Introduction
Colorectal cancer (CRC) is the fourth most commonly
diagnosed cancer and the third leading cause of cancer-associated
mortality in men and women (1). A
number of patients are diagnosed in the advanced disease stage,
despite efforts and improvements in early diagnosis (2). Although a variety of therapeutic options
are available for CRC patients, including surgery, chemotherapy and
radiotherapy, the five-year survival rate of CRC has not
significantly improved (3). Previous
studies have demonstrated that genetic and epigenetic alterations
are involved in the tumorigenesis of CRC, including the activation
of oncogenes and/or the suppression of tumor suppressor genes.
Increasing evidence suggests that microRNAs (miRNAs/miRs) may serve
key roles in the development of CRC (4–6).
miRNAs, a class of endogenous single-stranded
non-coding RNAs, have been associated with various types of cancer
(7). miRNAs serve an essential role
in the regulation of gene expression and are involved in numerous
important physiological and pathological processes, including
development, differentiation and tumorigenesis (8–10). By
binding the 3′-untranslated region (3′-UTR) of mRNA, miRNA
suppresses protein synthesis through mRNA degradation or
translational repression. As a result, miRNAs may act as either
tumor suppressors or oncogenes (11–13). It is
becoming increasingly evident that miRNAs serve important roles in
cancer etiology. As a tumor suppressor gene in several cancer
types, miR-497 is able to affect tumor cell growth, migration,
invasion and apoptosis (14,15). To date, certain genes have been
identified as miR-497 targets, including Nrdp1, Cyclin E1, B-cell
lymphoma 2 and insulin-like growth factor 1 receptor (IGF1R)
(14,16–18).
However, the role and underlying mechanism of miR-497 in regulating
tumorigenesis remains to be further elucidated. Notably, it has
been reported that miR-497 regulates malignant behavior of CRC
cells by targeting IGF1R (16). In
the present study, it was observed that miR-497 targeted insulin
receptor substrate 1 (IRS1), which is characterized as a typical
cytosolic adaptor protein in both insulin receptor (IR) and IGF1R
signaling. Studies have indicated that nuclear IRS1 is able to
participate in modulating the transcriptional activity of genes
involved in cell growth and proliferation (19,20).
Nuclear IRS1 binds β-catenin and works as a transcriptional
modulator to stimulate cyclin D1 and c-myc promoter activities a
number of cancer types, where it acts as an oncogene, including in
pancreatic (21) and breast cancer
(22). Epidemiological investigation
has revealed that IRS1 is important in the etiology of CRC
(23).
It has been reported that miR-497 may serve roles in
CRC via affecting various signaling pathways (16). In the present study, the aim was to
identify the roles of miR-497 and its molecular and cellular
mechanisms in CRC. Ectopic expression of miR-497 inhibited
proliferation, migration and invasion of CRC cells by suppressing a
key target, IRS1. Furthermore, the present study defined the
molecular mechanism of the tumor suppressive function of miR-497 by
inhibiting both phosphoinositide 3-kinase (PI3K)/AKT and
mitogen-activated protein kinase (MAPK)/extracellular
signal-regulated kinase (ERK) signaling pathways via IRS1
suppression. The results of the present study revealed that miR-497
expression was significantly downregulated in human CRC tissues
compared with adjacent paired normal controls. IRS1 expression in
CRC tumors was negatively correlated with miR-497 expression. The
results of the present study revealed a novel mechanism of miR-497
in CRC, and demonstrate its potential to be used as a novel
strategy to develop miR-497-based therapeutics.
Materials and methods
Cell culture and clinical tissues
Human CRC cell lines SW1116 and SW480 (purchased
from Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), and HEK-293T cells (American Type Culture
Collection, Manassas, VA, USA) were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific,
Inc.), 100 IU/ml penicillin and 100 mg/ml streptomycin. All cells
were incubated at 37°C in an atmosphere of 5% CO2.
Colon cancer tissues and adjacent normal tissues
were collected from clinical patients undergoing colon cancer
resection. All tissue samples were immediately snap-frozen in
liquid nitrogen following surgery. All human CRC samples were
divided into Grade I, Grade II and Grade III–IV according to the
WHO classification (24). In total,
50 pairs of CRC tissues and adjacent normal tissues from patients
who underwent surgical operations at The Third Affiliated Hospital
of Soochow University (Changzhou, China) from August 1, 2013 to
July 31, 2014, were obtained for the study. Written informed
consent was obtained from all patients. The present study was
approved by the review board and ethics committee of The Third
Affiliated Hospital of Soochow University.
Lentiviral packaging and stable cell
line establishment
The Lentiviral Packaging kit was used (Thermo Fisher
Scientific, Inc.) for stably overexpressing miR-497 in CRC cells.
Lentivirus carrying miR-497 or negative control (miR-NC) was
packaged following according to the manufacturer's protocol.
Lentivirus was packaged in HEK-293T cells and secreted into the
medium. Cells were transfected with lentivirus carrying miR-497 or
miR-NC in the presence of polybrene (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and selected by puromycin (Sigma-Aldrich; Merck
KGaA) for 2 weeks to obtain stable cell lines.
miRNA mimic transfection
Cells were seeded into 6, 12, 24, or 96-well plates
and incubated at 37°C and 5% CO2 overnight. miR-497
mimics and miR-NC were chemically synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). Cells at 50–70% confluence
were transfected with miR-497 or miR-NC using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Transfected cells were harvested at 24 or
48 h following transfection.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells using
1.0 ml of Trizol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol, and purified RNA
was stored at −80°C prior to further analysis. RT-qPCR analysis for
mature miR-497 was performed in triplicate using the PrimeScript RT
Reagent kit (Takara Biotechnology Co., Ltd, Dalian, China)
according to the manufacturer's protocol. Briefly, 500 ng total RNA
was reverse transcribed into cDNA, and qPCR was performed using
SYBR Premix DimerEraser (Takara Biotechnology Co., Ltd.) on a
7900HT system. The thermocycling conditions were as follows:
Pre-denaturation at 95°C for 30 sec, followed by 40 cycles of 95°C
for 5 sec, 55°C for 30 sec and 72°C for 31 sec. The sequences of
the primers used for RT-qPCR were as follows: miR-497 RT,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACA-3′; miR-497-forward
(F), 5′-ACACTCCAGCTGGGCAGCAGCACACTGTGG-3′; miR-497-reverse (R),
5′-TGGTGTCGTGGAGTCG-3′; U6 RT, 5′-AACGCTTCACGAATTTGCGT-3′; U6-F,
5′-CTCGCTTCGGCAGCACA-3′; and U6-R, 5′-TGGTGTCGTGGAGTCG-3′. The
miR-497 expression in each group was determined relative to that of
U6, and fold changes were calculated by relative quantification
(2−ΔΔCq) (25).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) assay was used to determine
cell viability. Cells were seeded at a density of 2,000 cells per
well in 96-well plates and cultured as described above for 48 h
following transfection. After 24, 48, 72 and 96 h incubation, CCK-8
was added into each well, followed by an additional 2 h incubation
at 37°C. Absorbance at a wavelength of 450 nm was subsequently
determined. Experiments were performed in triplicate.
Wound healing assay
Cells were transfected with miR-497 or miR-NC
according to the manufacturer's protocol, and subsequently cultured
to 95% confluence in 6-well plates. Cell monolayers were scratched
using a 20 µl tip to form wound gaps and washed twice with PBS to
remove the detached cells. After 24 h, the wound healing was
photographed at various time points. The cell migration distances
were measured in three different areas to indicate the migration
ability of various cell treatments.
Invasion assay
The effect of miR-497 on tumor invasion was
investigated using 24-well BD Matrigel invasion chambers (BD
Biosciences, Franklin Lakes, NJ, USA) according to the
manufacturer's protocol. The transfected cells (5×104)
were seeded in the upper well of the invasion chamber containing
serum-free RPMI-1640, and RPMI-1640 containing 10% FBS was applied
to the lower chamber. After 24 h, any non-invading cells on the top
well were removed with a cotton swab, while cells in the bottom
well were fixed with 3% paraformaldehyde and stained with 0.1%
crystal violet. Images were captured in three independent fields
(magnification, ×10). The membrane was air-dried, soaked with 33%
acetic acid (200 µl/well) at room temperature for 15 min and
subsequently transferred to a 96-well plate. The absorbance at a
wavelength of 570 nm was recorded (Synergy 2; BioTek Instruments,
Inc., Winooski, VT, USA). Results were obtained from three
independent experiments.
Western blotting
Cells were treated as previously described, and were
harvested after 24 h and lyzed in radioimmunoprecipitation assay
buffer supplemented with protease inhibitors (100 mM Tris-HCl, pH
7.4, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 2 mM
phenylmethylsulfonyl fluoride, 1% deoxycholate acid, 0.1% SDS, 2 mM
DTT, 1 mM sodium orthovanadate, 2 mM leupeptin and 2 mM pepstatin)
on ice for 30 min (26). Following
centrifugation, protein concentrations were determined by the
bicinchoninic acid method (Beyotime Institute of Biotechnology,
Haimen, China), and 20 µg protein was then separated by 10%
SDS-PAGE. Subsequently, protein was electrically transferred onto a
nitrocellulose membrane (Whatman GmbH, Dassel, Germany). The
membrane was incubated with anti-IRS1 (1:1,000; catalog no. CST
2382; Cell Signaling Technology, Inc., Danvers, MA, USA) and
anti-GAPDH (1:5,000; catalog no. MB001; Bioworld Technology, Inc.,
St. Louis Park, MN, USA) antibodies at 4°C overnight, followed by
incubation at room temperature for 2 h with a secondary antibody
(catalog no. 31460; Thermo Fisher Scientific, Inc.) diluted 1:2,000
for IRS1 detection and 1:5,000 for GAPDH detection. Antibodies
against phosphorylated (p)-AKT (Ser473) (1:1,000; catalog no. CST
4060), AKT (1:2,000; catalog no. CST 9272), p-ERK1/2 (1:1,000;
catalog no. CST 14474) and ERK1/2 (1:2,000; catalog no. CST 4348)
were purchased from Cell Signaling Technology, Inc., and were
incubated at 4°C overnight, followed by incubation at room
temperature for 2 h with the aforementioned secondary antibody a
1:2,000 dilution. ECL Detection System (Thermo Fisher Scientific,
Inc.) was used for protein signal detection. The density of the
signals was quantified using ImageJ software with the ChemiDoc
Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
GAPDH was used as a control for normalization.
Luciferase reporter assay
Prediction of miR-497 binding sites was performed
using TargetScan software using the key words ‘IRS1’ and ‘human
species’. TargetScan (www.targetscan.org) and miRanda (www.microrna.org) predict biological targets of miRNAs
by searching for the presence of conserved 8mer, 7mer and 6mer
sites that match the seed region of each miRNA (27). A fragment of 3′-UTR of IRS1 containing
the putative miR-497 binding site was amplified by PCR. To generate
a construct containing the mutant miR-497 binding site, two
nucleotides corresponding to the 5′-seeding region of the miR-497
binding site on the wild type fragment were substituted. Its
complementary sequence in the 3′-UTR of IRS1 (UGCUGCU) was replaced
by UCCACCA. The PCR products were digested using SacI and
HindIII, inserted into pMIR-REPORTER (Promega Corporation,
Madison, WI, USA) and validated by DNA sequencing. Constructs were
transfected into HEK-293 cells in 24-well plates and co-transfected
with miR-497 or miR-NC. Luciferase assays were performed 24 h
post-transfection using the Dual Luciferase Reporter Assay system
(Promega Corporation).
Xenograft studies
For tumor growth assay, male nude mice [BALB/cA-nu
(nu/nu), 6-week-old, weighting 20–25 g] were purchased from
Shanghai Laboratory Animal Center (Chinese Academy of Sciences,
Shanghai, China), and animals were maintained under special
pathogen-free (SPF) conditions. Mice were housed at a temperature
of 26–28°C, relative humidity of 40–60% and 10 h light/14 h dark
cycle, with access to food and water ad libitum. Animal
protocols were approved by the Animal Welfare Committee of Soochow
University (The Third Affiliated Hospital of Soochow University).
Aliquots of cells (5×106) were suspended in 150 µl of
FBS-free RPMI-1640 medium and subcutaneously injected into each
side of the posterior flank of nude mice. Tumor size was measured
using vernier calipers every 2 days when they became visible, and
the tumor volume was calculated according to the formula: Volume =
0.5 × length × width2. Mice were sacrificed on day 22
following injection of tumor cells, and xenografts were collected.
The animals were euthanized by cervical dislocation (28).
Statistical analysis
All experiments were performed in triplicate, and
data were analyzed with GraphPad Prism 5 (GraphPad Software, Inc.,
La Jolla, CA, USA). The correlation between the expression of
miR-497 and IRS1 in CRC tissues was analyzed using Spearman's rank
test. Statistical evaluation for data analysis was determined by
the Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-497 is significantly
downregulated in CRC tissue
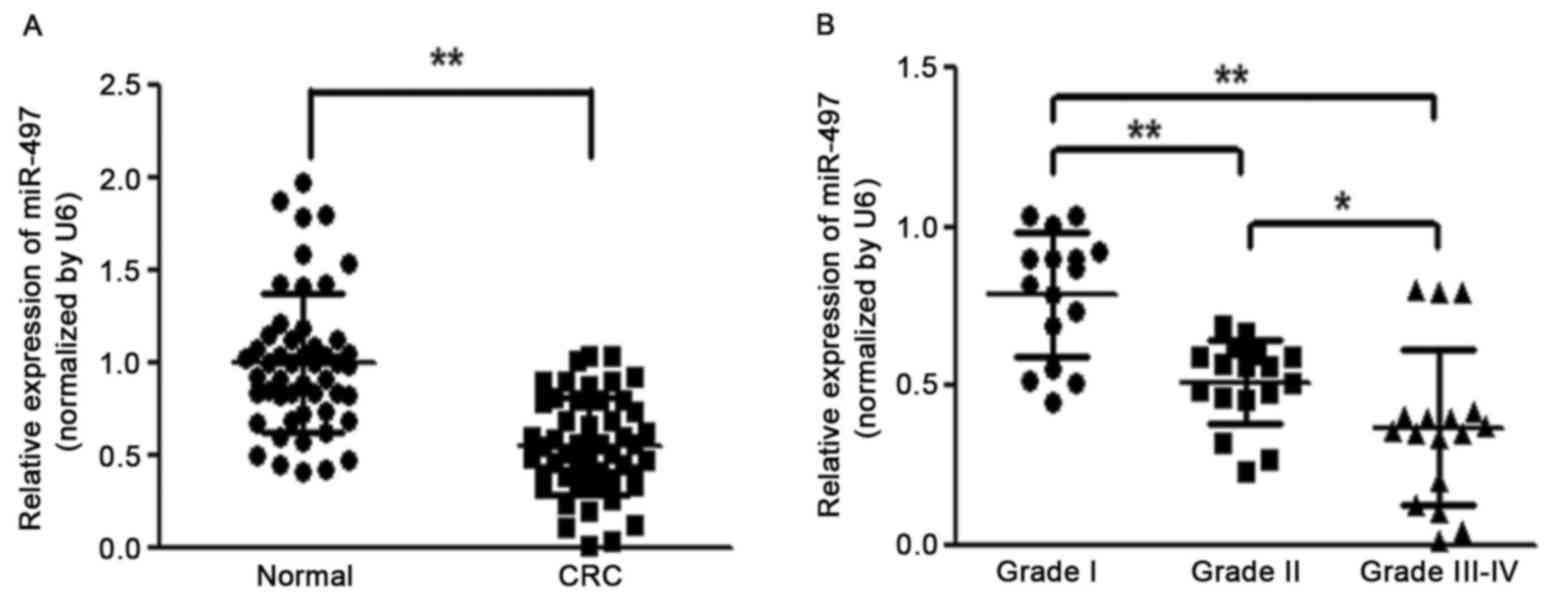
To identify the role of miR-497 in the development
of colorectal cancer, the expression level of miR-497 was analyzed
in 50 pairs of CRC tissues and adjacent normal tissues. RT-qPCR
analysis revealed that the miR-497 level was significantly
downregulated in CRC tissues (Fig.
1A). It was also observed that reduced levels of miR-497 in CRC
patients were positively correlated with the status of pathology
classification (Fig. 1B). These
results indicated that the progressive loss of miR-497 may be
associated with CRC disease progression.
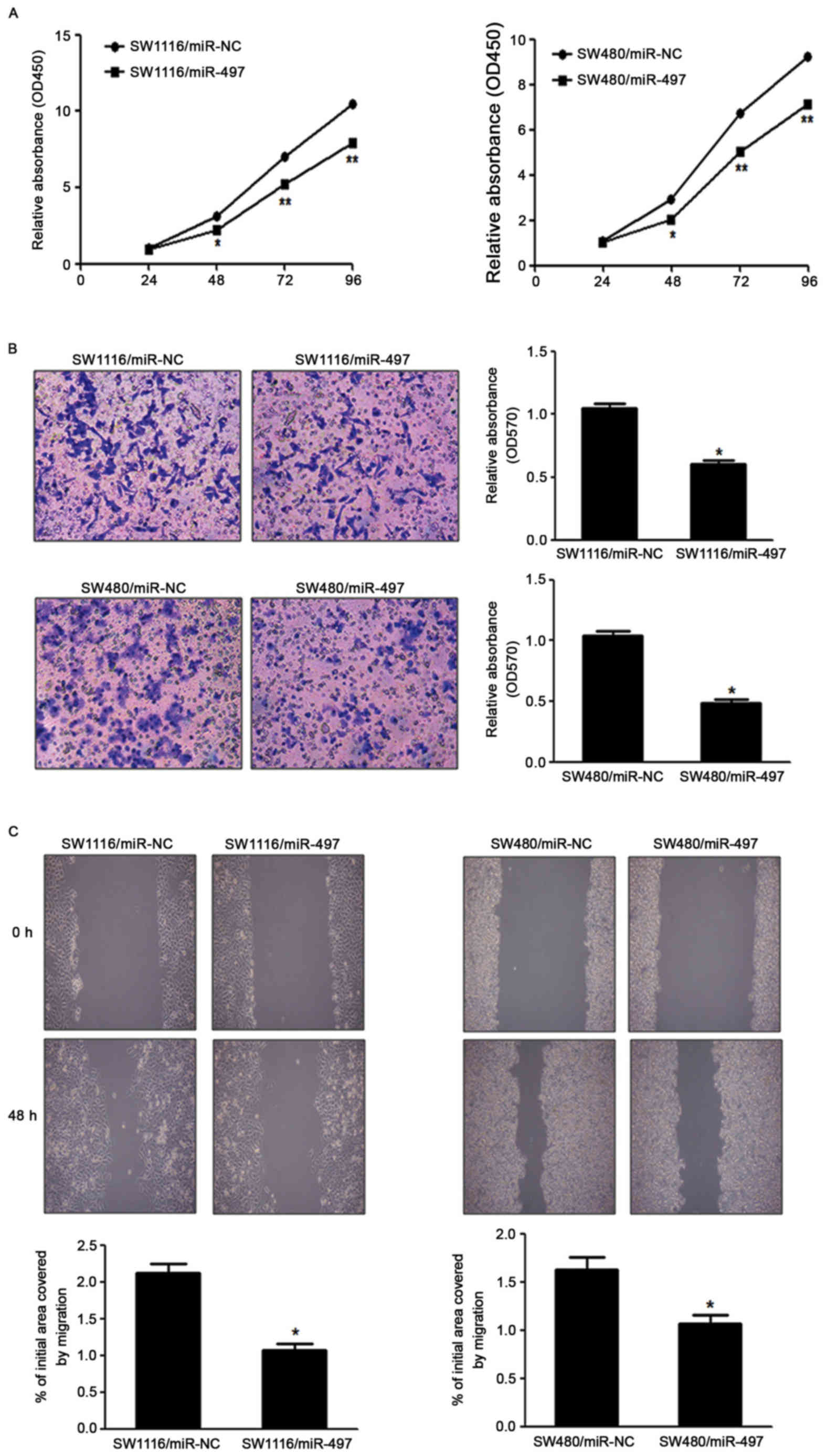
miR-497 suppresses CRC cell growth,
migration and invasion in vitro
To investigate the direct role of miR-497 in CRC
cells, the present study established stable cell lines by
transfecting SW1116 and SW480 cells with miR-497 overexpressing
lentiviral vector (Lv-miR-497) or a control lentiviral empty vector
(Lv-miR-NC), followed by puromycin selection. Subsequently, CRC
cell proliferation was detected in vitro. Cell viability
assay indicated that overexpression of miR-497 significantly
reduced the cell proliferation rate at 48 h following cell seeding
in SW1116 and SW480 cells, compared with the LV-miR-NC group
(Fig. 2A). Since invasion and
migration are key characteristics of malignant tumors, the present
study investigated the effects of miR-497 on invasion and migration
in vitro. Forced expression of miR-497 also markedly
suppressed the invasion of SW1116 and SW480 cells in migration
assays, as well as wound healing assays (Fig. 2B and C).
miR-497 inhibits AKT and ERK1/2
signaling pathways via targeting IRS1
To investigate the underlying mechanism of miR-497
in CRC, the present study analyzed TargetScan and miRanda
databases. It was observed that miR-497 likely regulates the IRS1
gene, since the 3′-UTR of IRS1 contained the binding site for the
seed region of miR-497. IRS1 is characterized as a typical
cytosolic adaptor protein in both IR and IGF1R signaling. According
to the putative binding site of miR-497 in the 3′UTR of the IRS1
gene, the present study initially constructed two types of plasmids
containing the luciferase reporting gene and wild-type or mutant
IRS1 3′UTR and cotransfected a miR-497 mimic into HEK-293T cells;
cells co-transfected with a miR-497 mimic and wild-type IRS1 3′UTR
demonstrated a significant decrease in luciferase activity.
However, in the mutant group, no detectable change in luciferase
activity was observed (Fig. 3A),
suggesting that miR-497 suppressed the transcription of the IRS1
gene by targeting the putative 3′UTR of IRS1 mRNA independently.
Western blotting analysis was conducted to determine IRS1 protein
expression. The results revealed that the IRS1 expression in SW116
and SW480 cells transfected with miR-497 mimics was downregulated
at the protein level, compared with cells transfected with negative
control (Fig. 3B). These data
demonstrated that miR-497 directly targeted IRS1 by binding its
seed region to the 3′-UTRs in CRC cells. The present study
additionally examined the IRS1 expression at the protein level in
human CRC specimens and normal tissues. The results demonstrated
that the average expression level of IRS1 was significantly
increased in tumor tissue compared with normal tissue (Fig. 3C). Further analysis revealed the
significant reciprocal association of expression levels of IRS1
with miR-497 in the same human CRC tissue (r=−0.6247; Fig. 3D).
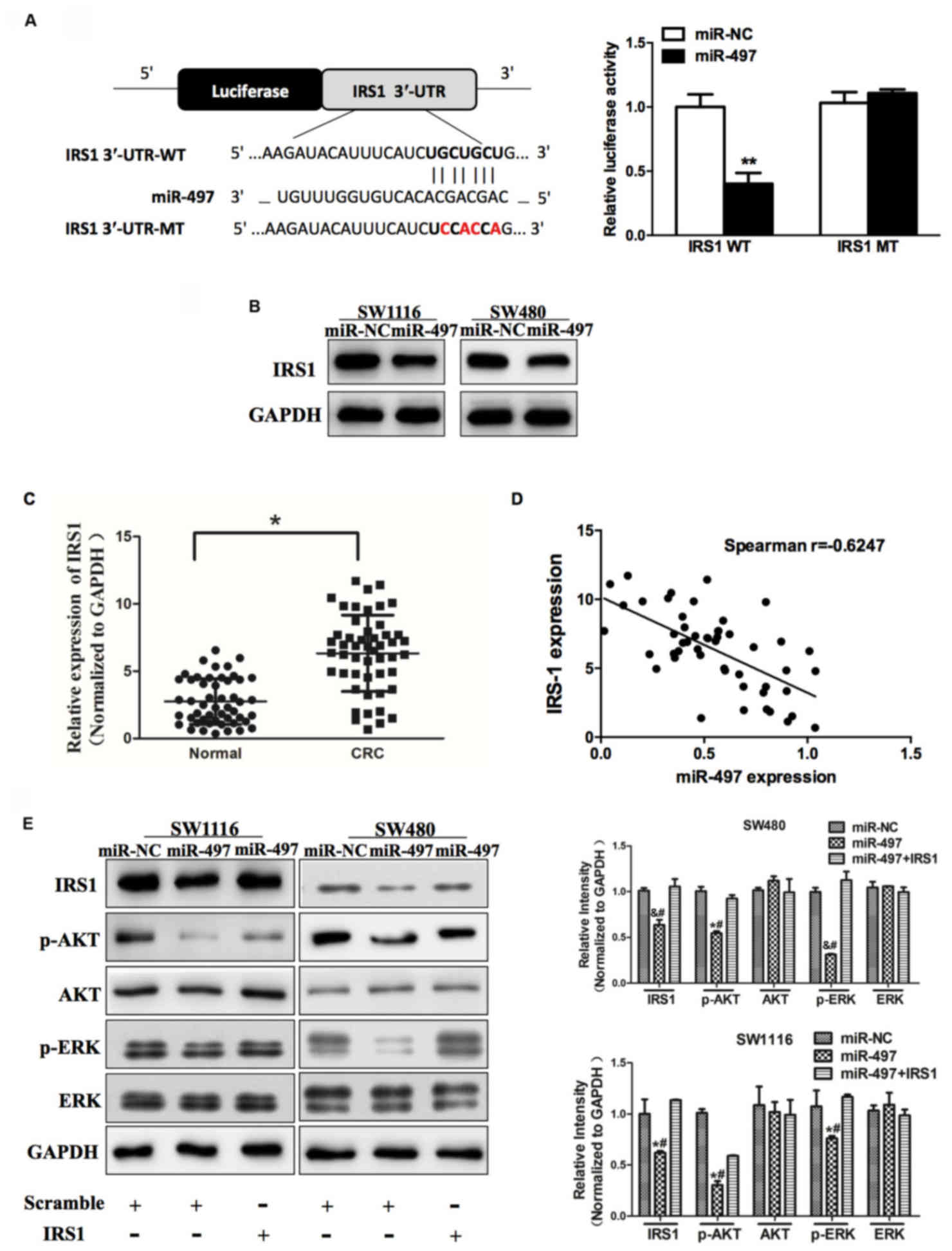 | Figure 3.miR-497 inhibits AKT and ERK1/2
signaling pathways via targeting IRS1. (A) Sequence of the miR-497
binding site within the human IRS1 3′-UTR and a schematic diagram
of the reporter construct showing the entire IRS1 3′-UTR sequence
and the mutant IRS1 3′-UTR sequence. The mutant nucleotides of the
IRS1 3′-UTR are labeled in red. Luciferase assay on SW1116 cells,
which were co-transfected with miR-NC or miR-497 and a luciferase
reporter containing the full length of IRS1 3′-UTR (WT) or a mutant
(MT) harboring four mutant nucleotides of the miR-497 binding site.
Luciferase activities were measured 24 h post-transfection. miR-497
markedly suppressed luciferase activity in IRS1 3′-UTR (WT)
reporter constructs. The data respresent the mean ± standard error
for separate transfections (n=4). (B) Immunoblotting revealed that
expression levels of IRS1 were decreased in cells with miR-497
overexpression. (C) Expression of IRS1 in adjacent normal tissues
and human CRC specimens was determined by western blot analysis,
and fold changes were obtained from the ratio of IRS1 to GAPDH
levels. (D) Spearman's correlation analysis was used to determine
the correlation between the expression levels of IRS1 and miR-497
in human CRC specimens. (E) Expression levels of p-AKT and p-ERK1/2
were decreased in cells with miR-497 overexpression, while AKT and
ERK1/2 protein levels remained unchanged. Overexpression of IRS1
restored miR-497-inhibited cellular protein levels of p-AKT and
p-ERK1/2. Data represent the mean ± standard deviation of three
replicates. *P<0.01 vs. control, **P<0.01,
#P<0.01 vs. miR-497+IRS1, &P<0.05
vs. control. miR, microRNA; ERK, extracellular signal-regulated
kinase; IRS1, insulin receptor substrate 1; UTR, untranslated
region; NC, negative control; CRC, colorectal cancer; p,
phosphorylated. |
AKT and ERK1/2 signaling pathways act as major
downstream regulators of IRS1 signaling, which are critical in
mitogenesis and oncogenesis. Cellular levels of p-AKT and p-ERK1/2
were significantly changed in SW1116 and SW480 cells stably
expressing miR-497 compared with miR-NC, but the changes in AKT and
ERK1/2 were not statistically significant (Fig. 3E). To additionally investigate whether
the overexpression of IRS1 affected the expression of p-AKT and
p-ERK1/2, cells were co-transfected with or without pCMV6-IRS1
cDNA. The results of the present study demonstrated that forced
expression of IRS1 restored miR-497-inhibited cellular levels of
p-AKT and p-ERK1/2. These data revealed that miR-497 inhibited AKT
and ERK1/2 signaling pathways via targeting IRS1 (Fig. 3E).
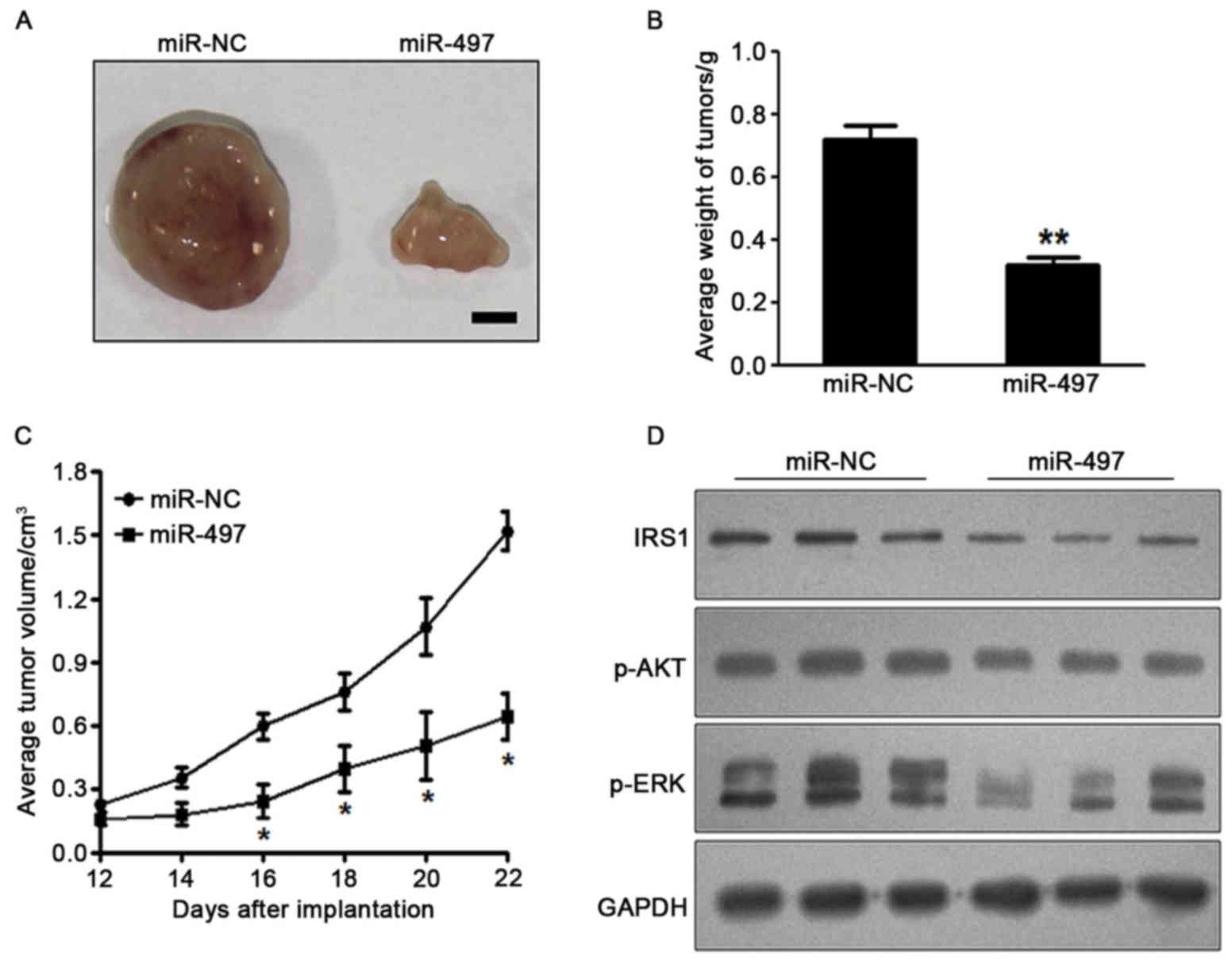
miR-497 inhibits tumor growth in
vivo
To investigate the effect of miR-497 on tumor
growth, SW1116 cells overexpressing miR-497 or miR-NC were
subcutaneously injected into the posterior flanks of nude mice
(n=6). Xenograft tumor volumes were determined every 2 days after
they had become visible. Nude mice were sacrificed on day 22
following injection of tumor cells, and xenografts were collected.
Fig. 4A shows representative
xenograft tumors. The average tumor weight of the miR-497
overexpression group was markedly reduced by 60% compared with that
of the control (Fig. 4B). On day 16
post-implantation, the tumor growth of the miR 497 overexpression
group was significantly reduced compared with that of the control
group (Fig. 4C). Total proteins from
representative tumor samples were analyzed by western blotting, and
the results demonstrated that miR-497 suppressed the expression of
IRS1 and p-AKT, as well as p-ERK1/2, in vivo (Fig. 4D). Taken together, these results
suggested that miR-497 inhibited tumor growth in vivo via
targeting IRS1 and other downstream signaling molecules.
Discussion
Previous studies have demonstrated that miRNAs serve
important roles in carcinogenesis by a number of mechanisms, and
certain miRNAs have been reported to be correlated with clinical
characteristics and outcomes (29,30). The
role of certain miRNAs in CRC has also been reported. For example,
miR-378 is frequently downregulated in CRC and colorectal cell
lines, and upregulation of miR-378 inhibits cell growth and
enhances oxaliplatin-induced apoptosis in human CRC (31). miR-194 functions as a tumor suppressor
in colorectal carcinogenesis via targeting
phosphoinositide-dependent kinase-1/AKT2/X-linked inhibitor of
apoptosis protein signaling pathway (32). Previous studies have demonstrated that
miR-497 is downregulated in several cancer types, including CRC.
Han et al (33) confirmed that
miR-497 suppresses the proliferation of human cervical carcinoma
HeLa cells by targeting cyclin E1. Another study demonstrated that
miR-497 targeted insulin-like growth factor 1 receptor and
inhibited proliferation and invasive behavior in colon cancer cells
(16). Consistent with previous
studies, the present study identified that miR-497 was
downregulated in CRC tissues compared with normal controls, and the
degree of miR-497 suppression was negatively correlated with
increased grades of human CRC malignancy. Notably, the present
study further predicted IRS1 as a target of miR-497 by
bioinformatic analysis. For the first time to the best of our
knowledge, it was demonstrated that IRS1 was upregulated in CRC
tissues and was inversely correlated with miR-497 levels. Thus, in
combination with previous research, the present study demonstrated
that miR-497 regulated the IGF1R/IRS1 signaling pathway, and may
provide novel therapeutic strategies for CRC prevention and
treatment.
IRS1 transmits signals from insulin or IGF receptors
to activate PI3K/AKT and MAPK pathways, both of which are critical
in mitogenesis and oncogenesis (34,35). It
has been observed that the expression of IRS1 may promote
proliferation in several cell lines (36–38). In
the present study, the IRS1 oncogene was experimentally validated
as a novel target of miR-497 in vitro and in vivo.
Initially, luciferase reporter assay confirmed that miR-497
directly recognized the 3′-UTR of IRS1 transcripts. Furthermore,
IRS1 expression was significantly abolished in CRC cells stably
expressing miR-497. In addition, a negative correlation was
observed between IRS1 protein and miR-497 in clinical samples.
Finally, inhibition of IRS1 expression by miR-497 suppressed
constitutive phosphorylation of AKT and ERK1/2. These results
demonstrate that miR-497 is a tumor suppressor that inhibits the
AKT and ERK1/2 signaling pathway through partly targeting IRS1.
In conclusion, the results of the present study
provide the first evidence, to the best of our knowledge, that
miR-497 serves a significant role in suppressing CRC cell growth
via inhibition of IRS1. Although the present study confirmed that
miR-497 was able to inhibit the phenotype of CRC by targeting IRS1,
there may be other targets of miR-497, which could also affect the
growth of CRC cells. However, the present study demonstrated that
such an effect was exerted through the suppression of IRS1.
Therefore, further studies are required to identify additional
targets and signaling pathways of miR-497.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81171653, 30972703,
81201741, 81301960 and 81302047); the Natural Science Foundation of
Jiangsu Province (grant no. BK2011246, BK2011247 and BK20130243);
The Project of Six Batch of Major Talent Summit (grant no.
BRA2010037); Society Development Plans, Department of Science and
Technology Changzhou (grant nos. CJ20112020, CZ20110024 and
CS20102020) and the Innovative Talents Training Project of
Changzhou Health Bureau (grant nos. 2016CZBJ001 and
2016CZLJ022).
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen J, Chen Y and Chen Z: MiR-125a/b
regulates the activation of cancer stem cells in
paclitaxel-resistant colon cancer. Cancer Invest. 31:17–23. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nie J, Liu L, Xing G, Zhang M, Wei R, Guo
M, Li X, Xie P, Li L, He F, et al: CKIP-1 acts as a colonic tumor
suppressor by repressing oncogenic Smurf1 synthesis and promoting
Smurf1 autodegradation. Oncogene. 33:3677–3687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Faltejskova P, Svoboda M, Srutova K,
Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K,
Kiss I, et al: Identification and functional screening of microRNAs
highly deregulated in colorectal cancer. J Cell Mol Med.
16:2655–2666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hüttenhofer A, Schattner P and Polacek N:
Non-coding RNAs: Hope or hype? Trends Genet. 21:289–297. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: MicroRNA pathways in flies and
worms: Growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carlsson J, Davidsson S, Helenius G,
Karlsson M, Lubovac Z, Andrén O, Olsson B and Klinga-Levan K: A
miRNA expression signature that separates between normal and
malignant prostate tissues. Cancer Cell Int. 11:142011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Caldas C and Brenton JD: Sizing up miRNAs
as cancer genes. Nat Med. 11:712–714. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kefas B, Godlewski J, Comeau L, Li Y,
Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S and
Purow B: microRNA-7 inhibits the epidermal growth factor receptor
and the Akt pathway and is down-regulated in glioblastoma. Cancer
Res. 68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo Q, Li X, Gao Y, Long Y, Chen L, Huang
Y and Fang L: MiRNA-497 regulates cell growth and invasion by
targeting cyclin E1 in breast cancer. Cancer Cell Int. 13:952013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao WY, Wang Y, An ZJ, Shi CG, Zhu GA,
Wang B, Lu MY, Pan CK and Chen P: Downregulation of miR-497
promotes tumor growth and angiogenesis by targeting HDGF in
non-small cell lung cancer. Biochem Biophys Res Commu. 435:466–471.
2013. View Article : Google Scholar
|
|
16
|
Guo ST, Jiang CC, Wang GP, Li YP, Wang CY,
Guo XY, Yang RH, Feng Y, Wang FH, Tseng HY, et al: MicroRNA-497
targets insulin-like growth factor 1 receptor and has a tumour
suppressive role in human colorectal cancer. Oncogene.
32:1910–1920. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang
B, Shu Y and Liu P: miR-497 modulates multidrug resistance of human
cancer cell lines by targeting BCL2. Med Oncol. 29:384–391. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Y, Meng Q, Qi J, Shen H and Sun S:
MiR-497 promotes metastasis of colorectal cancer cells through
Nrdp1 inhibition. Tumour Biol. 36:7641–7647. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen J, Wu A, Sun H, Drakas R, Garofalo C,
Cascio S, Surmacz E and Baserga R: Functional significance of type
1 insulin-like growth factor-mediated nuclear translocation of the
insulin receptor substrate-1 and beta-catenin. J Biol Chem.
280:29912–29920. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu A, Chen J and Baserga R: Nuclear
insulin receptor substrate-1 activates promoters of cell cycle
progression genes. Oncogene. 27:397–403. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bergmann U, Funatomi H, Kornmann M, Beger
HG and Korc M: Increased expression of insulin receptor substrate-1
in human pancreatic cancer. Biochem Biophys Res Commun.
220:886–890. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang Q, Li Y, White MF, Fletcher JA and
Xiao S: Constitutive activation of insulin receptor substrate 1 is
a frequent event in human tumors: Therapeutic implications. Cancer
Res. 62:6035–6038. 2002.PubMed/NCBI
|
|
23
|
Slattery ML, Samowitz W, Curtin K, Ma KN,
Hoffman M, Caan B and Neuhausen S: Associations among IRS1, IRS2,
IGF1, and IGFBP3 genetic polymorphisms and colorectal cancer.
Cancer Epidemiol Biomarkers Prev. 13:1206–1214. 2004.PubMed/NCBI
|
|
24
|
Fléjou JF: WHO classification of digestive
tumors: The fourth edition. Ann Pathol. 31:S27–31. 2011.(In
French). View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi ZM, Wang XF, Qian X, Tao T, Wang L,
Chen QD, Wang XR, Cao L, Wang YY, Zhang JX, et al: MiRNA-181b
suppresses IGF-1R and functions as a tumor suppressor gene in
gliomas. RNA. 19:552–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peterson SM, Thompson JA, Ufkin ML,
Sathyanarayana P, Liaw L and Congdon CB: Common features of
microRNA target prediction tools. Front Genet. 5:232014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang
C, Liu X, Wang X, Li H, Kang C, Jiang T, Liu L, You Y, Liu N and
Jiang B: MiR-124 governs glioma growth and angiogenesis and
enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro
Oncol. 16:1341–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu J, Wang T, Cao Z, Huang H, Li J, Liu W,
Liu S, You L, Zhou L, Zhang T and Zhao Y: MiR-497 downregulation
contributes to the malignancy of pancreatic cancer and associates
with a poor prognosis. Oncotarget. 5:6983–6993. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Majid S, Dar AA, Saini S, Deng G, Chang I,
Greene K, Tanaka Y, Dahiya R and Yamamura S: MicroRNA-23b functions
as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS
One. 8:e676862013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang KY, Ma J, Zhang FX, Yu MJ, Xue JS and
Zhao JS: MicroRNA-378 inhibits cell growth and enhances
L-OHP-induced apoptosis in human colorectal cancer. IUBMB Life.
66:645–654. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao HJ, Ren LL, Wang ZH, Sun TT, Yu YN,
Wang YC, Yan TT, Zou W, He J, Zhang Y, et al: MiR-194 deregulation
contributes to colorectal carcinogenesis via targeting AKT2
pathway. Theranostics. 4:1193–1208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han J, Huo M, Mu M, Liu J and Zhang J:
miR-497 suppresses proliferation of human cervical carcinoma HeLa
cells by targeting cyclin E1. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi.
30:597–600. 2014.(In Chinese). PubMed/NCBI
|
|
34
|
Scharf JG and Braulke T: The role of the
IGF axis in hepatocarcinogenesis. Horm Metab Res. 35:685–693. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Scharf JG, Dombrowski F and Ramadori G:
The IGF axis and hepatocarcinogenesis. Mol Pathol. 54:138–144.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang LM, Myers MG Jr, Sun XJ, Aaronson SA,
White M and Pierce JH: IRS-1: Essential for insulin- and
iL-4-stimulated mitogenesis in hematopoietic cells. Science.
261:1591–1594. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Taouis M, Dupont J, Gillet A, Derouet M
and Simon J: Insulin receptor substrate 1 antisense expression in
an hepatoma cell line reduces cell proliferation and induces
overexpression of the Src homology 2 domain and collagen protein
(SHC). Mol Cell Endocrinol. 137:177–186. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Waters SB, Yamauchi K and Pessin JE:
Functional expression of insulin receptor substrate-1 is required
for insulin-stimulated mitogenic signaling. J Biol Chem.
268:22231–22234. 1993.PubMed/NCBI
|