Introduction
Gastric carcinoma (GC) is one of the most common
types of gastrointestinal cancers (1). Local and systemic metastasis mainly
decided its poor prognosis (2). At
present, identifying the potential molecular target of the
metastatic process is critical for the diagnosis and treatment of
gastric carcinoma.
MicroRNAs (miRNAs) are a subset of noncoding
transcripts shorter than 25 nucleotides (3). Despite of their short length, miRNAs
have been confirmed to act vital important roles in various
biological processes including glycometabolism (4), angiogenesis (5), cell growth (6) and movement (7).
MicroRNA-381 (miR-381) was shown to serve as a tumor
suppressor in breast cancer (8),
chondrosarcoma (9), osteosarcoma
(10) and ovarian cancer (11). In these cancers, the expression levels
of miR-381 were commonly downregulated; over-expression of miR-381
in vitro could play anti-cancer roles by growth arrest and
metastasis inhibition through targeting Cx43, VEGF-C, LRRC4 and
YY1. However, the expression and functions of miR-381 in gastric
carcinoma are still unclear. Recently, long noncoding RNAs
(lncRNAs) were found to act as a sponge of microRNAs and regulate
the expression and function of microRNAs (12), but there is still no study to
investigate whether miR-381 can be regulated by lncRNAs.
In this study, we revealed that the expression of
miR-381 was significantly downregulatedted in GC tissues,
especially in metastatic patients. Functionally, we found that
over-expression of miR-381 could inhibit the metastatic ability
including migration and invasion, as well as epithelial-mesenchymal
transition (EMT) of GC cells through targeting SOX4. Furthermore,
the present study elucidated that long noncoding RNA TUG1 was a
direct negative regulator of miR-381 expression in gastric
carcinoma.
Materials and methods
Patients and cell culture
All protocols were approved by the Huaxi Hospital
Ethics Committee (Chengdu, Sichuan, China). Gastric carcinoma
tissues and matched tumor-adjacent tissues were collected from 60
GC patients including 36 males and 24 females who received surgical
resection between April 2013 and June 2015. The age range was
between 45 and 73 years, and the median age was 54 years.
GES-1, BGC-823, MGC-803, SGC-7901 and MKN28 cells
were bought from American Type Culture Collection and cultured in
Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Waltham, MA,
USA) containing 10% fetal bovine serum (FBS, Gibco, Grand Island,
NY, USA), 1% penicillin and 1% streptomycin.
Real time quantitative reverse
transcription-PCR (qRT-PCR)
TRIzol (Life Technologies, Grand Island, NY, USA)
regant was used to isolate total RNA from tissues and cells
according to the instructions. To determine the expression levels
of miR-381, SOX4 mRNA and lncRNA-TUG1, a quantitative one-step
Perfect Real Time RT-qPCR (SYBR-Green I) kit (Takara Biotechnology
Co., Ltd., Dalian, China) was used to synthetize cDNA and amplify
target genes. MiR-381 and RNU6 Bulge-Loop™ primers were purchased
from RiboBio Co., Ltd. (Guangzhou, China). SOX4, lncRNA-TUG1 and
GAPDH primers were synthetized by Sangon Co., Ltd. (Shanghai,
China). Sequences of these primers were shown as follow. SOX4
primers: 5′-AGCGACAAGATCCCTTTCATTC-3′ (forward) and
5′-CGTTGCCGGACTTCACCTT-3′ (reverse); lncRNA-TUG1:
5′-TAGCAGTTCCCCAATCCTTG3-3′ (forward) and
5′-CACAAATTCCCATCATTCCC-3′ (reverse); GAPDH:
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse). GAPDH was used as an
endogenous control for SOX4 and lncRNA-TUG1. RNU6 served as an
endogenous control for miR-381.
Cell transfection
The miR-381 mimics, negative control mimics
(miR-NC), siRNAs against lncRNA-TUG1 (forward:
5′-CCCUCCAUGAAUACCUGAATT-3′, reverse: 5′-UUCAGGUAUUCAUGGAGGGTT-3′)
and the negative control siRNA (si-NC) were bought from RiboBio
Co., Ltd. These mimics and siRNAs mentioned above were transfected
into GC cells with Lipofectamine 3000 according to the
manufacturer's instructions (Invitrogen, Waltham, MA, USA).
Wound healing assay
SGC-7901 cells were transfected with miR-381 mimics
or NC mimics for 24 h. Cells were seeded onto six-well plate until
the fusion reached above 90%. 100 µl tip was used to make a wound
at the middle of the well. Remnant cells were washed away by PBS.
Serum-free DMEM medium was used to culture cells for 48 h. The
healing of the wound was observed under the inverted
microscope.
Transwell assay
GC cells were re-suspended with serum-free DMEM
medium at a concentration of 2.5×105/ml. 200 µl cells
were seeded onto the upper well of Matrigel-coated (BD Biosciences,
Franklin Lakes, NJ, USA) 8-µm pore Transwell inserts (Nalge Nunc
International, Penfield, NY, USA) for invasion assay or uncoated
transwell inserts for migration assay. 750 µl DMEM medium with 10%
FBS was added to lower chamber. Cells were cultured 24 h for
invasion assay or 48 h for migration assay. After that, cells in
the lower surface were stained with 0.1% crystal violet, and cell
numbers were counted from 10 random fields of the lower surface of
the filter.
IHC staining
SP link IHC Detection Kits (Biotin-Streptavidin HRP
Detection Systems, SP-9001, including goat-rabbit secondary
antibodies) was purchased from Zhongshan Golden Bridge
Biotechnology, Inc. (Beijing, China). In brief, rabbit anti-human
polyclonal SOX4 (ab80261, dilution, 1:100) antibodies were
purchased from Abcam Biotechnology, Inc. (Cambridge, MA, USA) and
used to detect the protein expression in GC tissues. Tissue
sections were incubated with primary antibodies at 4°C overnight.
Goat-rabbit secondary antibodies were used to combine the primary
antibodies. The protein location and expression intensity were
visualized by 3,3-Diaminobenzidine tetrahydrochloride (DAB;
Zhongshan Golden Bridge Biotechnology, Inc.). The staining results
for the SOX4 were semi-quantitatively calculated by multiplying the
staining intensity and the percentage of positive normal cells, as
previously reported (13).
Luciferase reporter assay
The 3′-untranslated region (UTR) sequence of SOX4
predicted to interact with miR-381 and a mutation type were
synthesized and inserted into the XbaI and FseI sites of the pGL3
control vector (Promega Corporation, Fitchburg, WI, USA). These
constructs were named as wild-type SOX4 3′-UTR and mutant SOX4
3′-UTR. The reporter luciferase activity assay was performed using
the Dual-Luciferase® Reporter Assay System (Promega
Corporation) according to the instructions.
Western blot analysis
A total of 40 µg protein of each sample isolated by
RIPA lysis buffer (BioMed, China) was separated on SDS-PAGE gel and
transferred onto a nitrocellulose membrane (Invitrogen). The
membranes were incubated with the following primary rabbit
anti-human antibodies purchased from Abcam Biotechnology, Inc. at
4°C overnight: SOX4 (ab80261, dilution at 1:1,000), E-cadherin
(ab15148, dilution at 1:1,000), N-cadherin (ab18203, dilution at
1:1,000), Vimentin (ab16700, dilution at 1:1,000) and GAPDH
(ab9485, dilution at 1:1,000). Then, the membranes were incubated
with HRP-conjugated goat anti-rabbit secondary antibodies (ASR1038,
ABGENT, WuXi, China, dilution at 1:5,000), and the signal were
detected using the Bio-Rad Gel imaging system.
Statistical analysis
All quantitative data are expressed as (mean ± SD).
The Statistical Product and Service Solutions v21.0 software (SPSS
21.0, IBM, Armonk, New York, USA) was used to perform statistical
analysis. The Pearson's Chi-square test and the Student's t-test
were performed in this study. P<0.05 showed statistically
significant difference.
Results
MiR-381 expression is downregulatedted
in GC tissues
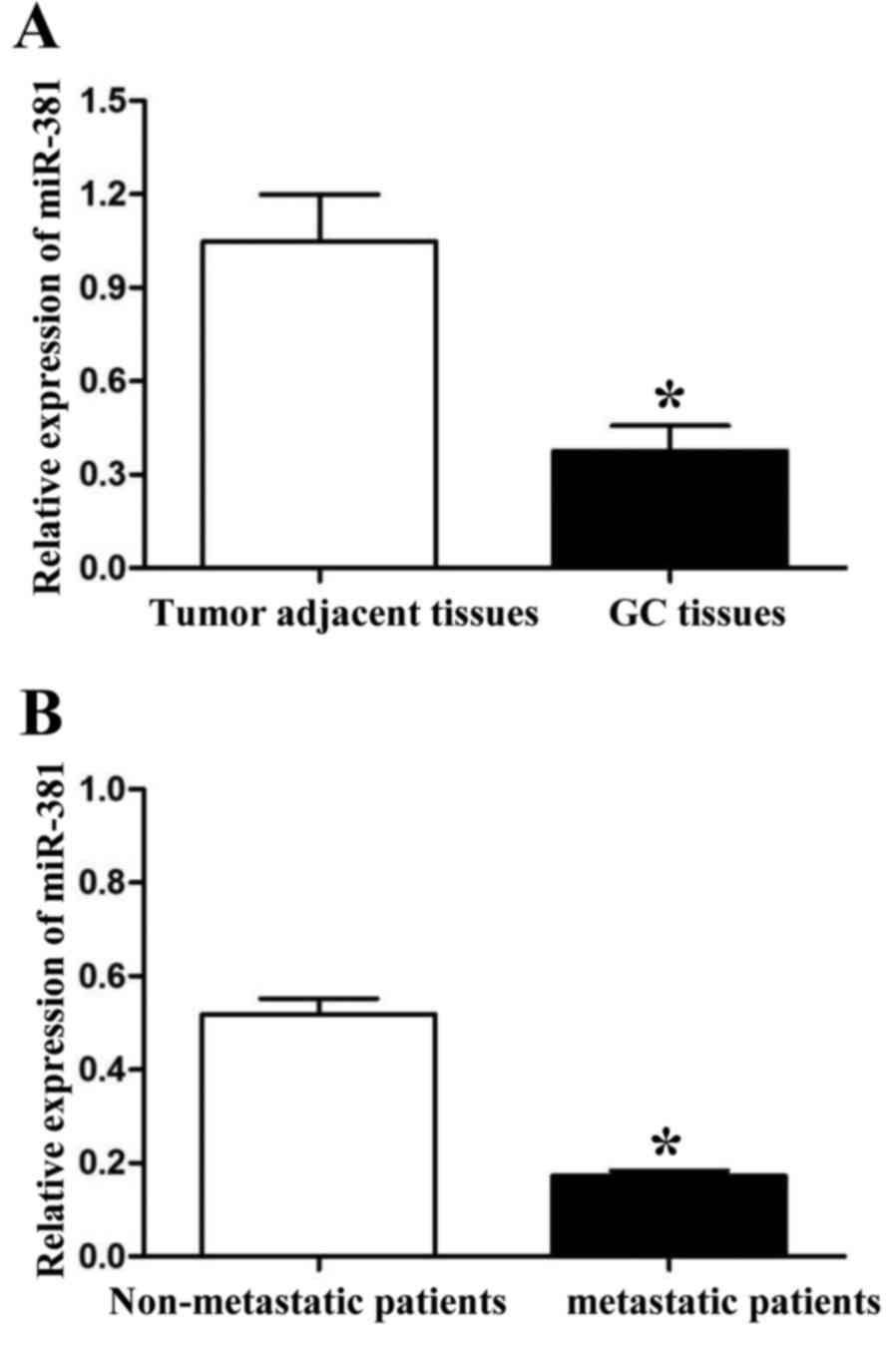
We detected the expression of miR-381 by qRT-PCR in
60 paired GC tissues and tumor adjacent tissues. Compared with
tumor adjacent tissues, lower expression level of miR-381 was
detected in GC tissues (P<0.05, Fig.
1A). Further, we divided these 60 GC patients into two groups
according to their metastatic status. PCR results showed that the
expression of miR-381 in tumors with metastasis was significantly
lower than those without metastasis (P<0.05, Fig. 1B). To determine the clinical values of
miR-381, we used the mean expression level of miR-381 as a cut-off
value to divide 60 GC tissues into two groups: miR-381 high
expression group (n=24) and miR-381 low expression group (n=36). As
shown in Table I, low expression of
miR-381 was associated with lymphatic metastasis (P=0.031) and
advanced TNM stage (III+IV, P=0.003). These results indicated that
miR-381 was related to tumor metastasis.
 | Table I.Clinical correlation of miR-381
expression in GC (n=60). |
Table I.
Clinical correlation of miR-381
expression in GC (n=60).
|
| miR-381 |
|
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristics | High expression
n=24 | Low expression
n=36 | χ2 | P-value |
|---|
| Gender |
|
| 0.104 | 0.793 |
| Male | 15 | 21 |
|
|
|
Female | 9 | 15 |
|
|
| Age (year) |
|
| 0.100 | 0.797 |
|
<50 | 11 | 18 |
|
|
| ≥50 | 13 | 18 |
|
|
| Hp infection |
|
| 3.665 | 0.068 |
| Yes | 10 | 24 |
|
|
| No | 14 | 12 |
|
|
| Lymphatic
metastasis |
|
| 5.602 | 0.031a |
| Yes | 10 | 26 |
|
|
| No | 14 | 10 |
|
|
| Histological
grade |
|
| 0.909 | 0.430 |
| I+II | 9 | 18 |
|
|
|
III+IV | 15 | 18 |
|
|
| TNM stage |
|
| 9.074 | 0.003a |
| I+II | 20 | 16 |
|
|
|
III+IV | 4 | 20 |
|
|
MiR-381 was associated with migration
and invasion of SGC-7901 cells
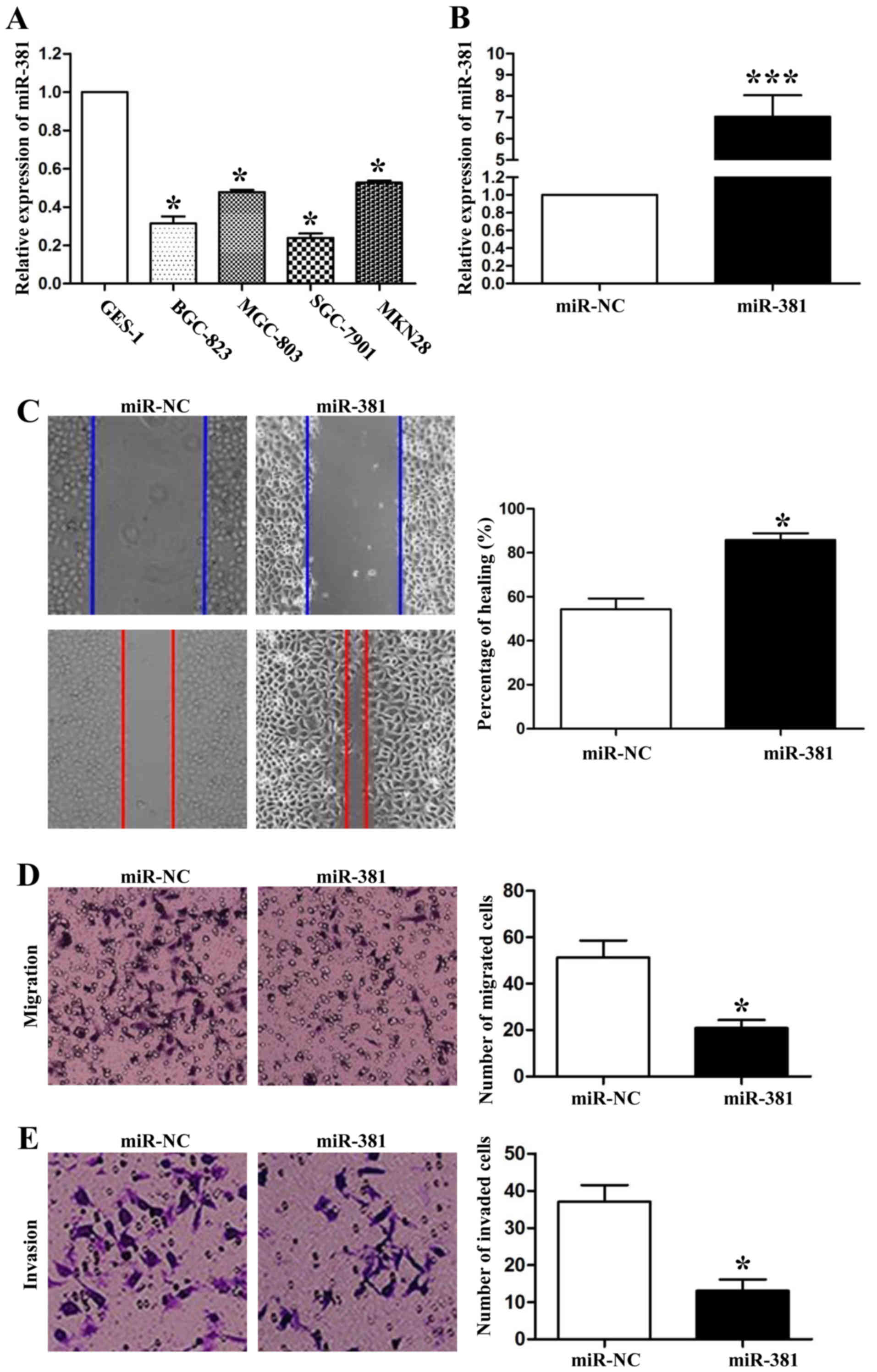
Because the expression of miR-381 was related to the
metastasis in gastric cancer, we wanted to confirm whether miR-381
influenced migration and invasion on GC cells. First, we detected
the expression levels of miR-381 in four GC cell lines (BGC-823,
MGC-803, SGC-7901, MKN28) and a normal gastric epithelial cell line
(GES-1). qRT-PCR data showed that the expression levels of miR-381
on these four GC cell lines, especially on SGC-7901 cells, were
lower than that in GES-1 cells (P<0.05, respectively, Fig. 2A). Next, we transfected miR-381 mimics
into SGC-7901 cells to successfully upregulatedte its expression
(P<0.001, Fig. 2B). Then, we used
wound healing assays to confirm that miR-381 over-expression
significantly inhibited the migration of SGC-7901 cells (P<0.05,
Fig. 2C). Transwell assays also
confirmed the effect of miR-381 for cell migration in vitro
(P<0.05, Fig. 2D). Moreover, we
still used the transwell assays to investigate the functions of
miR-381 for cell invasion. As shown in Fig. 3E, compared with the negative control
mimics group, upregulatedtion of miR-381 expression in SGC-7901
cells significantly decreased the invaded cell numbers (P<0.05).
Taken together, these data suggested that miR-381 could attenuate
the metastatic ability of SGC-7901 cells.
MiR-381 inhibited the
epithelial-mesenchymal transition through downregulatedting SRY-Box
4
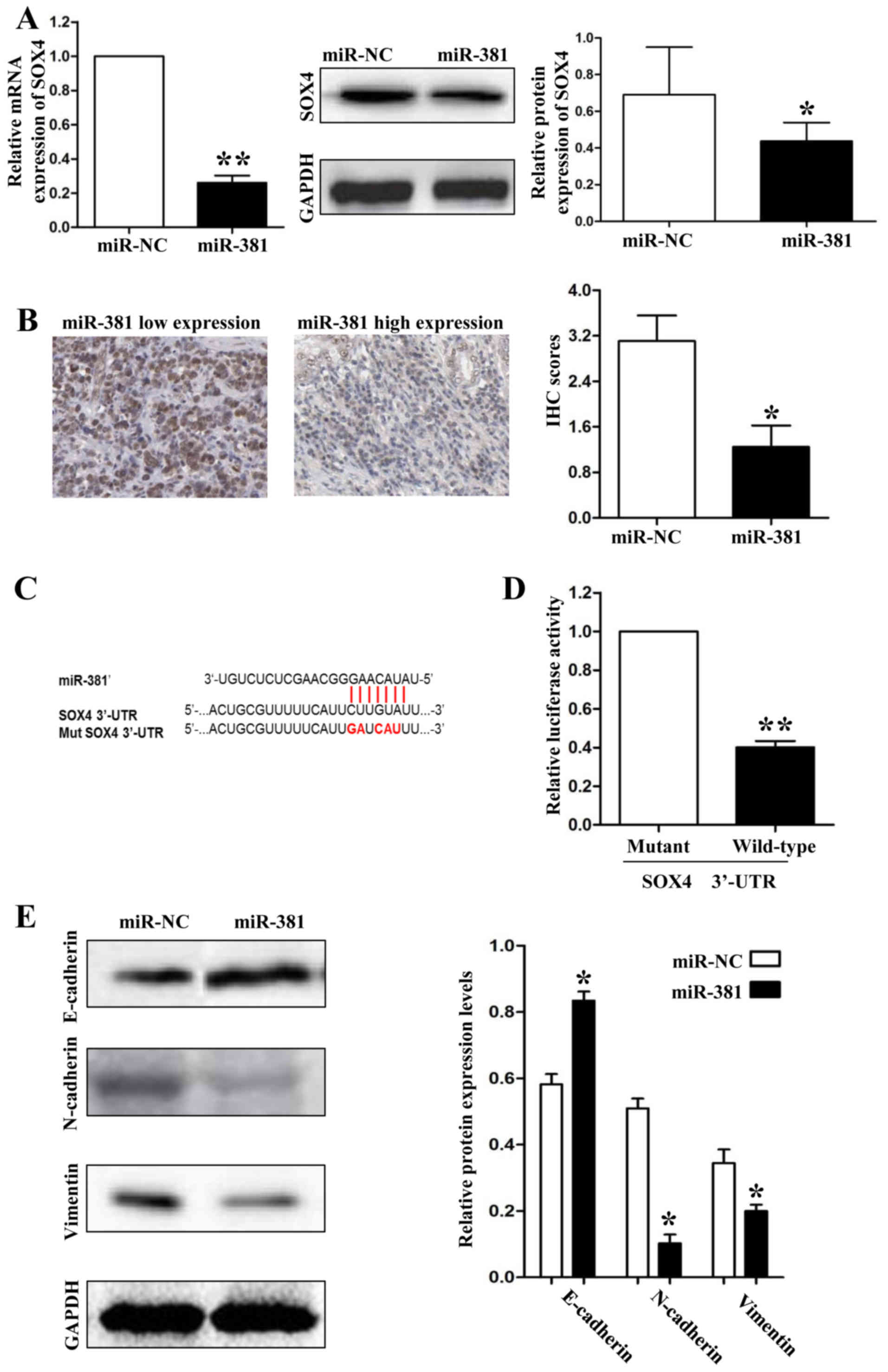
To investigate the mechanisms by which miR-381
inhibits GC metastasis, we screened the target genes of miR-381
through online databases (TargetScan: http://www.targetscan.org/and MiRanda: http://www.microrna.org/). SOX4 was predicted as a
potential downstream target of miR-381 according to the
bioinformation analysis. To confirm the relationship between
miR-381 and SOX4, first, we detect the expression of SOX4 in
SGC-7901 cells which were transfected with miR-381 mimics. As shown
in Fig. 3A, both the mRNA and protein
expression levels were downregulatedted when we overexpressed
miR-381 in SGC-7901 cells (P<0.05, respectively). Second, we
detected the protein expression levels of SOX4 in GC tissues by IHC
and demonstrated that the protein expression of SOX4 was higher in
miR-381 low-expression group than in miR-381 high-expression group
(P<0.05, Fig. 3B). Finally, we
used a Dual-Luciferase® Reporter Assay System to confirm
that miR-381 decreased the luciferase activity of SOX4 containing a
wild-type (wt) 3′-UTR but did not suppress the activity of SOX4
with a mutant (mt) 3′-UTR (P<0.01, Fig. 3C and 3D). Epithelial-mesenchymal
transition (EMT) was characterized as an important mechanism for
human cancer metastasis. Because Sox4 has been considered as a
master regulator of EMT in human cancers, we hypothesized that
miR-381 could attenuate EMT of SGC-7901 cells through inhibiting
SOX4 expression. So, we detected the protein expression levels of
epithelial marker (E-cadherin) and mesenchymal markers (N-cadherin
and vimentin) by immunoblotting. As shown in Fig. 3E, upregulatedtion of miR-381 in
SGC-7901 cells resulted in significantly increase of E-cadherin
expression (P<0.05) and decrease of N-cadherin and vimentin
expression (P<0.05, respectively).
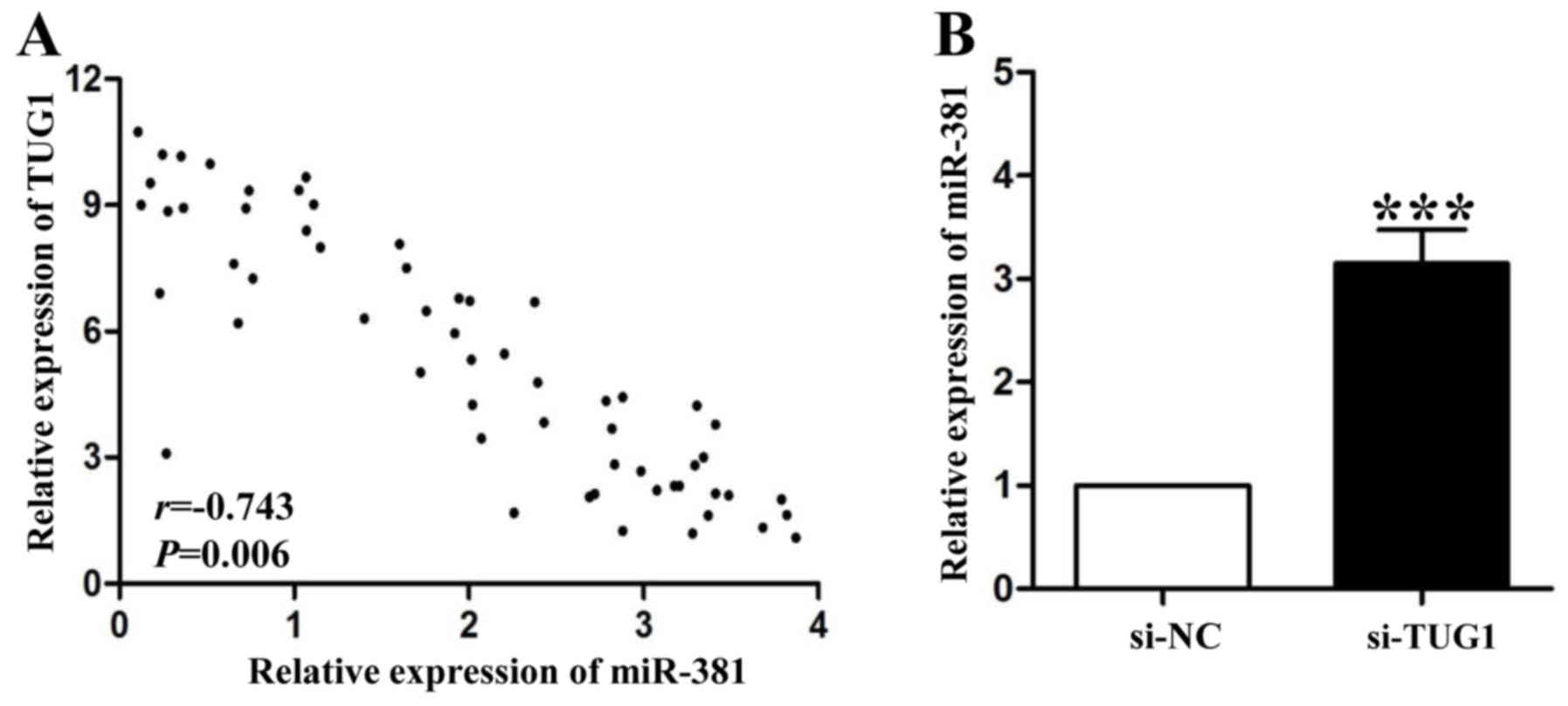
MiR-381 was negatively regulated by
long noncoding RNA TUG1
It has been demonstrated that miRNAs can be
downregulatedted through complementing with long noncoding RNAs
(lncRNAs) (12). In this study, we
found a negative relationship between long noncoding RNA TUG1
(lncRNA-TUG1) and miR-381 in GC tissues (P<0.01, Fig. 4A). LncRNA-TUG1 knockdown by siRNA
could significantly increase miR-381 expression in SGC-7901 cells
(P<0.001, Fig. 4B). These results
partly explained the reason of low miR-381 expression in GC
tissues.
Discussion
Metastasis including lymphatic metastasis and
distant metastasis are the important risk factors for the poor
prognosis of gastric carcinoma patients (7). Recently, microRNAs were demonstrated to
play an important role in cancer metastasis (14). In breast cancer, miR-181b could
inhibit CCL-18 induced cell migration and invasion through
targeting NF-κB (15). In endometrial
cancer, over-expression of microRNA-205 predicted tumor lymph node
metastasis and poor prognosis (16).
These evidences indicated that microRNAs might become a potential
target for tumor metastasis control.
In the present study, we focused on miR-381, which
exerted anti-cancer roles in breast cancer, osteosarcoma,
colorectal cancer and hepatocellular carcinoma. However, the
expression and biological functions in gastric carcinoma are still
unclear. In order to elucidate these questions, first, we detected
the expression levels of miR-381 in 60 paired gastric carcinoma
tissues and matched tumor adjacent tissues, and found that miR-381
was downregulatedted in cancer tissues. Clinical data showed that
low expression of miR-381 are correlated to lymphatic metastasis
and advanced TNM stage. Interestingly, we demonstrated that gastric
carcinoma tissues with metastasis had lower expression levels of
miR-381 than those without metastasis. These results indicated that
miR-381 could be served as a metastasis suppressor in gastric
carcinoma.
Previous studies have shown that miR-381 has various
biological functions in human cancers. For tumor growth, miR-381
could inhibit cell proliferation in colon cancer through targeting
LRH-1 (17), in glioma through
targeting BRD7 (17) and in renal
cancer through targeting WEE1 (18).
MiR-381 also could sensitize glioblastoma cells to temozolomide by
targeting neurofilament light polypeptide (19). In the present study, the results of
wound healing assays showed that over-expression of miR-381
inhibited the migration of gastric carcinoma cells. The transwell
chamber assays further confirmed this effect. Moreover, the
transwell results also showed that upregulatedtion of miR-381
suppressed cell invasion remarkably. Therefore, our results showed
that miR-381 could impair the metastatic ability of gastric
carcinoma cells.
Online databases result showed that SOX4, a member
of the SOX (SRY-related HMG-box) family of transcription factors,
might be a direct target of miR-381. In vitro, we
demonstrated that miR-381 inhibited SOX4 expression through binding
to its 3′-UTR in SGC-7901 cells. Moreover, patients with low
expression levels of miR-381 exerted high SOX4 protein expression
levels. EMT caused tumor metastasis has been well established
(20). Even, in general, SOX4 didn't
bind to the promoters of EMT markers to regulate their expression
(21), but SOX4 has been demonstrated
to promote the EMT in lung cancer (22) and prostate cancer (23). Data showed that a SOX4/EZH2 protein
complex could be formed to bind to promote serval EMT related
transcription factors expression, such as Snail, Zeb, and Twist
(20). In order to know whether
miR-381/SOX4 axis could regulate the EMT in gastric cancer, we
detected three markers of EMT and found that miR-381 overexpression
increased the endothelial marker's (E-cadherin) expression whereas
inhibited mesenchymal makers' (N-cadherin and vimentin) expression.
These results may explain why miR-381 could inhibit migration and
invasion of SGC-7901 cells.
LncRNA TUG1 has been demonstrated to be
overexpressed in gastric carcinoma and acts a metastasis inducer
(24). In the present study, we
confirmed a negative correlation between lncRNA TUG1 and miR-381 in
their expression levels. The expression of miR-381 can be rescued
after inhibition of lncRNA TUG1 in vitro. These demonstrated
that lncRNA TUG1 could inhibit the expression of miR-381 in gastric
cancer cells.
In conclusion, the present study demonstrates for
the first time that the expression of miR-381 is significantly
decreased in gastric cancer tissues, especially in cancer tissues
with metastasis. MiR-381 can inhibit the metastatic behaviors and
EMT of SGC-7901 cells. Furthermore, lncRNA TUG1 can inhibit the
expression of miR-381 in gastric carcinoma.
References
|
1
|
Zeng YJ, Zhang CD and Dai DQ: Impact of
lymph node micrometastasis on gastric carcinoma prognosis: A
meta-analysis. World J Gastroenterol. 21:1628–1635. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vilaseca Cabo A, Musquera Felip M, Ribal
Caparros MJ and Alcaraz Asensio A: Metastasis of gastric carcinoma
simulating a urothelial tumor. Case report and review of the
literature. Arch Esp Urol. 66:885–889. 2013.(In English, Spanish).
PubMed/NCBI
|
|
3
|
Micolucci L, Akhtar MM, Olivieri F, Rippo
MR and Procopio AD: Diagnostic value of microRNAs in asbestos
exposure and malignant mesothelioma: Systematic review and
qualitative meta-analysis. Oncotarget. 7:58606–58637. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang L, Huang J, Chen Y, Yang Y, Li R, Li
Y, Chen X and Yang D: Identification of several circulating
microRNAs from a genome-wide circulating microRNA expression
profile as potential biomarkers for impaired glucose metabolism in
polycystic ovarian syndrome. Endocrine. 53:280–290. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hansen TF, Andersen CL, Nielsen BS,
Spindler KL, Sørensen FB, Lindebjerg J, Brandslund I and Jakobsen
A: Elevated microRNA-126 is associated with high vascular
endothelial growth factor receptor 2 expression levels and high
microvessel density in colorectal cancer. Oncol Lett. 2:1101–1106.
2011.PubMed/NCBI
|
|
6
|
Liu C, Wang C, Wang J and Huang H:
miR-1297 promotes cell proliferation by inhibiting RB1 in liver
cancer. Oncol Lett. 12:5177–5182. 2016.PubMed/NCBI
|
|
7
|
Mataki H, Enokida H, Chiyomaru T, Mizuno
K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T,
Nakagawa M, et al: Downregulation of the microRNA-1/133a cluster
enhances cancer cell migration and invasion in lung-squamous cell
carcinoma via regulation of Coronin1C. J Hum Genet. 60:53–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ming J, Zhou Y, Du J, Fan S, Pan B, Wang
Y, Fan L and Jiang J: miR-381 suppresses C/EBPα-dependent Cx43
expression in breast cancer cells. Biosci Rep. 35:pii: e00266.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tzeng HE, Chang AC, Tsai CH, Wang SW and
Tang CH: Basic fibroblast growth factor promotes VEGF-C-dependent
lymphangiogenesis via inhibition of miR-381 in human chondrosarcoma
cells. Oncotarget. 7:38566–38578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Zhao C, Yu Z, Chen J, She X, Li P,
Liu C, Zhang Y, Feng J, Fu H, et al: Low expression of miR-381 is a
favorite prognosis factor and enhances the chemosensitivity of
osteosarcoma. Oncotarget. 7:68585–68596. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia B, Li H, Yang S, Liu T and Lou G:
MiR-381 inhibits epithelial ovarian cancer malignancy via YY1
suppression. Tumour Biol. 37:9157–9167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bayoumi AS, Sayed A, Broskova Z, Teoh JP,
Wilson J, Su H, Tang YL and Kim IM: Crosstalk between long
noncoding RNAs and MicroRNAs in health and disease. Int J Mol Sci.
17:3562016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li C, Yang W, Zhang J, Zheng X, Yao Y, Tu
K and Liu Q: SREBP-1 has a prognostic role and contributes to
invasion and metastasis in human hepatocellular carcinoma. Int J
Mol Sci. 15:7124–7138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei WJ, Shen CT, Song HJ, Qiu ZL and Luo
QY: MicroRNAs as a potential tool in the differential diagnosis of
thyroid cancer: A systematic review and meta-analysis. Clin
Endocrinol (Oxf). 84:127–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Wang YX, Chen LP and Ji ML:
Upregulation of microRNA-181b inhibits CCL18-induced breast cancer
cell metastasis and invasion via the NF-κB signaling pathway. Oncol
Lett. 12:4411–4418. 2016.PubMed/NCBI
|
|
16
|
Ma YJ, Ha CF, Bai ZM, Li HN, Xiong Y and
Jiang J: Overexpression of microRNA-205 predicts lymph node
metastasis and indicates an unfavorable prognosis in endometrial
cancer. Oncol Lett. 12:4403–4410. 2016.PubMed/NCBI
|
|
17
|
Liang Y, Zhao Q, Fan L, Zhang Z, Tan B,
Liu Y and Li Y: Downregulatedtion of MicroRNA-381 promotes cell
proliferation and invasion in colon cancer through upregulatedtion
of LRH-1. Biomed Pharmacother. 75:137–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen B, Duan L, Yin G, Tan J and Jiang X:
miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer
cells to 5-FU by upregulatedtion of Cdc2 activities in 786-O. J
Chemother. 25:229–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Z, Yang J, Xu G, Wang W, Liu C, Yang
H, Yu Z, Lei Q, Xiao L, Xiong J, et al: Targeting miR-381-NEFL axis
sensitizes glioblastoma cells to temozolomide by regulating
stemness factors and multidrug resistance factors. Oncotarget.
6:3147–3164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yeung KT and Yang J:
Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol.
11:28–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Parvani JG and Schiemann WP: Sox4, EMT
programs, and the metastatic progression of breast cancers:
Mastering the masters of EMT. Breast Cancer Res. 15:R722013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Chen P, Zu L, Liu B, Wang M and Zhou
Q: MicroRNA-338-3p suppresses metastasis of lung cancer cells by
targeting the EMT regulator Sox4. Am J Cancer Res. 6:127–140.
2016.PubMed/NCBI
|
|
23
|
Fu W, Tao T, Qi M, Wang L, Hu J, Li X,
Xing N, Du R and Han B: MicroRNA-132/212 upregulation inhibits
TGF-β-Mediated epithelial-mesenchymal transition of prostate cancer
cells by targeting SOX4. Prostate. 76:1560–1570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ji TT, Huang X, Jin J, Pan SH and Zhuge
XJ: Inhibition of long non-coding RNA TUG1 on gastric cancer cell
transference and invasion through regulating and controlling the
expression of miR-144/c-Met axis. Asian Pac J Trop Med. 9:508–512.
2016. View Article : Google Scholar : PubMed/NCBI
|