Introduction
Lung adenocarcinoma is the most common form of lung
cancer and it belongs to the histologic subgroup of non-small cell
lung cancer (1,2). This cancer type grows slowly but can
undergo hematogenous metastasis at an early stage. The prognosis
for survival is poorer for lung adenocarcinoma than squamous
carcinoma, and its 5-year survival rate after surgical removal is
less than 10% (3–6). In the United States, almost 40% of lung
cancers are adenocarcinoma and usually originate from the
surrounding lung tissue. The incidence of lung adenocarcinoma
varies with age, and is more common among female subjects. The
number of newly diagnosed cases has been on the increase in many
western developed countries in recent decades, and it now
constitutes the most common type of lung cancer among smokers,
replacing squamous cell lung cancer (7–9).
A microRNA (miRNA) is a short single
highly-conserved non-coding RNA that is important in the expression
and functional regulation of eukaryotic genomes (e.g.,
proliferation, apoptosis, migration and angiogenesis) and these
biological processes are integral for tumor formation and
development (10–12). miR-378 (11) inhibits migration and invasion of
prostate cancer cell, and promotes apoptosis. In addition, Zhou
et al (13) found that
miR-590-5p inhibited breast cancer cells, thereby providing a novel
therapeutic approach for breast cancer patients. The Cancer Genome
Atlas (TCGA), is an existing relatively authoritative sequencing
database, that contains a variety of common tumors (14). The purpose of this database is to
better understand the molecular basis of cancer through the
application of genome analysis technologies, identifying mutations
in DNA sequence, copy number variation and alterations in
methylation status.
The aim of the present study was to combine partial
genomic data to determine the prognosis of patients with lung
adenocarcinoma.
Materials and methods
miRNA and patient data
Level 3 data of miRNA expression profile and the
corresponding clinical data were downloaded from the TCGA (14). All the data were publically available.
The downloaded miRNA expression data and clinical data were
integrated, and patients with lung adenocarcinoma were selected for
inclusion. Patients with lung adenocarcinoma whose age was unknown
and those whose survival time was <30 days were excluded.
The downloaded data from TCGA included tissue miRNA
data from 521 patients with lung adenocarcinoma and miRNA
expression data of 46 cases of para-carcinoma tissue. To screen
differentially expressed miRNA, the selection criteria were set to
be log fold change >1 and P<0.05. The up- and downregulated
expression of miRNA in lung adenocarcinoma tissue was screened, and
standardized treatment was conducted on the miRNA expression. In
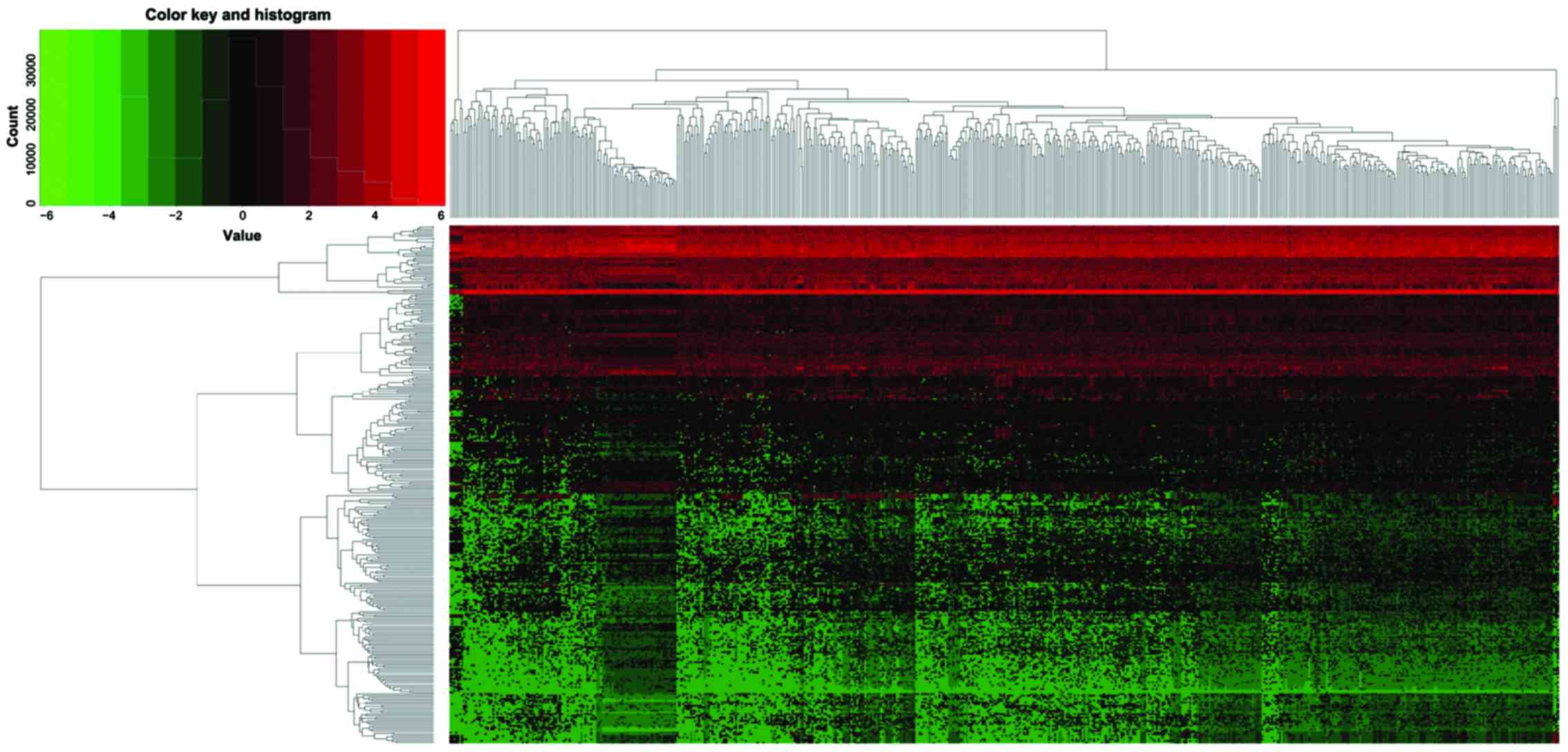
addition, the thermograph of tissue miRNA expression of patients
with lung adenocarcinoma was drawn through pheatmap R of software R
(15).
Establishment of prognostic model
To construct the prognosis model focusing on miRNA
for lung adenocarcinoma, a log2 transform was conducted on the
expression level of miRNA. A total of 478 cases with lung
adenocarcinoma were randomly divided into either the training or
test set, and the Chi-square test was applied to test whether there
was statistical significance at each stage from the two sets
(P<0.05). The miRNA that was closely associated with survival
time of the patients in the training set were selected through
univariate Cox proportional hazards regression (P<0.001). Next,
miRNA was constructed into the prognostic model with the
multivariate Cox regression. The optimal prognostic model was
selected based on the Akaike information criterion (16). The median value of the model of the
training set was regarded as the cut-off value, and the patients
with lung adenocarcinoma in the training set were divided into
either the high-risk or low-risk group. A Kaplan-Meier (KM)
survival plot was constructed and if P<0.05 (the model was
established in the training set) the model was entered into the
verification set. The median value of the training set model was
regarded as the cut-off value. In addition, the patients with lung
adenocarcinoma in the verification set were divided into high- or
low-risk groups. KM survival curve was constructed and if
P<0.05, it showed that the prognosis model was established.
After the model was established, KM survival curves were
respectively drawn for various subgroups (e.g., male vs. female,
age >65 or ≤65 years, type of treatment). Then, receiver
operating characteristic (ROC) curve of 5-year survival rate of
subgroups was drawn using the ROC package of software R (17).
Cox regression
A single factor Cox regression was used to evaluate
the relationship between clinical characteristics and survival time
of patients with lung adenocarcinoma. The clinical features that
were closely related to the survival were screened, and they,
together with prognosis model were analyzed using multivariate Cox
regression model to explore whether the prognosis model could be
used as an independent predictor of patients prognosis with lung
adenocarcinoma.
Results
A total of 1,881 miRNA expression spectrum (level 3)
data of 521 patients with lung adenocarcinoma tissue and 46 cases
of para-carcinoma tissue were downloaded from TCGA. A total of 309
differentially expressed miRNAs were screened, including 188
upregulated miRNAs and 121 downregulated miRNAs (Fig. 1). hsa-miR-210, hsa-miR-708 and
hsa-miR-96 were the three most significantly upregulated miRNAs,
while hsa-miR-486-1, hsa-miR-486-2 and hsa-miR-4732 were the three
most significantly downregulated miRNAs. According to the inclusion
criteria, the data from 478 patients with lung adenocarcinoma were
analyzed. These patients were sub-divided into the training (n=239)
or test set (n=239), based on the principle of random distribution
(Table I). There was no statistical
difference between the stages from the training or validation set
(P>0.05).
 | Table I.Patient demographics and tumor
characteristics from the training and validation set. |
Table I.
Patient demographics and tumor
characteristics from the training and validation set.
| Covariates | Total (n=478) | Training set
(n=239) | Testing set
(n=239) | P-value |
|---|
| Age (years) |
|
|
| 0.833 |
| ≤65 | 232 | 121 | 111 |
|
|
>65 | 246 | 118 | 128 |
|
| Pathological
stages |
|
|
| 0.90 |
| I | 259 | 134 | 125 |
|
| II | 111 | 53 | 58 |
|
| III | 78 | 33 | 45 |
|
| IV | 24 | 17 | 7 |
|
| NA | 6 | 2 | 4 |
|
| Pathology T
stages |
|
|
| 0.87 |
| T1 | 163 | 85 | 78 |
|
| T2 | 250 | 121 | 129 |
|
| T3 | 44 | 22 | 22 |
|
| T4 | 18 | 9 | 9 |
|
| Tx | 3 | 2 | 1 |
|
| Pathology N
stages |
|
|
| 0.83 |
| N0 | 309 | 161 | 148 |
|
| N1 | 87 | 40 | 47 |
|
|
N2-N3 | 70 | 32 | 38 |
|
|
NX-NA | 12 | 6 | 6 |
|
| Pathology M
stages |
|
|
| 0.91 |
| M0 | 315 | 155 | 160 |
|
| M1 | 23 | 17 | 6 |
|
| MX | 136 | 65 | 71 |
|
| NA | 4 | 2 | 2 |
|
| Sex |
|
|
| 0.86 |
| Male | 222 | 113 | 109 |
|
|
Female | 256 | 126 | 130 |
|
| Radiation
therapy |
|
|
| 0.75 |
| No | 374 | 192 | 182 |
|
| Yes | 59 | 24 | 35 |
|
| NA | 45 | 23 | 22 |
|
| Ethnicity |
|
|
| 0.86 |
|
Asian | 7 | 4 | 3 |
|
|
Caucasian | 373 | 191 | 182 |
|
| African
or African-American | 52 | 23 | 29 |
|
| NA | 46 | 21 | 25 |
|
| Status |
|
|
| 0.79 |
| Dead | 169 | 83 | 86 |
|
|
Surviving | 309 | 156 | 153 |
|
For the prognosis model constructed in the present
study, the prognostic score = 44.488 × (expression quantity of
hsa-miR-101-1) - 1.673 × (expression quantity of hsa-miR-200a) +
0.428 × (expression quantity of hsa-miR-4661) + 0.515 × (expression
quantity of hsa-miR-450a-2). Of the four miRNAs, hsa-miR-4661 and
hsa-miR-4661 had an upregulated expression, while hsa-miR-101-1 and
hsa-miR-200a had a downregulated expression. The significance
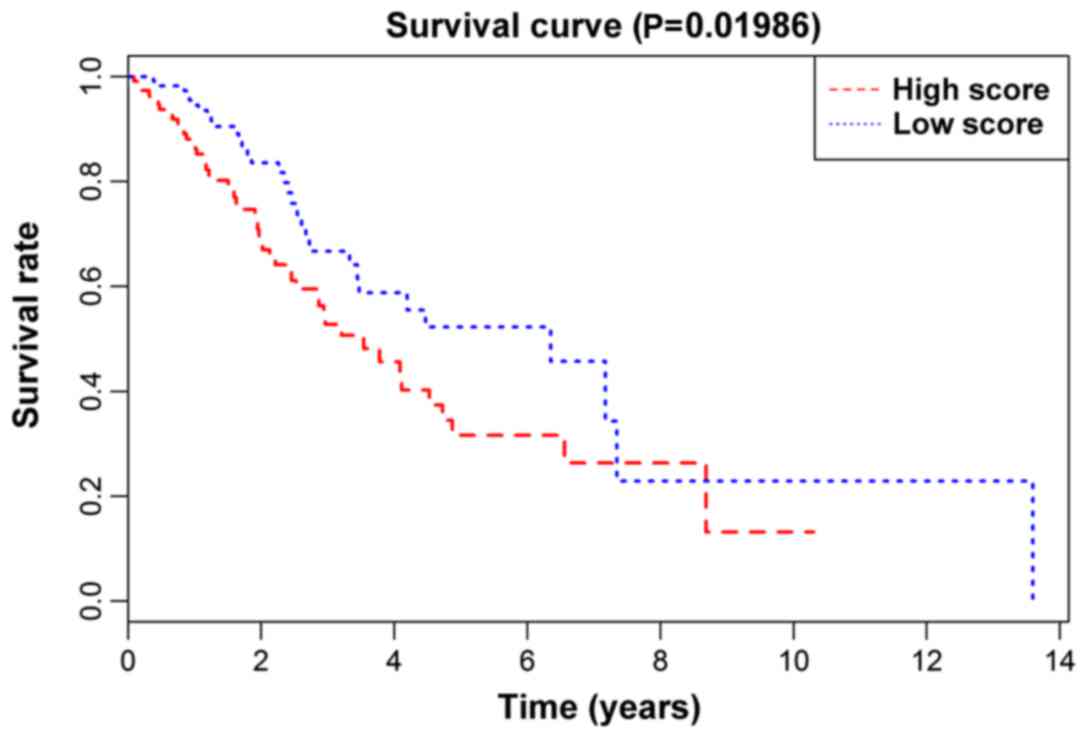
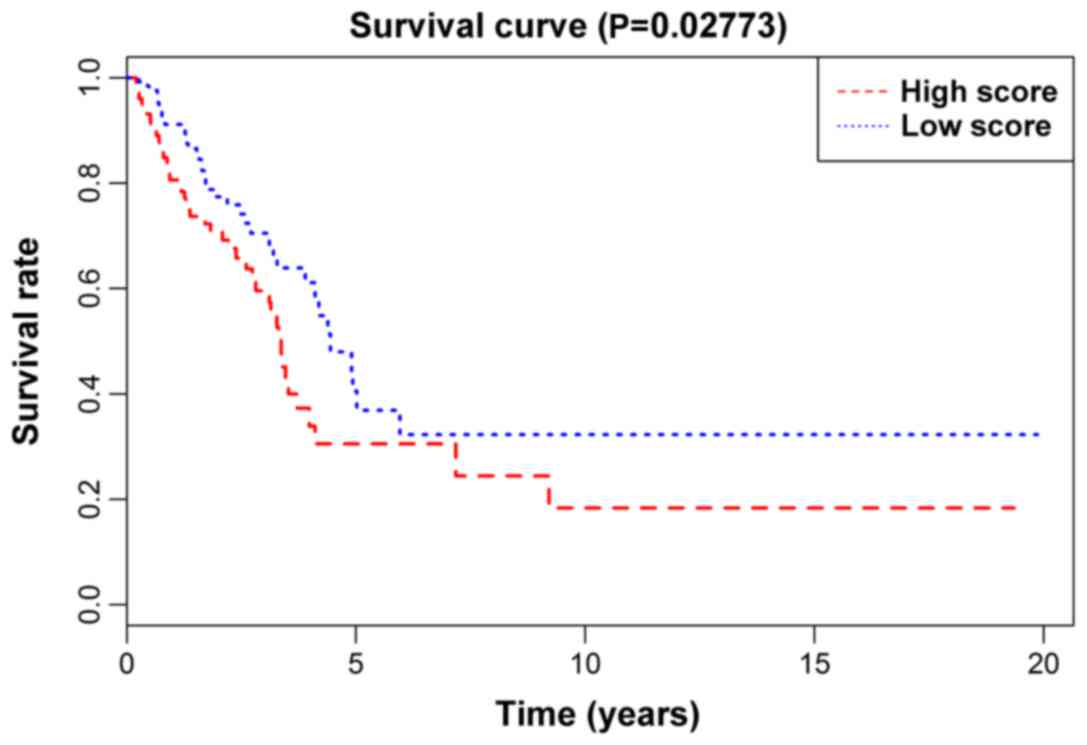
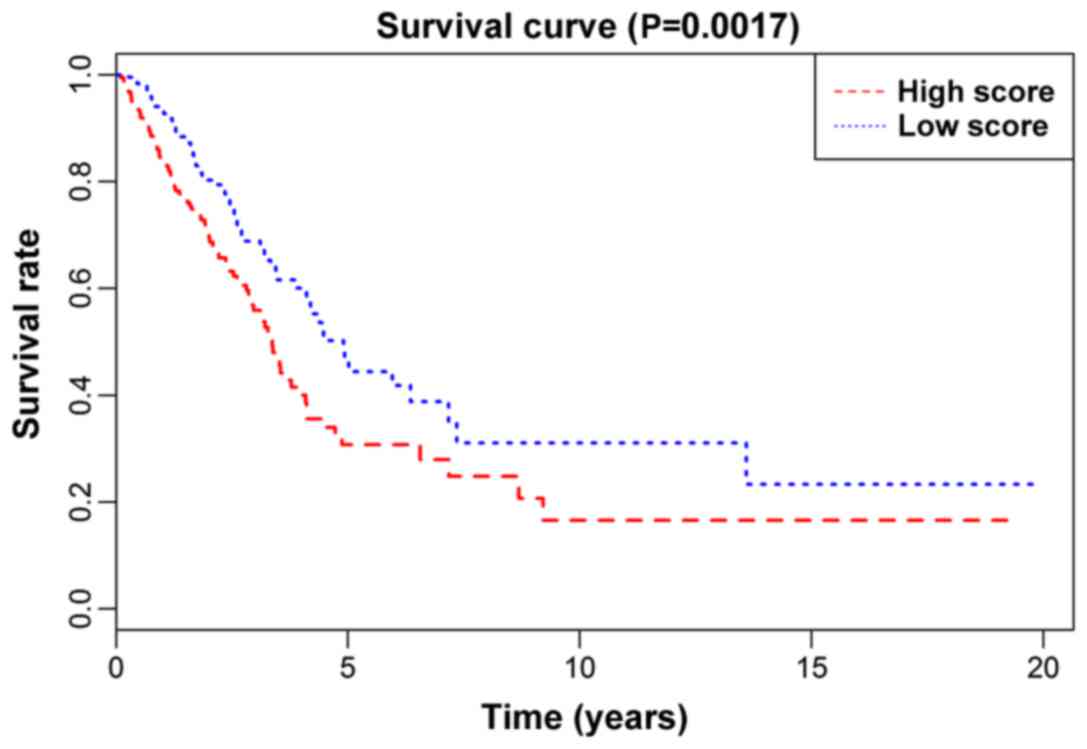
(P-value) from the KM survival curve of the prognostic model in the
training set, the validation set, and the total number of patients,
was 0.01986 (Fig. 2), 0.02773
(Fig. 3) and 0.0017 (Fig. 4), respectively, showing that the
high-risk group of lung adenocarcinoma patients in the model had a
shorter survival time than low-risk group. In addition, the model
assessed each subgroup. The statistical significance (P-values)
from the KM survival curve of the male, female, treatment,
non-treatment, Caucasian group and non-Caucasian groups was
0.04055, 0.01487, 0.65091, 0.00088, 0.00043 and 0.60825,
respectively (Figs. 2–4). In the present study, only the ROC curve
of the 5-year survival rate of the subgroups which KM survival
curve P<0.05 was utilized. The results showed that the prognosis
model had the best diagnostic performance in the Caucasian group,
with the area under the curve AUC=0.629; AUC of male group, female
group and non-treatment group was 0.595, 0.592 and 0.579
respectively.
In addition, the analysis through the single factor
Cox regression showed that pathologic stages (P=2.22E-08),
pathology N stages (P=2.52E-07), and pathology T stages (P=0.0007)
were closely related to the survival of patients with lung
adenocarcinoma (Table II).
Multivariable Cox regression showed both the prognosis model
(P=0.005) and age (P=0.03) were independent prognostic variable
models of lung adenocarcinoma (Table
II). However, the prognosis model constructed in the study was
superior to the assessment of the survival of patients with lung
adenocarcinoma with age.
 | Table II.Association of clinical factors and
the miRNA signature score with survival time from lung
adenocarcinoma patients. |
Table II.
Association of clinical factors and
the miRNA signature score with survival time from lung
adenocarcinoma patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (>65 vs.
≤65) | 1.18 | 0.307 | 1.31 | 0.03 |
| Pathologic stages
(IV vs. III vs. II vs. I) | 1.56 | 2.22E-08 |
|
|
| Pathology T stages
(T4 vs. T3 vs. T2 vs. T1 vs. T0) | 1.42 | 0.0007 | 1.19 | 0.157 |
| Pathology N stages
(N3 vs. N2 vs. N1 vs. N0) | 1.65 | 2.52E-07 | 1.17 | 0.15 |
| Sex (male vs.
female) | 1.12 | 0.498 |
|
|
| Ethnicity (white
vs. non-white) | 1.74 | 0.211 |
|
|
| miRNA model scores
(high vs. low score) | 2.87 | 0.0002 | 1.62 | 0.005 |
Discussion
By constructing a prognosis model focusing on four
miRNAs, patients with lung adenocarcinoma were divided into a high-
or low-risk group, and the survival time of the high-risk group was
shown to be less than that of low-risk group. After constructing a
multivariable Cox regression model using patient clinical
characteristics, the results showed that the prognosis model serves
as a potential independent prognostic model to assess the survival
time of patients with lung adenocarcinoma (P=0.005). The prognostic
model had a general assessment effect on the treatment (P=0.65091)
and non-Caucasian (P=0.60825) groups, but were underpowered due to
too few patients in the two groups. The prognostic model showed a
good assessment effect on the non-treatment and Caucasian groups,
and it could also predict the 5-year survival rate of patients with
lung adenocarcinoma of the Caucasian group (AUC=0.629). As the
majority of the data in TCGA included Caucasian patients, and
contained data from only a relatively smaller proportion from other
ethnicities, the conclusions drawn from the present study are more
applicable to Caucasian patients.
The prognostic model from the present study utilized
the miRNAs hsa-miR-101-1, hsa-miR-200a, hsa-miR-4661 and
hsa-miR-450a-2. Chang et al (18) showed that hsa-miR-200a was
downregulated in patients with gastric cancer, and could be used as
a potential biomarker to predict the survival and prognosis of
patients with gastric cancer. In the present study, there was an
upregulated expression of hsa-miR-200a miRNA in lung adenocarcinoma
tissue samples, indicating a differential regulation of the same
miRNA from these different tumors. Liu et al (19) demonstrated that the expression of
miR-101 in breast cancer could lead to E-cadherin downregulation,
and miR-101 may inhibit the expression of DNMT3A and the
proliferation and migration of human breast adenocarcinoma
MDA-MB-231 cells. Furthermore, hsa-miR-101 is involved in a wide
variety of other tumor processes [e.g., pancreatic cancer (20) and hepatocellular carcinoma (21)]. Other findings showed that the
downregulated expression of miRNA-450b-3p could lead to the
upregulated expression of HER3, thereby affecting the prognosis of
patients with breast cancer (22).
The prognosis model focusing on the four miRNAs constructed in the
present study confirmed that the expression of these tissue miRNAs
correlated with the survival time of patients with lung
adenocarcinoma, and it can be used as an independent prognostic
model for prognosis assessment of patients with lung
adenocarcinoma.
Although the independent prognosis model for
evaluating the patients with lung adenocarcinoma was established in
the study, there are limitations. Firstly, all the data in the
study were from one database, the TCGA, and the conclusions could
be more reliable if they were verified using other independent
databases. In addition, the study only included 478 patients with
lung adenocarcinoma, and some of the subgroups were underpowered.
In conclusion, the prognostic model developed in the present study,
focusing on miRNAs, can be used as an independent prognostic model
for survival time of patients with lung adenocarcinoma.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng F, Li H, Ning Z, Yang Z, Li H, Wang
Y, Chen F and Wu Y: CD147 and prostate cancer: a systematic review
and meta-analysis. PLoS One. 11:e0163672016. View Article : Google Scholar
|
|
7
|
Toh CK, Gao F, Lim WT, Leong SS, Fong KW,
Yap SP, Hsu AA, Eng P, Koong HN, Thirugnanam A and Tan EH:
Never-smokers with lung cancer: epidemiologic evidence of a
distinct disease entity. J Clin Oncol. 24:2245–2251. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Subramanian J and Govindan R: Lung cancer
in never smokers: a review. J Clin Oncol. 25:561–570. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gavelli G and Giampalma E: Sensitivity and
specificity of chest X-ray screening for lung cancer: review
article. Cancer. 89 Suppl:2453–2456. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Z, Li S, Song E and Liu S: The roles
of ncRNAs and histone-modifiers in regulating breast cancer stem
cells. Protein Cell. 7:89–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
OKelly F, Marignol L, Meunier A, Lynch TH,
Perry AS and Hollywood D: MicroRNAs as putative mediators of
treatment response in prostate cancer. Nat Rev Urol. 9:397–407.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou L, Zhao LC, Jiang N, Wang XL, Zhou
XN, Luo XL and Ren J: MicroRNA miR-590-5p inhibits breast cancer
cell stemness and metastasis by targeting SOX2. Eur Rev Med
Pharmacol Sci. 21:87–94. 2017.PubMed/NCBI
|
|
14
|
Feng Y, Liu J, Kang Y, He Y, Liang B, Yang
P and Yu Z: miR-19a acts as an oncogenic microRNA and is
up-regulated in bladder cancer. J Exp Clin Cancer Res. 33:672014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: a Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Posada D and Buckley TR: Model selection
and model averaging in phylogenetics: advantages of Akaike
information criterion and Bayesian approaches over likelihood ratio
tests. Syst Biol. 53:793–808. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang L, Guo F, Huo B, Lv Y, Wang Y and
Liu W: Expression and clinical significance of the microRNA-200
family in gastric cancer. Oncol Lett. 9:2317–2324. 2015.PubMed/NCBI
|
|
19
|
Liu J, Pang Y, Wang H, Li Y, Sun X, Xu F,
Ren H and Liu D: miR-101 inhibits the proliferation and migration
of breast cancer cells via downregulating the expression of DNA
methyltransferase 3a. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi.
32:299–303. 2016.(In Chinese). PubMed/NCBI
|
|
20
|
Fan P, Liu L, Yin Y, Zhao Z, Zhang Y,
Amponsah PS, Xiao X, Bauer N, Abukiwan A, Nwaeburu CC, et al:
MicroRNA-101-3p reverses gemcitabine resistance by inhibition of
ribonucleotide reductase M1 in pancreatic cancer. Cancer Lett.
373:130–137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao K, Li J, Zhao Y, Wang Q, Zeng Q, He S,
Yu L, Zhou J and Cao P: miR-101 inhibiting cell proliferation,
migration and invasion in hepatocellular carcinoma through
downregulating Girdin. Mol Cell. 39:96–102. 2016. View Article : Google Scholar
|
|
22
|
Zhao Z, Li R, Sha S, Wang Q, Mao W and Liu
T: Targeting HER3 with miR-450b-3p suppresses breast cancer cells
proliferation. Cancer Biol Ther. 15:1404–1412. 2014. View Article : Google Scholar : PubMed/NCBI
|