Biol. 19:1–11.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kalra N and Kumar V: c-Fos is a mediator
of the c-Myc-induced apoptotic signaling in serum-deprived hepatoma
cells via the p38 mitogen-activated protein kinase pathway. J Biol
Chem. 279:25313–25329. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo H, Li Q, O'Neal J, Kreisel F, Le Beau
MM and Tomasson MH: c-Myc rapidly induces acute myeloid leukemia in
mice without evidence of lymphoma-associated antiapoptotic
mutations. Blood. 106:2452–2461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davis AC, Wims M, Spotts GD, Hann SR and
Bradley A: A null c-myc mutation causes lethality before 10.5 days
of gestation in homozygotes and reduced fertility in heterozygous
female mice. Genes Dev. 7:671–682. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yasuda SY, Tsuneyoshi N, Sumi T, Hasegawa
K, Tada T, Nakatsuji N and Suemori H: NANOG maintains self-renewal
of primate ES cells in the absence of a feeder layer. Genes Cells.
11:1115–1123. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Loh YH, Nq JH and Nq HH: Molecular
framework underlying pluripotency. Cell Cycle. 7:885–891. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han DW, Greber B, Wu G, Tapia N,
Araúzo-Bravo MJ, Ko K, Bernemann C, Stehling M and Schöler HR:
Direct reprogramming of fibroblasts into epiblast stem cells. Nat
Cell Biol. 13:66–71. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jackstadt R and Hermeking H: MicroRNAs as
regulators and mediators of c-MYC function. Biochim Biophys Acta.
1849:544–553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paddison PJ, Caudy AA, Bernstein E, Hannon
GJ and Conklin DS: Short hairpin RNAs (shRNAs) induce
sequence-specific silencing in mammalian cells. Genes Dev.
16:948–958. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Steinberg F, Gerber SD, Rieckmann T and
Trueb B: Rapid fusion and syncytium formation of heterologous cells
upon expression of the FGFRL1 receptor. J Biol Chem.
285:37704–37715. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hornik TC, Neniskyte U and Brown GC:
Inflammation induces multinucleation of Microglia via PKC
inhibition of cytokinesis, generating highly phagocytic
multinucleated giant cells. J Neurochem. 128:650–661. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang HY, Gu YY, Li ZG, Jia YH, Yuan L, Li
SY, An GS, Ni JH and Jia HT: Exposure of human lung cancer cells to
8-chloro-adenosine induces G2/M arrest and mitotic catastrophe.
Neoplasia. 6:802–812. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
An J, Yang DY, Xu QZ, Zhang SM, Huo YY,
Shang ZF, Wang Y, Wu DC and Zhou PK: DNA-dependent protein kinase
catalytic subunit modulates the stability of c-Myc oncoprotein. Mol
Cancer. 7:322008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee KJ, Lin YF, Chou HY, Yajima H, Fattah
KR, Lee SC and Chen BP: Involvement of DNA-dependent protein kinase
in normal cell cycle progression through mitosis. J Biol Chem.
286:12796–12802. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang B, Shang ZF, Li B, Wang Y, Liu XD,
Zhang SM, Guan H, Rang WQ, Hu JA and Zhou PK: DNA-PKcs associates
with PLK1 and is involved in proper chromosome segregation and
cytokinesis. J Cell Biochem. 115:1077–1088. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shang ZF, Huang B, Xu QZ, Zhang SM, Fan R,
Liu XD, Wang Y and Zhou PK: Inactivation of DNA-dependent protein
kinase leads to spindle disruption and mitotic catastrophe with
attenuated checkpoint protein 2 phosphorylation in response to DNA
damage. Cancer Res. 70:3657–3666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eberhardy SR and Farnham PJ: c-Myc
mediates activation of the cad promoter via a post-RNA polymerase
II recruitment mechanism. J Biol Chem. 276:48562–48571. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
El Sayegh TY, Kapus A and McCulloch CA:
Beyond the epithelium: Cadherin function in fibrous connective
tissues. FEBS Lett. 581:167–174. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu YN, Lee WW, Wang CY, Chao TH, Chen Y
and Chen JH: Regulatory mechanisms controlling human E-cadherin
gene expression. Oncogene. 24:8277–8290. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fransvea E, Angelotti U, Antonaci S and
Giannelli G: Blocking transforming growth factor-beta up-regulates
E-cadherin and reduces migration and invasion of hepatocellular
carcinoma cells. Hepatology. 47:1557–1566. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Amatangelo MD, Goodyear S, Varma D and
Stearns ME: c-Myc expression and MEK1-induced Erk2 nuclear
localization are required for TGF-beta induced
epithelial-mesenchymal transition and invasion in prostate cancer.
Carcinogenesis. 33:1965–1975. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim BY, Kwak SY, Yang JS and Han YH:
Phosphorylation and stabilization of c-Myc by NEMO renders cells
resistant to ionizing radiation through up-regulation of γ-GCS.
Oncol Rep. 26:1587–1593. 2011.PubMed/NCBI
|
|
36
|
Skvortsova I, Popper BA, Skvortsov S,
Saurer M, Auer T, Moser R, Kamleitner H, Zwierzina H and Lukas P:
Pretreatment with rituximab enhances radiosensitivity of
non-Hodgkin's lymphoma cells. J Radiat Res. 46:241–248. 2005.
View Article : Google Scholar : PubMed/NCBI
|