Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant tumors, with the third highest tumor-associated
mortality rate worldwide (1). Given
that the majority of patients with HCC are in the terminal stage of
the disease when diagnosed, numerous strategies, including surgical
exeresis, transarterial chemoembolization and liver transplantation
cannot not be applied (2).
Consequently, this leads to reduced survival time due to high
proliferation rate and migration ability of HCC (3).
Esophageal cancer-related gene 4 (ECRG4) was first
clonally identified as an esophageal cancer-associated tumor
suppressor gene in 1998 (4). The
ECRG4 gene is 12,500 base pairs (bp) in length and is localized on
chromosome 2ql2.2. The 444 bp open reading frame of ECRG4 comprises
four exons that encode a 148-amino acid peptide named ECRG4 protein
with a size of 17 κDa. ECRG4 is widely expressed in normal tissues,
including the heart, brain, lung, liver, placenta, skeletal muscle,
pancreas, kidney and prostate (5). In
recent years, ECRG4 expression, which serves a crucial role in the
progression of the malignant biological phenotype, has reported to
be suppressed in esophageal squamous, breast, colorectal cancers
and neurogliocytoma (6–9). However, few studies have reported ECRG4
expression in HCC. The present study aimed to investigate ECRG4
expression levels in HCC in patient samples and in HCC cell lines.
Furthermore, the roles of ECRG4 in HCC initiation and progression
as well as its underlying mechanism were examined.
Materials and methods
Patients and samples
Pathological samples from 56 patients with HCC were
collected during radical surgery from the Department of
Hepatobiliary Surgery, Shandong Provincial Hospital (Jinan, China)
from January 2008 to December 2010. Of the 56 patients, 42 were
male and 14 were female, with a median age of 55.68 years (range,
35–77 years). All patient samples were histologically graded
according to the World Health Organization Classification of Tumors
(10). The major clinicopathological
parameters are presented in Table I.
The tumors were graded into three types according to their degree
of differentiation; well-differentiated, moderately differentiated
and poorly differentiated. The number of patients for each tumor
type were 8, 31 and 17, respectively. Normal hepatic tissues
distant from tumor tissues that had no hepatic cirrhosis, adenoma
or focal nodular hyperplasia were also obtained. Tumor staging was
performed according to the recommendations of the International
Union Against Cancer (11). None of
the patients received any treatment prior to surgery, including
radiotherapy or chemotherapy. Follow-up sessions were performed on
all of the patients and the median follow-up duration was 43.4±3.61
months after the patients who succumbed to the disease were
excluded. Written informed consent was obtained from all patients
prior to enrollment in the present study. The use of the tissue
specimens was approved by the Research Ethics Committee of Shandong
Medical University (Jinan, Shandong).
 | Table I.Association of ECRG4 expression level
with clinicopathological factors (n=56). |
Table I.
Association of ECRG4 expression level
with clinicopathological factors (n=56).
|
|
| ECRG4 expression
level |
|
|
|---|
|
|
|
|
|
|
|---|
|
| Total (n=56) | Low (n=49) | High (n=7) | χ2 | P-value |
|---|
| Age at surgery
(years) |
|
|
| 7.439 | 0.024a |
| ≤35 | 1 | 0 | 1 |
|
|
|
36–50 | 13 | 11 | 2 |
|
|
| ≥51 | 42 | 38 | 4 |
|
|
| Differentiation |
|
|
| 1.828 | 0.401 |
| Well | 17 | 16 | 1 |
|
|
|
Moderately | 31 | 27 | 4 |
|
|
|
Poorly | 8 | 6 | 2 |
|
|
| Tumor size |
|
|
| 0.280 | 0.870 |
| <2
cm | 6 | 5 | 1 |
|
|
| 2–5
cm | 21 | 18 | 3 |
|
|
| >5
cm | 29 | 26 | 3 |
| PVTT |
|
|
| 1.377 | 0.241 |
|
Absent | 37 | 31 | 6 |
|
|
|
Present | 19 | 18 | 1 |
|
|
| Metastasis |
|
|
| 4.800 | 0.028a |
|
Absent | 35 | 28 | 7 |
|
|
|
Present | 21 | 21 | 0 |
|
|
| Satellite
lesions |
|
|
| 1.956 | 0.162 |
|
Absent | 45 | 38 | 7 |
|
|
|
Present | 11 | 11 | 0 |
|
|
| Cirrhosis |
|
|
| 0.242 | 0.622 |
|
Absent | 12 | 11 | 1 |
|
|
|
Present | 44 | 38 | 6 |
|
|
| Progression |
|
|
| 0.369 | 0.543 |
|
Absent | 26 | 22 | 4 |
|
|
|
Present | 30 | 27 | 3 |
|
|
| Mortality |
|
|
| 3.046 | 0.081 |
|
Absent | 23 | 18 | 5 |
|
|
|
Present | 33 | 31 | 2 |
|
|
| Ki67 status |
|
|
| 6.383 | 0.012a |
|
Low | 17 | 12 | 5 |
|
|
|
High | 39 | 37 | 2 |
|
|
Immunohistochemical procedures and
evaluation
Immunohistochemical Envision method was performed on
tissues sections (thickness, 4 µm) cut from formalin-fixed,
paraffin-embedded blocks. The samples were deparaffinized in xylene
and rehydrated through a graded series of ethanol washes. Following
inhibition of the endogenous peroxidase and antigen retrieval using
microwave irradiation in 0.01 M citrate buffer at pH 6.0), the
tissue sections were blocked with a 3%
H2O2-methanol solution for 10 min at room
temperature and then incubated with primary antibodies at 4°C
overnight, and then with horseradish peroxidase (HRP)-conjugated
secondary antibodies (Dako; Agilent Technologies, Inc., Santa
Clara, CA, USA) for 4 min at 4°C. All cases were investigated for
the presence of a rabbit polyclonal antibody against ECRG4 (cat.
no., sc-135139; 1:100; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) and a monoclonal antibody against Ki-67 (cat. no., 9449;
1:500; Cell Signaling Technology, Inc., Danvers, MA, USA).
Subsequent to washing, sections were stained with
3,30-diaminobenzidine (DAB) chromogen for 5 min and counterstained
with hematoxylin (Zhongshan Golden Bridge, Inc.) at room
temperature, then dehydrated and coverslips placed on top of the
samples. Colon carcinoma tissues were used for the positive
controls, and the negative control sections were incubated with PBS
instead of the primary antibody.
The immunostaining results were examined
independently by two clinical pathologists of the Department of
Pathology, Shandong Provincial Hospital using a light microscope
(Scope.A1; Carl Zeiss AG, Oberkochen, Germany) at a magnification
of ×400. For ECRG4, the results were classified as negative (score
0), weak (score 1–3), moderate (score 4–6) and intense (score 7–9),
based on the percentage of positive cells and staining intensity by
multiplying the two scores. To specify, a sample was scored 0–3
according to the percentage of positive cells (0, 0%; 1, 1–10%; 2,
11–50%; 3, 51–100%) and the staining intensity (0, negative; 1,
light brown; 2, moderate brown; 3, dark brown). All cases were
divided into two groups: Low (score 0 or 1+) and high (2+ or 3+)
expression. The Ki67 labeling index was scored by counting 500
cells and evaluating the percentage of cells that stained
positively in the nucleus: Score 1, 0–10%; score 2, 10–30%; score
3, 30–70%; score 4, >70%. For the evaluation of ECRG4 expression
level, a score of 1 or 2 were considered as the low expression,
whereas scores of 3 or 4 were classified as high expression.
Cell culture
SMMC-7721, HepG2, BEL-7404 and L02 cell lines were
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). All cell lines were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (PAA Laboratories; GE
Healthcare Life Sciences, Chalfont, UK), 100 U/ml penicillin and
100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). The
cells were cultured at 37°C in a humidified atmosphere containing
5% CO2.
Plasmid transfection
SMMC-7721 cells were transfected with recombinant
eukaryotic expression vector pcDNA3.1-ECRG4 (purchased from
Transheep Biotech, Shanghai, China) or pcDNA3.1 vector using
Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol: The cells were grown to
60% confluence in a 6-well dish prior to plasmid transfection, 4 µg
plasmid DNA and 10 µl Lipofectamine 2000 complexes were added in 2
ml of Opti-MEM medium (Gibco; Thermo Fisher Scientific, Inc.).
Following a 6 h incubation at 37°C, culture medium was changed to
usual complete medium, and the cells were subsequently cultured at
37°C for another 42 h until harvested.
Protein extraction and western blot
analysis
The cells were collected and lysed in modified
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Haimen, China), supplemented with Complete Protease
Inhibitor Cocktail (1 tablet/50 ml; Roche Molecular Diagnostics,
Pleasanton, CA, USA). Total cell lysate (50 µg protein) was
resolved by 10% SDS-PAGE and electrophoretically transferred onto
polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica,
MA, USA). Following blocking in 5% non-fat milk for 40 min at room
temperature, PVDF membranes were incubated overnight with primary
antibodies against ECRG4 (cat. no., sc-135139, 1:1,000; Santa Cruz
Biotechnology, Inc.), β-actin (cat. no., A1978, 1:2,000;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), Bax (cat. no.,
2774, 1:1,000; Cell Signaling Technology, Inc.), B cell lymphoma-2
(cat. no., 2872, Bcl-2; 1:1,000; Cell Signaling Technology, Inc.),
E-cadherin (cat. no., 3195, 1:1,000; Cell Signaling Technology,
Inc.), N-cadherin (cat. no., 13116, 1:1,000; Cell Signaling
Technology, Inc.) and Snail (cat. no., 3879, 1:1,000; Cell
Signaling Technology, Inc.) at 4°C, respectively. Subsequently, the
membranes were incubated for 1 h at 4°C with the appropriate
horseradish peroxidase-conjugated goat anti-mouse IgG (cat. no.,
31430) and goat anti-rabbit IgG (H+L, cat. no., 31460, Invitrogen;
Thermo Fisher Scientific, Inc.) secondary antibodies. Specific
protein bands were detected by enzyme-linked chemiluminescence kit
(ECL; Pierce; Thermo Fisher Scientific, Inc.) and protein
concentration was estimated relative to β-actin using Quantity
one® 1-D software (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Cell proliferation assay
Cell proliferation was determined using a modified
MTT assay (Roche Applied Science, Penzberg, Germany). Transfected
and control cells (7×103 cells/well) were seeded into
96-well plates. Following 1–7 days culture with 5% CO2,
20 µl 1 mg/ml MTT was added to each well and incubated for a
further 4 h at 37°C, and was subsequently replaced with 150 µl
dimethyl sulfoxide for 10 min at 37°C. Absorbance was determined at
490 nm using an ELISA multi-well spectrophotometer (Molecular
Devices, LLC, Sunnyvale, CA, USA). Each group contained 6 wells,
and the experiments were repeated three times.
Flow cytometry for cell apoptosis
detection
A total of 5×104 SMMC-7721 human
hepatocellular carcinoma cells were cultured in RPMI-1640
supplemented with 10% fetal bovine serum in 5% CO2 at
37°C. The SMMC-7721 cells were then transfected with pcDNA3.1-ECRG4
constructs as aforementioned. Following a 48 h incubation at 37°C,
the cells were collected by adding 0.25% pancreatic enzyme at room
temperature for 1 min and then 1 ml cold phosphate buffered
solution at room temperature for 1 min. Subsequently, the cells
were transferred to 1.5 ml cold Eppendorf microcentrifuge tubes and
centrifuged at 4°C for 5 min at 600 × g. The supernatant was then
discarded, and the cells were resuspended in 1 ml cold 70% ethanol.
The cells were stored overnight at −20°C. Flow cytometry was
subsequently performed to detect cell apoptosis. The un-transfected
SMMC-7721 cells were used as the control. All the protocols were
performed repeated in triplicate.
Transwell invasion and migration
assays
Cell invasion ability was evaluated in Boyden dual
chambers with 8 µm pore size membranes (BD Biosciences, Franklin
Lakes, NJ, USA). The membranes were coated with 40 µl Matrigel (BD
Biosciences) for 4 h at 37°C. A density of 2×104 cells
were suspended in 0.7 ml serum-free media (Gibco; Thermo Fisher
Scientific, Inc.) and added to the upper chamber, and the lower
chamber contained serum-positive media as a chemoattractant.
Following incubation for 48 h at 37°C with 5% CO2, the
media and cells remaining in the upper chamber were removed using a
cotton swab. The insert was fixed in 75% methanol and stained with
hematoxylin and eosin for 5 min at room temperature. The number of
invading cells was counted in five random high-power fields
(magnification, ×400) using the inverted microscope (Nikon Eclipse
TE300, Tokyo, Japan) and the mean number of invading cells was
evaluated for analysis. The procedure of cell migration assays
followed the aforementioned steps, with the exception of not adding
Matrigel to the membranes. For each assay, three identical
replicates were performed.
Statistical analysis
Statistics were calculated by SPSS software v.19
(IBM SPSS, Armonk, NY, USA). All data are presented as the mean ±
standard error of the mean. The differences were analyzed by using
the ANOVA, Student's t-test, as appropriate. Dunnett's-test was
used to compare the migration and invasion abilities. Kaplan-Meier
survival analysis was used to estimate the prognostic relevance of
ECRG4 and the survival difference between groups was assessed by
the log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
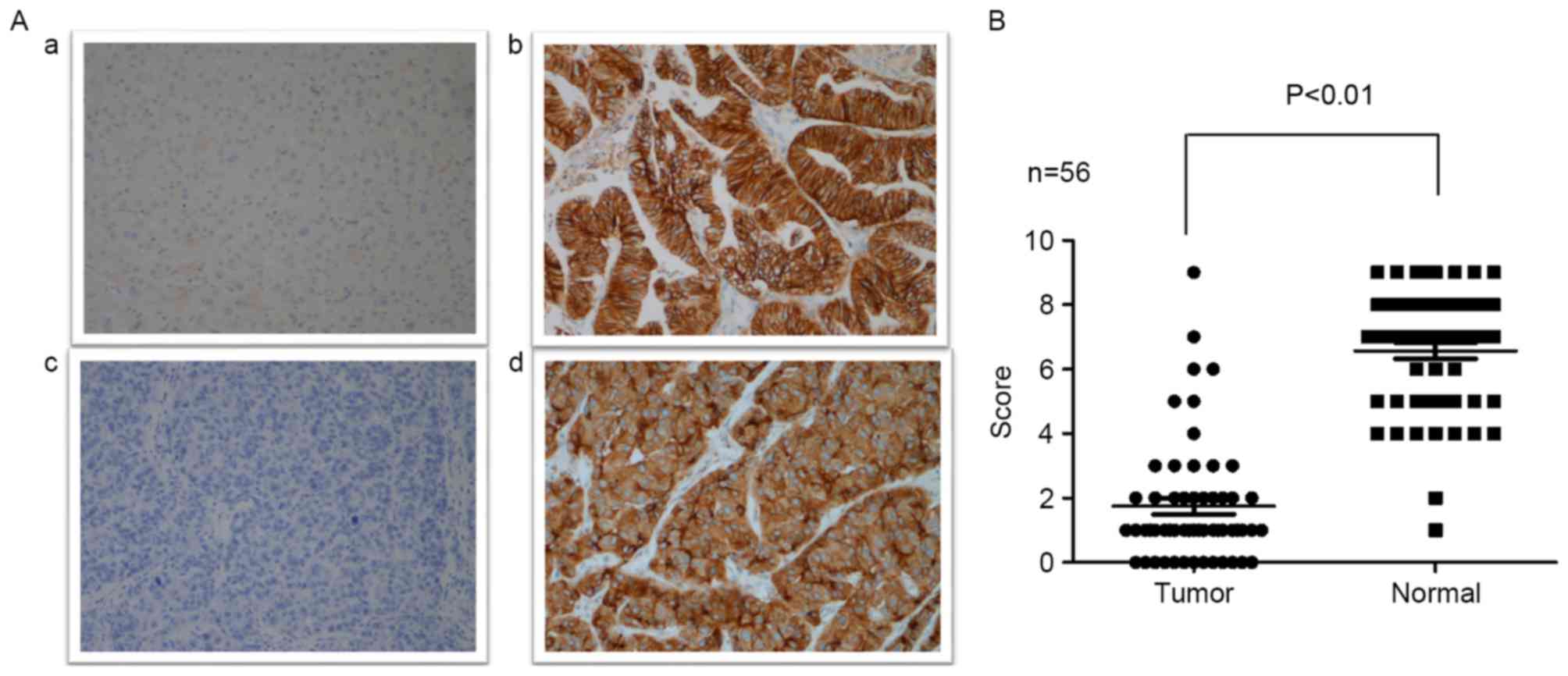
ECRG4 expression level in HCC samples
and cell lines
Immunohistochemistry was performed to evaluate ECRG4
expression level in 56 HCC samples and normal hepatic tissues.
Moderate to strong positive ECRG4 expression was detected in the
majority of the normal hepatic tissues, with only 2 samples
revealing weak positive expression. By contrast, ECRG4 expression
was significantly downregulated in HCC tissues compared with normal
hepatic tissues. Of the 56 HCC tissue samples, 7 samples
demonstrated high expression levels, which was reduced compared
with normal hepatic tissues (Fig. 1;
P<0.01).
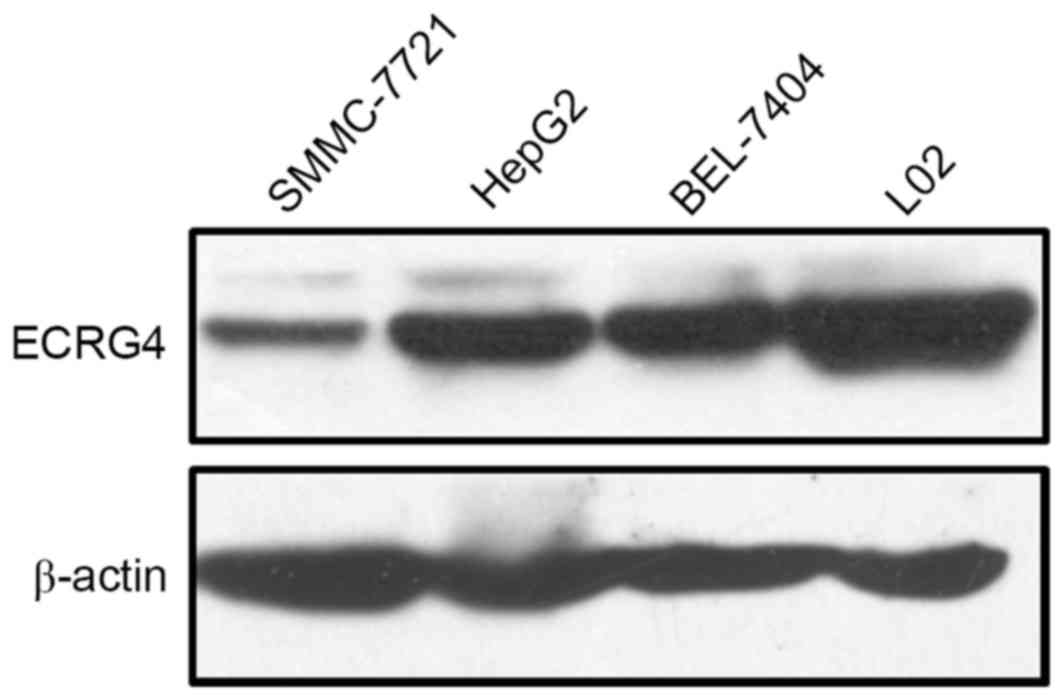
Furthermore, western blot analysis was performed to
assess ECRG4 protein expression levels in HCC and normal hepatic
cell lines. The HepG2, SMMC-7721, and BEL-7404 human HCC cell lines
exhibited lower ECRG4 expression levels compared with the L02
normal hepatic cell line (Fig. 2).
These results were consistent with the immunohistochemistry results
in patient samples.
Association of ECRG4 expression level
with clinicopathologic parameters
As presented in Table
I, there was no significant association between levels of ECRG4
expression and histological degree of differentiation, tumor size,
presence of portal vein tumor thrombosis, satellite lesions, tumor
relapse or mortality rate (P>0.05). However, there were
significant negative associations between levels of ECRG4
expression and age, metastasis and Ki-67 proliferation index
(P<0.05; Table I).
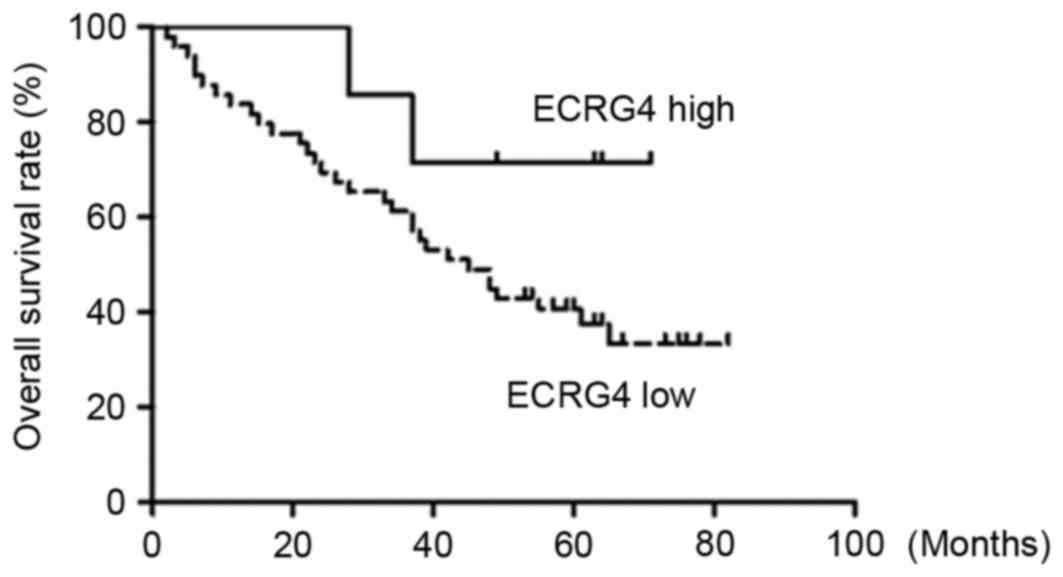
Log-rank test results demonstrated that the overall
survival rate was significantly higher in the high ECRG4 expression
level group compared with in the low ECRG4 expression level group
(Table II). Kaplan-Meier survival
curves also revealed the overall survival time of the low ECRG4
expression group was much shorter compared with the high ECRG4
expression group (Fig. 3). However,
the difference between the two groups was not statistically
significant due to the small number of the samples enrolled in the
present study (P>0.05).
 | Table II.Log-rank test of overall survival
time of patients with hepatocellular carcinoma. |
Table II.
Log-rank test of overall survival
time of patients with hepatocellular carcinoma.
| A, Mean survival
time |
|---|
|
|---|
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| ECRG4 expression
level | Average | SE | Lower | Upper |
|---|
| Low | 47.231 | 4.197 | 39.005 | 55.458 |
| High | 60.000 | 6.636 | 46.993 | 73.007 |
| Total | 49.796 | 3.932 | 42.089 | 57.503 |
|
| B, Median survival
time |
|
| Low | 45.000 | 6.999 | 31.283 | 58.717 |
| High | – | – | – | – |
| Total | 48.000 | 9.759 | 28.872 | 67.128 |
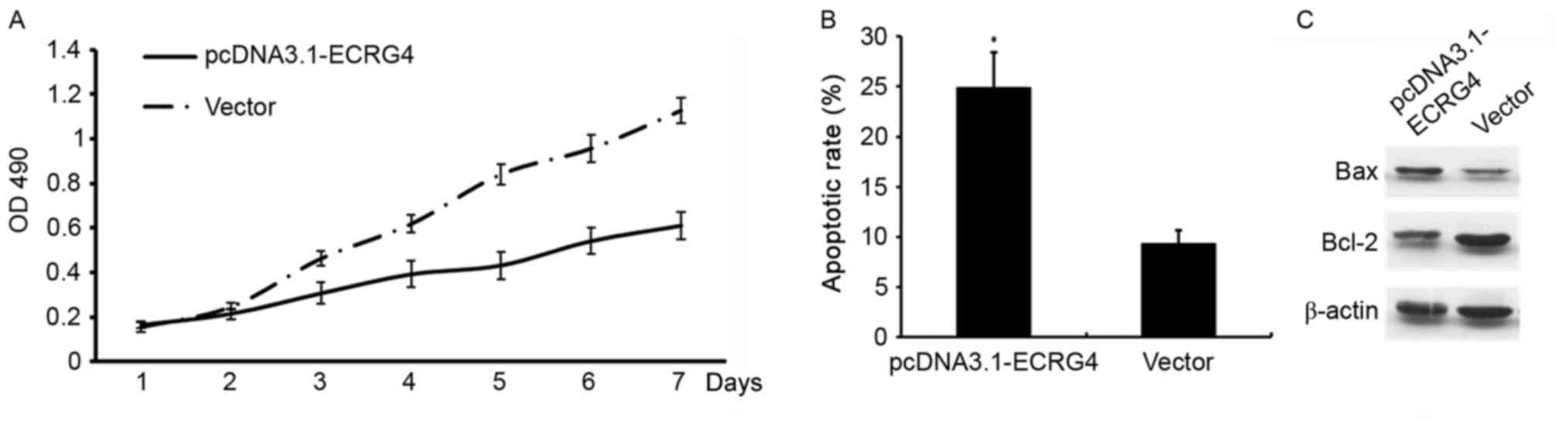
Elevated ECRG4 may inhibit HCC cell
proliferation and promote apoptosis
In order to determine whether ECRG4 affects HCC cell
growth, ECRG4 expression was upregulated by constructing a
pcDNA3.1-ECRG4 overexpression plasmid and transfecting the
construct into SMMC-7721 cells. Cell proliferation and apoptosis
was determined using MTT assays and flow cytometry, respectively.
Compared with the non-transfected cells, the proliferation rate was
decreased (P<0.05; Fig. 4A), and
the apoptotic rate was significantly increased in SMMC-7721 cells
transfected with the pcDNA3.1-ECRG4 plasmid (P<0.05; Fig. 4B).
The mitochondria signaling pathway is a classical
cell apoptosis pathway (12). Western
blotting was used to investigate the potential mechanisms
underlying ECRG4 in the induction of apoptosis of SMMC-7721 cells.
Western blotting revealed that when ECRG4 expression was
upregulated, BAX expression level decreased. By contrast, Bcl-2
expression level was increased, which may effectively promote
apoptosis in SMMC-7721 cells (Fig.
4C).
Upregulation of ECRG4 may increase
migration ability of HCC
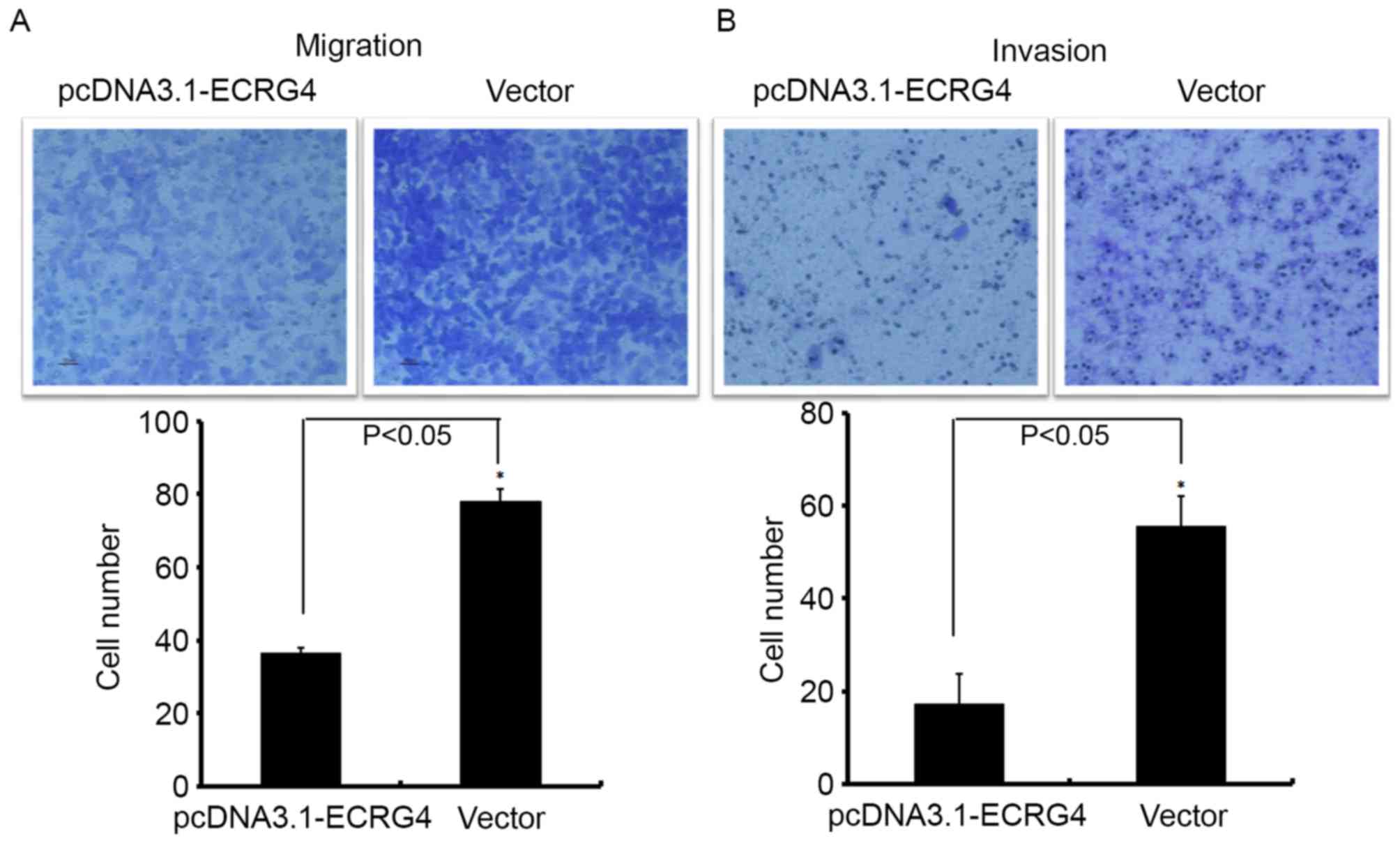
A Transwell chamber assay was used to investigate
the role of ECRG4 in HCC metastasis. The number of
SMMC-7721-ECRG4+ cells that invaded into the Matrigel
filter membrane (or without Matrigel) was significantly decreased
compared with the control (Fig. 5),
suggesting that upregulation of ECRG4 may effectively inhibit the
migration ability of HCC cells.
ECRG4 may affect the migration ability
of HCC cells by reversing epithelial-mesenchymal transition
(EMT)
EMT is one of the most important factors in
promoting tumor cell migration, which serves a key role in the
development of an organism and participates in tissue healing and
oncogenesis (13). To explore the
potential mechanism underlying the effect of ECRG4 on HCC cells
migration ability, ECRG4 expression was upregulated in SMMC-7721
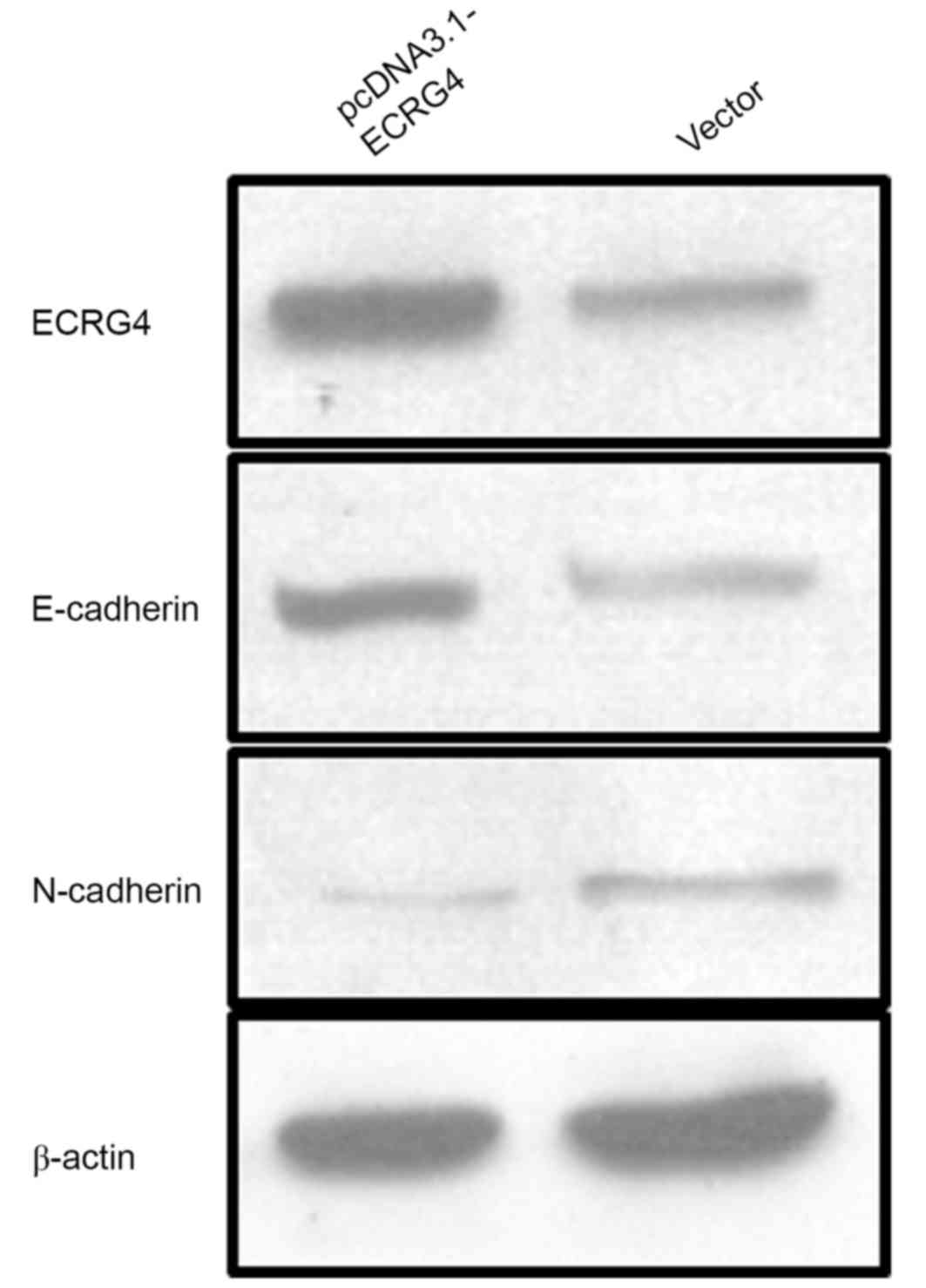
cells by transfecting pcDNA3.1-ECRG4 plasmid constructs. Western
blot analysis was performed to determine the changes in
EMT-associated phenotypic markers E-cadherin and N-cadherin
(Fig. 6). The results demonstrated
that E-cadherin expression levels were increased in the
pcDNA3.1-ECRG4-transfected SMMC-7721 cells compared with the cells
transfected with the vector alone. These findings indicate that
ECRG4 may affect HCC cell migration ability by reversing EMT.
Discussion
HCC is one of the most common malignant types of
tumor worldwide (14). Given that
effective measures for early discovery and diagnosis are lacking,
most patients with HCC are diagnosed at the end stage of the
disease, resulting in high mortality rates (15). Therefore, the development of a
biomarker for HCC diagnosis and prognosis is urgently required.
ECRG4 (C2ORF40; GenBank accession no. 325503) was
first identified as a differentially expressed gene through
comparison of normal esophageal tissues and familial esophageal
cancer tissues by the Cancer Research Institute of Chinese Academy
of Medical Sciences (4). The
downregulation of ECRG4 expression in esophageal cancer tissues
indicated that it may be a candidate tumor suppressor gene
(16,17). Thereafter, more studies demonstrated
that ECRG4 was involved in the initiation and progression of
numerous types of cancer, including breast carcinoma, squamous cell
carcinoma of the head and neck, gastric cancer and neurogliocytoma
(6,18–20). In
particular, low ECRG4 expression level was reported to
significantly promote tumor cell migration and inhibit apoptosis
(9,17,21). The
present study investigated ECRG4 expression level in HCC. To the
best of our knowledge, the present study is the first to
investigate the underlying mechanisms of ECRG4 in HCC cell
proliferation and migration.
A total of 56 HCC patient samples were used in the
present study. Of note, in 49 of the HCC tissue samples, ECRG4
expression was low or even undetectable. By contrast, ECRG4
expression in normal hepatic tissues was positive in all cases.
Similar levels of ECRG4 expression were demonstrated in the
SMMC-7721, HepG2, BEL-7404 HCC cell lines compared with the L02
normal hepatic cell line. Although there was no statistically
significant associations between the levels of ECRG4 expression and
histological degree of differentiation, tumor sizes, presence of
presence of portal vein tumor thrombosis and satellite lesions,
downregulated ECRG4 expression was significantly associated with
older age and tumor metastasis. These findings indicate that ECRG4
may affect tumor migration as well as invasion. Ki-67 expression, a
nuclear marker of cell proliferation, was associated with a poorer
outcome in predication of prognosis of patients with HCC (22). The present study demonstrated that HCC
tissue samples with decreased ECRG4 expression were more likely to
express high Ki-67 expression levels, suggesting that ECRG4
depletion may serve important roles in acquiring biological
malignant potential in HCC. Sabatier et al (6) observed a similar phenomenon in patients
with breast cancer, they revealed that ECRG4 was overexpressed in
smaller, early-stage tumors and significantly underexpressed in
later-stage tumors or positive sentinel lymph node status.
Furthermore, downregulated ECRG4 expression was closely associated
with poorer overall survival and disease-free survival (6). The survival analysis of the present
study also demonstrated that the 5-year survival rate in patients
underexpressing ECRG4 was lower compared with patients with high
levels of ECRG4 expression (36.7 vs. 71.4%). However, the number of
patients enrolled in the present study was insufficient to obtain
statistical significance. Future studies involving a larger group
of patients are required to validate the prognostic efficacy of
ECRG4 and reveal its pathophysiologic relevance in HCC.
A previous study by Xu et al (15) reported that upregulation of ECRG4 in
M2 cells, a head and neck squamous carcinoma cell line, resulted in
a significant decrease in proliferation rate. To determine whether
ECRG4 serves a similar role in the proliferation of HCC cells,
ECRG4 expression was upregulated in SMMC-7721 cells by transfecting
with pcDNA-3.1-ECRG4 plasmids to observe alterations in
proliferation and apoptosis. Upregulation of ECRG4 inhibited
SMMC-7721 cell proliferation and increased the rate of apoptosis.
Furthermore, the present study observed a significant change in the
Bax/Bcl-2 ratio, suggesting that ECRG4 may induce cell apoptosis by
activating a mitochondria-dependent apoptosis pathway via Bax/Bcl-2
changes. Another ECRG4-induced apoptosis pathway reported in
neurogliocytoma indicated that other pathways, including the
nuclear factor-κb signaling pathway, were also involved in
ECRG4-induced apoptosis (19).
Promotion of tumor cell migration ability is the
initiating factor for tumor migration, invasion and metastasis. The
Transwell chamber analysis of the present study indicated that
overexpressed ECRG4 resulted in a decrease in SMMC-7721 cell
migration ability. This finding partially explained the observation
in patient samples that those with low ECRG4 expression were more
vulnerable to metastasis. EMT is a biological process during which
epithelial cells transform into mesenchymal cells. This transition
is important in multiple physiological and pathological processes,
including embryonic development, tissue reconstruction and tumor
metastasis (23,24). Notably, high expression levels of
E-cadherin (an epithelial biomarker) and decreased expression
levels of N-cadherin (a mesenchymal biomarker), were observed in
SMMC-7721 cells with overexpressed ECRG4. These findings indicated
that ECRG4 may inhibit tumor migration and metastasis by reversing
EMT.
In conclusion, the present study investigated ECRG4
expression level in HCC tissue samples and cell lines. The present
study demonstrated that the downregulation of ECRG4 expression may
induce inhibition of apoptosis and promote migration, which
participated in the oncogenesis, development and metastasis of HCC.
Although marked differences of 5-year survival rates between
patients with high and low ECRG4 expression levels were observed,
the sample size was too small to obtain statistical significance.
Consequently, the conclusion that ECRG4 is an independent
prognostic factor for 5-year survival rates was not drawn. However,
ECRG4 is a candidate clinical biomarker for patients with HCC.
Therefore, an intensive investigation into the biological
mechanisms of ECRG4 in HCC should be performed in the future.
Acknowledgements
The present study was supported by the National
Nature Science Foundation for Young Scientists of China (grant no.
81202092), the Key and Development Program of Shandong Province
(grant no. 2015GSF118015) and the China Postdoctoral Science
Foundation (grant no. 2015M572053).
References
|
1
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akoad ME and Pomfret EA: Surgical
resection and liver transplantation for hepatocellular carcinoma.
Clin Liver Dis. 19:381–399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mlynarsky L, Menachem Y and Shibolet O:
Treatment of hepatocellular carcinoma: Steps forward but still a
long way to go. World J Hepatol. 7:566–574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Su T, Liu H and Lu S: Cloning and
identification of cDNA fragments related to human esophageal
cancer. Zhonghua Zhong Liu Za Zhi. 20:254–257. 1998.(In Chinese).
PubMed/NCBI
|
|
5
|
Matsuzaki J, Torigoe T, Hirohashi Y,
Tamura Y, Asanuma H, Nakazawa E, Saka E, Yasuda K, Takahashi S and
Sato N: Expression of ECRG4 is associated with lower proliferative
potential of esophageal cancer cells. Pathol Int. 63:391–397. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sabatier R, Finetti P, Adelaide J, Guille
A, Borg JP, Chaffanet M, Lane L, Birnbaum D and Bertucci F:
Down-regulation of ECRG4, a candidate tumor suppressor gene, in
human breast cancer. PLoS One. 6:e276562011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang YB and Ba CF: Promoter methylation of
esophageal cancer-related gene 4 in gastric cancer tissue and its
clinical significance. Hepatogastroenterology. 59:1696–1698.
2012.PubMed/NCBI
|
|
8
|
Matsuzaki J, Torigoe T, Hirohashi Y,
Kamiguchi K, Tamura Y, Tsukahara T, Kubo T, Takahashi A, Nakazawa
E, Saka E, et al: ECRG4 is a negative regulator of
caspase-8-mediated apoptosis in human T-leukemia cells.
Carcinogenesis. 33:996–1003. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Götze S, Feldhaus V, Traska T, Wolter M,
Reifenberger G, Tannapfel A, Kuhnen C, Martin D, Müller O and
Sievers S: ECRG4 is a candidate tumor suppressor gene frequently
hypermethylated in colorectal carcinoma and glioma. BMC Cancer.
9:4472009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND: World Health Organization Classification of Tumours of
the Digestive System. 3. 4th. IARC Press; Lyon: 2010
|
|
11
|
Xu HB, Xu LZ, Li L, Fu J and Mao XP:
Reversion of P-glycoprotein-mediated multidrug resistance by
guggulsterone in multidrug-resistant human cancer cell lines. Eur J
Pharmacol. 694:39–44. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weinberg SE and Chandel NS: Targeting
mitochondria metabolism for cancer therapy. Nat Chem Biol. 11:9–15.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu Z, Jiao D, Qiao J, Yang S, Yan M, Cui S
and Liu Z: Restin suppressed epithelial-mesenchymal transition and
tumor metastasis in breast cancer cells through upregulating
mir-200a/b expression via association with p73. Mol Cancer.
14:1022015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu F, Deng X, Yang X, Jin H, Gu D, Lv X,
Wang C, Zhang Y, Huo X, Shen Q, et al: Hypoxia upregulates
Rab11-family interacting protein 4 through HIF-1α to promote the
metastasis of hepatocellular carcinoma. Oncogene. 34:6007–6017.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu H, Li P, Zhai Y, Qu CF, Zhang LJ, Tan
YF, Li N and Ding HG: Diagnostic value of glypican-3 in serum and
liver for primary hepatocellular carcinoma. World J Gastroenterol.
16:4410–4415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mori Y, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Kurehara H, Mori R, Tomoda K, Ogawa R, Katada T, et al:
Expression of ECRG4 is an independent prognostic factor for poor
survival in patients with esophageal squamous cell carcinoma. Oncol
Rep. 18:981–985. 2007.PubMed/NCBI
|
|
17
|
Li L, Zhang C, Li X, Lu S and Zhou Y: The
candidate tumor suppressor gene ECRG4 inhibits cancer cells
migration and invasion in esophageal carcinoma. J Exp Clin Cancer
Res. 29:1332010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang CP, Wu BH, Wang BQ, Fu MY, Yang M,
Zhou Y and Liu F: Overexpression of ECRG4 enhances chemosensitivity
to 5-fluorouracil in the human gastric cancer SGC-7901 cell line.
Tumour Biol. 34:2269–2273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li W, Liu X, Zhang B, Qi D, Zhang L, Jin Y
and Yang H: Overexpression of candidate tumor suppressor ECRG4
inhibits glioma proliferation and invasion. J Exp Clin Cancer Res.
29:892010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu T, Xiao D and Zhang X: ECRG4 inhibits
growth and invasiveness of squamous cell carcinoma of the head and
neck in vitro and in vivo. Oncol Lett. 5:1921–1926.
2013.PubMed/NCBI
|
|
21
|
Jia J, Dai S, Sun X, Sang Y, Xu Z, Zhang
J, Cui X, Song J and Guo X: A preliminary study of the effect of
ECRG4 overexpression on the proliferation and apoptosis of human
laryngeal cancer cells and the underlying mechanisms. Mol Med Rep.
12:5058–5064. 2015.PubMed/NCBI
|
|
22
|
Chen HW, Huang XD, Li HC, He S, Ni RZ,
Chen CH, Peng C, Wu G, Wang GH, Wang YY, et al: Expression of FOXJ1
in hepatocellular carcinoma: Correlation with patients' prognosis
and tumor cell proliferation. Mol Carcinog. 52:647–659. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang JY, Zhang K, Chen DQ, Chen J, Feng
B, Song H, Chen Y, Zhu Z, Lu L, De W, et al: MicroRNA-451:
Epithelial-mesenchymal transition inhibitor and prognostic
biomarker of hepatocelluar carcinoma. Oncotarget. 6:18613–18630.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo Y, He DL, Jiang YG, Ning L, Shen SL,
Zhao JH and Cui XH: Role of beta-catenin signaling pathway in EMT
of human prostate cancer induced by HIF-1alpha. Zhonghua Yi Xue Za
Zhi. 90:1131–1136. 2010.(In Chinese). PubMed/NCBI
|