Introduction
In males over 50 years of age, prostate cancer is
the most common form of malignancy in industrialized countries
(1). In the United States of America
(USA), it is the most commonly diagnosed type of cancer and the
second most common cause of cancer-associated mortality (1). Annually, ~23,000 males in the USA are
diagnosed with the condition and almost 30,000 succumb to the
disease (1). In its early stages,
prostate cancer may be treated through androgen depletion,
radiation or chemotherapy (2).
However, the disease is often problematic to control long term;
20–25% of those affected will eventually become
androgen-independent and the disease will metastasize; advanced
stage prostate cancer is incurable. In addition, the side effects
of current treatment approaches mandate innovative and improved
strategies to treat patients with prostate cancer.
One promising approach is gene therapy (3–5). In
contrast to more conventional modalities, it may be tailored to the
individual malignancy (4,6). RNA interference (RNAi) is a method of
utilizing post-transcriptional gene silencing, and has been
demonstrated to be effective for gene knockdown; it appears to
exhibit marked potential for patient therapy (7–9).
In prior studies, there was a focus on the
transfection of synthetic small interfering RNA (siRNA) or plasmids
in cancer cell research in order to induce the expression of short
hairpin RNAs (shRNAs), making use of RNA polymerase III promoters
(8,10,11). A
number of studies have investigated the silencing of specific
cancer genes by RNAi in cell lines from various tissues types,
resulting in a significant inhibition of cancer cell growth
(9). In addition, this group has
successfully used this knockdown technology to inhibit the
expression of Ki-67 in renal cancer cell lines.
The nuclear factor special AT-rich sequence binding
protein 1 (SATB1) is the global chromatin organizer whose function
is to regulate the structure of chromatin and gene expression
(12). Its expression is abnormal in
a number of cancer types. It has been suggested that SATB1 is an
oncogene that promotes malignancy (13,14). It
has previously been suggested that it may be overexpressed in
metastatic prostatic cancer, and its ability to promote cancer cell
growth and invasion has been demonstrated (15). The present study reports the
production of a novel plasmid containing shRNA against SATB1,
pSilencer-SATB1-shRNA, and aimed to determine its antitumor
efficacy in xenograft prostate tumors in nude mice.
Materials and methods
Cell lines and vectors
The DU145 prostate cancer cell line was obtained
from the Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China), and cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), to which
10% heat-inactivated fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 4 mM glutamine, 50 U/ml penicillin and 50 µg/ml
streptomycin had been added. The culture was performed at 37°C in a
humidified atmosphere with 5% CO2. The cells were
routinely screened to verify they were free of mycoplasma
contamination, and were used for experiments in the logarithmic
phase of growth. The pSilencer-SATB1-shRNA plasmid and pSilencer
plasmid were kindly provided by Professor Li Jun Mao. pSilencer
plasmids were dissolved in K-PBS medium from KeyGen Biotech Co.,
Ltd (Nanjing, China) and used at concentrations of 1 g/l.
Xenograft tumor model in nude
mice
The Shanghai Experimental Animal Center of the
Chinese Academy of Sciences (Shanghai, China) provided 24 (4–5
week-old) male BALB/c nude mice weighing ~20 g. Prior to
implantation of the tumor cells, all mice were quarantined under
the condition of constant temperature and humidity for 1 week. Mice
had ad libitum access to food and water. Animal welfare and
experimental procedures were carried out strictly in accordance
with the Guide for the Care and Use of Laboratory Animals. A total
of 2×106 DU145 cells was intraperitoneally injected into
the right flank of the mice to establish the xenograft tumor model.
The present study was approved by the Ethics Committee of The
Affiliated Hospital of Xuzhou Medical University (Xuzhou,
China).
Once the tumors reached a volume of 100–150
mm3, the mice were randomly divided into three groups,
with 8 mice in each group. By group, the tumors were subsequently
injected with 0.1 ml pSilencer vector or pSilencer-SATB-1-shRNA for
three consecutive days, or with 0.1 ml PBS as a control. Over the
following 30 days, calipers were used weekly to assess the size of
the tumors. The volume of each tumor was then determined according
to the following formula: Volume (mm3)=length ×
width2×1/2. Data obtained for each group of 8 mice were
expressed as the mean ± standard error of the mean and assessed for
statistical significance. BALB/c nude mice (8 per group) were
sacrificed after 30 days by cervical dislocation, and the tumors
were removed and fixed in 10% neutral formalin at room temperature
for 24 h and embedded in paraffin for hematoxylin and eosin
(H&E) staining and immunohistochemistry.
Immunohistochemical staining
The harvested tumors were fixed in 10% formalin for
24 h at room temperature, embedded in paraffin and cut into 4-mm
sections. Subsequent to the samples being deparaffinized with
dimethylbenzene and rehydrated using a graduated alcohol series,
endogenous peroxidase activity was blocked using 3%
H2O2 for 10 min at room temperature, and the
sections were then incubated for 30 min at room temperature with
goat serum (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) to block membranes.
Immunohistochemistry was performed using an
anti-SATB1 antibody (cat. no. 611182; GenHunter Corporation,
Nashville, TN, USA). Following incubation with an anti-mouse
secondary antibody (cat. no. PA174460; dilution, 1:1,000; Abcam
Company) at 37°C for 30 min, SATB1 expression was determined using
3,3′-diaminobenzidine (DAB; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) and enhanced using an Avidin-Biotin Reaction ABC kit
(Vector Laboratories, Inc., Burlingame, CA, USA). The negative
controls were tissue sections that had been stained without the
primary antibody. The proportion of cells positive for SATB1 was
determined using optical microscope by counting a minimum of 200
cells from 6 distinct areas, and then calculating the mean.
H&E staining and the terminal dUTP
nick-end labelling (TUNEL) assay
Tumors were harvested and fixed in 4%
paraformaldehyde for 30–50 min at room temperature, embedded in
paraffin and cut into 4-mm sections. For histopathological
analysis, the paraffin-embedded tissue sections were stained with
H&E at room temperature for 10–30 min. Following
deparaffinization with xylene and rehydration using a graded
alcohol series, apoptotic cells in the tumor tissue sections were
quantified using the in situ Annexin V-FITC/PI Apoptosis
Detection kit (Roche Diagnostics, Indianapolis, IN, USA). Following
deparaffinization in xylene, sections were rinsed twice with PBS
and permeabilized using 15 µg/ml proteinase K (Yeasen, Shanghai,
China) in 10 mM Tris/HCl, pH 7.4–8.0 for 15 min at room
temperature. Endogenous peroxidase activity was blocked using 3%
H2O2 for 10 min at room temperature. The
sections were then incubated with an equilibration buffer and a
terminal deoxynucleotidyl transferase enzyme, and then with an
anti-digoxigenin-peroxidase conjugate. Negative controls were
prepared by omitting the terminal transferase. Incubation with 10
mg/ml DAB (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30
min at room temperature was used to visualize peroxidase activity.
Under light microscopy (magnification, ×400), 6 fields were
randomly selected from each sample and 100 cells were randomly
selected from each field. The apoptotic rate was determined using
the following formula: Number of apoptotic cells/100×100%.
Western blot analysis
The cells were harvested from the plates, then the
protein concentration was calculated by BCA method, and buffers
containing 100ug protein were prepared per lane. Aliquots of the
cell extracts were separated by 12% SDS-PAGE. The proteins were
then transferred onto a nitrocellulose membrane. The membrane was
then incubated overnight at 4°C with the following rabbit
polyclonal antibodies: Anti-SATB1 (cat. no. 611182; GenHunter
Corporation, Nashville, TN, USA), anti-MMP2 (cat. no. 2270S; Cell
Signaling Technology, Inc., Danvers, MA, USA) and anti-β-actin
(cat. no. sc-47778; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA). The membrane was then washed and incubated with alkaline
phosphatase-conjugated secondary antibodies in TBS containing
Tween-20 for 2 h at room temperature, and developed using the
NBT/BCIP color substrate (Promega Corporation, Madison, WI, USA).
The densities of the bands on the membrane were scanned and
analyzed with ImageJ 1.48 (National Institutes of Health, Bethesda,
MD, USA) and LabWorks (UVP LLC, Upland, CA, USA).
Statistical analysis
Data are expressed as the mean ± standard deviation
and analyzed using one-way analysis of variance followed by
Duncan's new multiple range test or the Newman-Keuls test as
appropriate by statistical software (SPSS Base 15.0 for Windows,
SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate
a statistically significant difference.
Results
SATB1 protein expression in tumor
tissues
The 8 control mice injected with PBS were designated
as group A, the 8 mice injected with the empty pSilencer vector as
group B and the 8 mice injected with the pSilencer-SATB1-shRNA as
group C. The expression of SATB1 protein in tumor tissue samples
obtained from each of the three groups was detected using
immunohistochemical staining and western blot analysis.
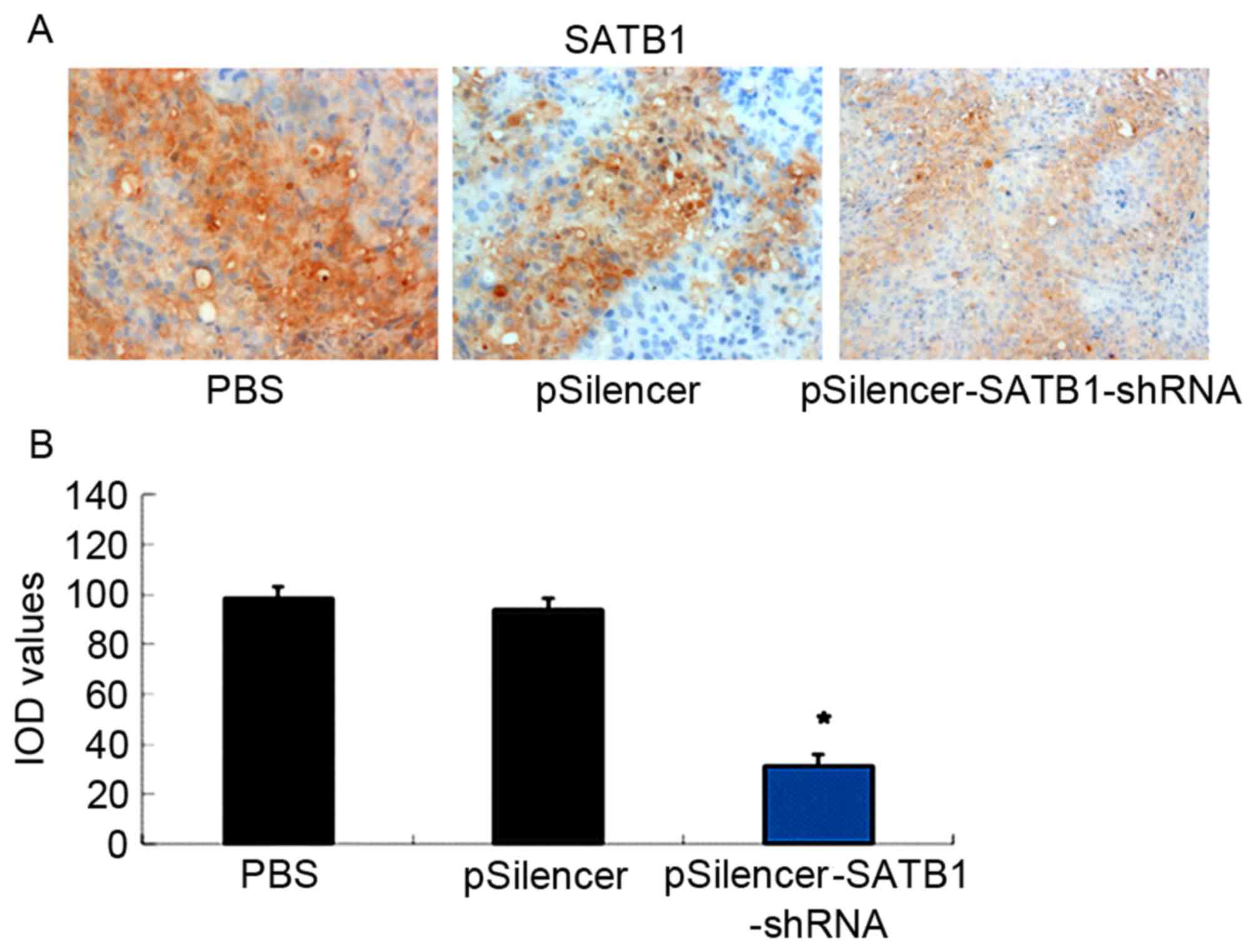
Immunohistochemical staining demonstrated that the SATB1 protein
was expressed primarily in the cytoplasm and the cell membrane
(brown staining, Fig. 1A). By using
the ImageJ analysis software, the integrated optical density (IOD)
absorbance values for PBS, pSilencer, and pSilencer-SATB1-shRNA
were 99.86±2.58, 93.83±2.15 and 32.78±0.53, respectively. Compared
with the other two groups, the SATB1 protein expression, based on
the IOD, in the pSilencer-SATB1-shRNA group was significantly low
(P<0.05; Fig. 1B). Additionally,
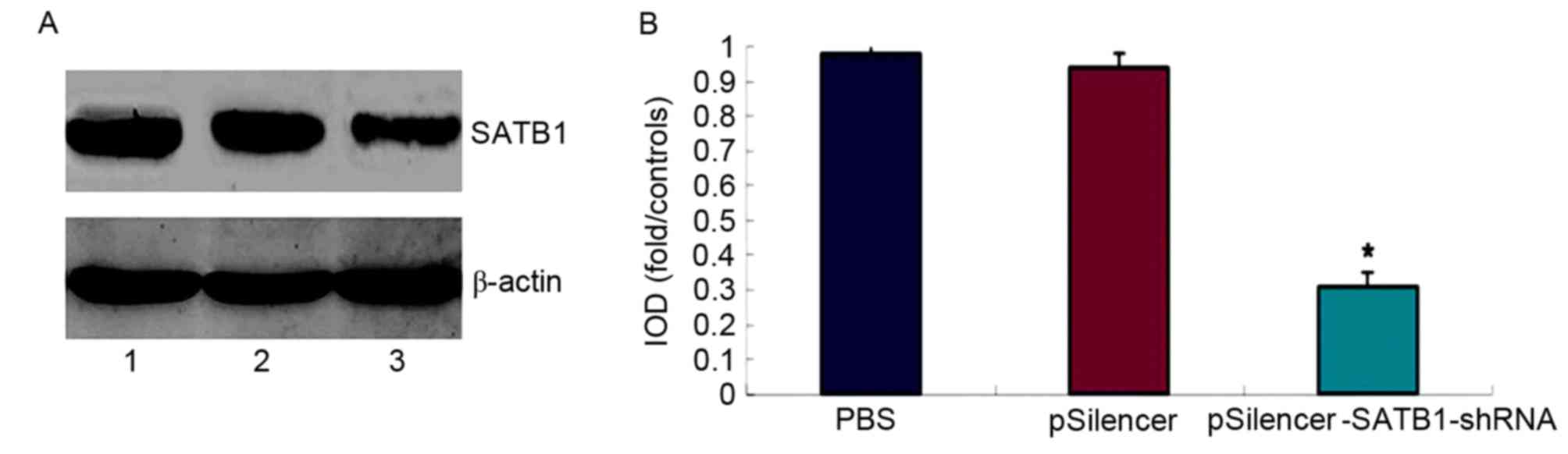
western blot analysis demonstrated that the proportion of SATB1
protein in group 1, group 2 and group 3 was 98.6±5.73, 95.73±4.52
and 31.4±5.78%, respectively (Fig.
2). Based on the data from the western blot analysis, the SATB1
protein expression in the pSilencer-SATB1-shRNA group was
significantly low when compared with the two control groups
(P<0.05; Fig. 2).
pSilencer-SATB1-shRNA inhibits tumor
growth
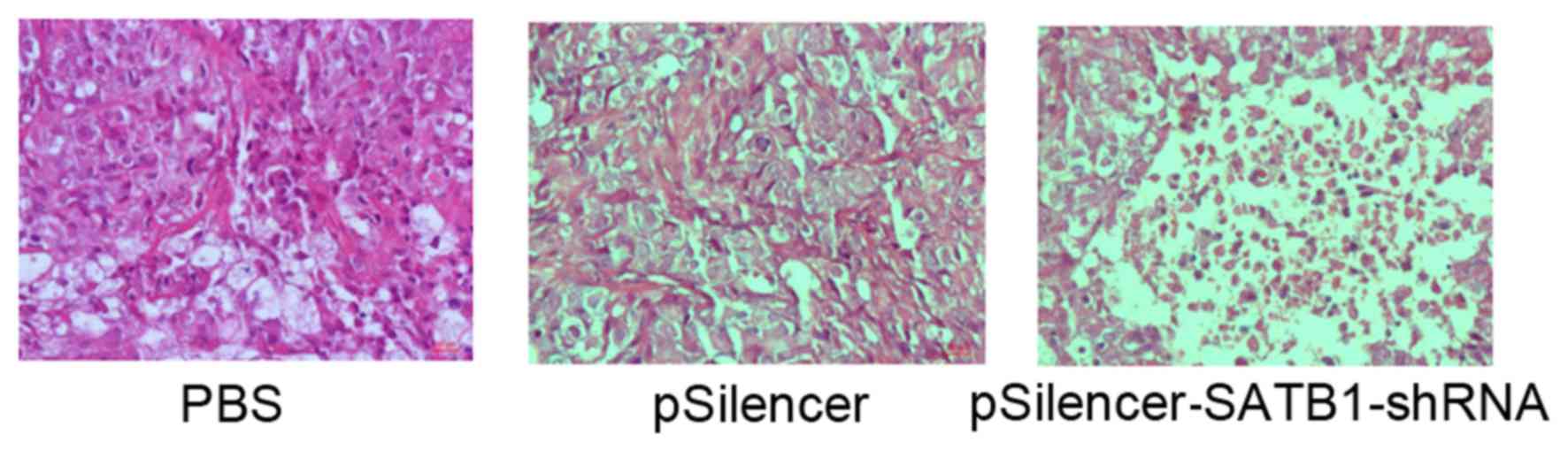
H&E staining was used to observe the tumor cell
growth. In the control (PBS) and empty vector groups (groups A and
B), marked tumor growth was observed (Table I). In addition, in the two control
samples, the cell nuclei were of various sizes and an irregular
cell shape was observed (Fig. 3).
Conversely, the pSilencer-SATB1-shRNA group demonstrated reduced
tumor growth (Table I). In addition,
numerous cells had died or were degraded; nuclear pyknosis was
observed and normal tissue structures had disappeared (Fig. 3). Together, these results suggested
that pSilencer-SATB1-shRNA inhibits tumor growth.
 | Table I.Calculated tumor volume in each group
at 1, 5, 9, 13, 17, 21 and 25 days. |
Table I.
Calculated tumor volume in each group
at 1, 5, 9, 13, 17, 21 and 25 days.
| Days | PBS group | pSilencer group | pSilencer-SATB1-shRNA
group |
|---|
| 1 |
96.42±12.04 |
98.64±11.53 |
100.31±13.25 |
| 5 |
682.34±198.02 |
593.31±184.56 |
508.90±139.78 |
| 9 |
1,248.23±185.73 |
1,129.39±180.36 |
1,045.32±189.56 |
| 13 |
1,528.34±190.13 |
1,498.32±179.38 |
1,228.23±179.83 |
| 17 |
1,923.37±281.04 |
1,837.57±174.48 |
1,148.78±137.45 |
| 21 |
2,310.98±202.56 |
2,194.67±187.12 |
703.16±110.34 |
| 25 |
2,713.38±276.99 |
2,397.39±178.63 |
519.03±110.83 |
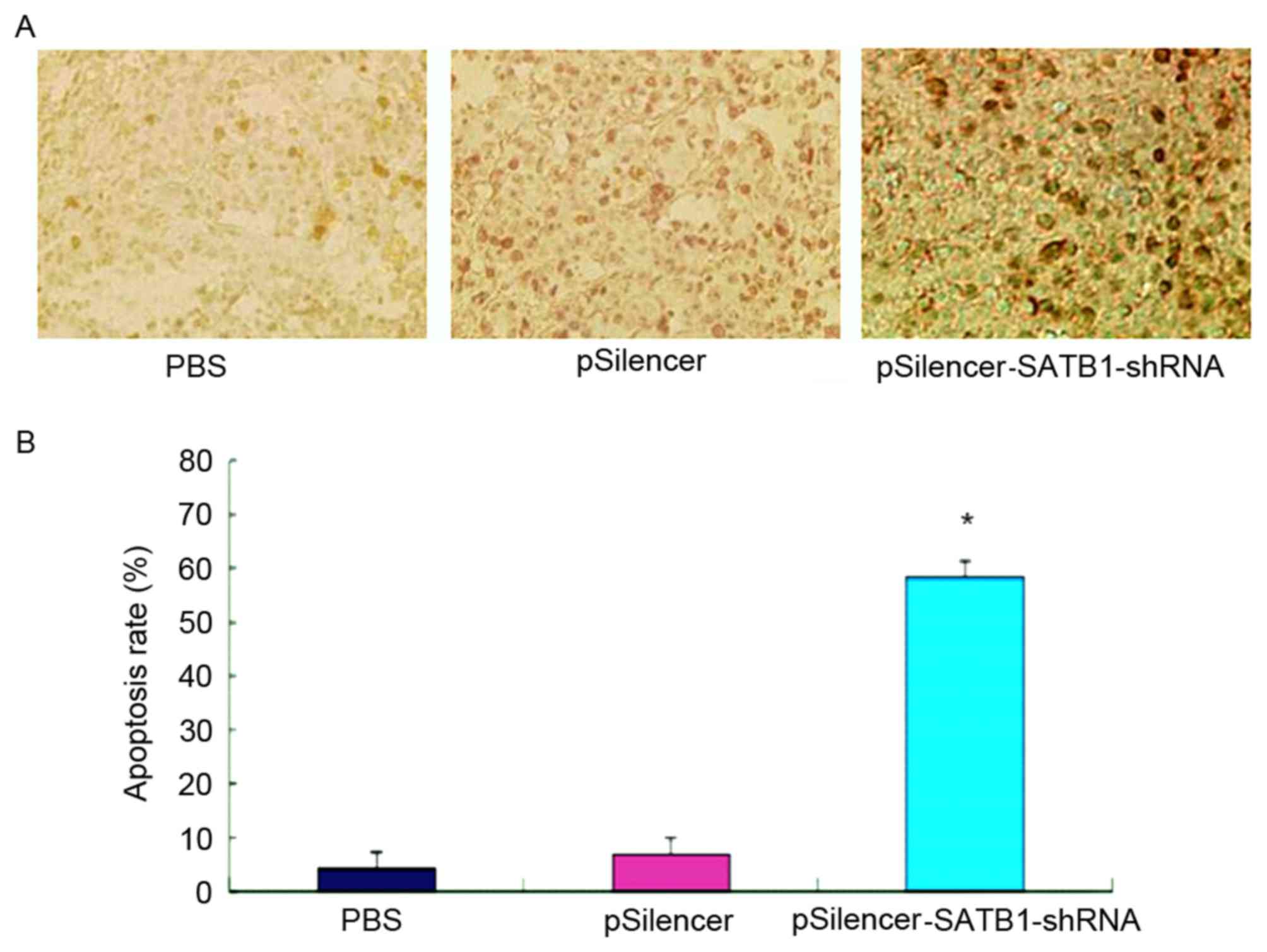
Subsequently, apoptosis in the tumor tissue samples
was evaluated using a TUNEL assay (Fig.
4A). The percentage of apoptotic cells in the
pSilencer-SATB1-shRNA sample (58.23±6.62%) was significantly
increased compared with in the PBS sample (4.16±1.83%) and the
empty plasmid sample (6.84±3.16%; P<0.05; Fig. 4B). Additionally, pSilencer-SATB1-shRNA
demonstrated an increased rate of tumor growth inhibition
(83.98±2.32%) compared with the controls (Table II).
 | Table II.Tumor volume and inhibitory rate in
each treatment group. |
Table II.
Tumor volume and inhibitory rate in
each treatment group.
| Groups | Tumor volume
(mm3) | Tumor inhibitory
rate, % |
|---|
| PBS group (A) |
2713.38±276.99 | – |
| pSilencer group
(B) |
2397.39±178.63 | 9.56±4.89 |
|
pSilencer-SATB-1-shRNA group (C) |
a519.03±110.83 | 83.98±2.32 |
Discussion
A prior study on prostate cancer demonstrated that
SATB1 staining was more marked if there were metastases compared
with if there were not, and that the staining was absent in benign
prostate hyperplasia (15). These
data suggested that SATB1 serves a crucial role in the metastasis
of prostate cancer.
The current study demonstrated that SATB1 protein
was produced lower expression in the pSliencer-SATB1-shRNA
treatment group compared to pSilencer or PBS-treated groups.
Additionally, tumor tissues from the pSilencer-SATB1-shRNA
treatment group exhibited reduced tumor growth, with an increased
rate of apoptosis, nuclear pyknosis and loss of normal tissue
structures. Together, these results indicate that treatment with
pSliencer-SATB1-shRNA inhibits xenograft tumor growth in nude
mice.
These data are concordant with the results of
previous in vivo studies of highly aggressive human prostate
cancer cells, which suggested that SATB1 knockdown inhibits tumor
growth and invasiveness (16,17). In addition, Mao et al (17) previously examined the effect of a
replicative oncolytic adenovirus expressing SATB1-shRNA in a mouse
model, and determined that this treatment exhibited a potent
antitumor effect against human prostate cancer cells.
In conclusion, the in vivo treatment of
prostate cancer with pSilencer-SATB1-shRNA led to a significant
reduction in tumor growth and an increased rate of apoptosis. These
data demonstrate that pSilencer-SATB1-shRNA is cytotoxic to
prostate cancer cells and inhibits tumor growth, indicating their
potential as a novel strategy for treating human prostate
cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shore N: Management of early-stage
prostate cancer. Am J Manag Care. 20 12 Suppl:S260–S272.
2014.PubMed/NCBI
|
|
3
|
Gridley DS and Slater JM: Gene therapy: A
possible aid to cancer radiotherapy. Discov Med. 4:408–414.
2004.PubMed/NCBI
|
|
4
|
Altaner C: Prodrug cancer gene therapy.
Cancer Lett. 270:191–201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tangney M: Gene therapy for cancer: Dairy
bacteria as delivery vectors. Discov Med. 10:195–200.
2010.PubMed/NCBI
|
|
6
|
Singh P, Yam M, Russell PJ and Khatri A:
Molecular and traditional chemotherapy: A united front against
prostate cancer. Cancer Lett. 293:1–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambesajir A, Kaushik A, Kaushik JJ and
Petros ST: RNA interference: A futuristic tool and its therapeutic
applications. Saudi J Biol Sci. 19:395–403. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bora RS, Gupta D, Mukkur TK and Saini KS:
RNA interference therapeutics for cancer: Challenges and
opportunities (review). Mol Med Rep. 6:9–15. 2012.PubMed/NCBI
|
|
9
|
Mansoori B, Shotorbani Sandoghchian S and
Baradaran B: RNA interference and its role in cancer therapy. Adv
Pharm Bull. 4:313–321. 2014.PubMed/NCBI
|
|
10
|
Agrawal N, Dasaradhi PV, Mohmmed A,
Malhotra P, Bhatnagar RK and Mukherjee SK: RNA interference:
Biology, mechanism, and applications. Microbiol Mol Biol Rev.
67:657–685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kohwi-Shigematsu T, Poterlowicz K,
Ordinario E, Han HJ, Botchkarev VA and Kohwi Y: Genome organizing
function of SATB1 in tumor progression. Semin Cancer Biol.
23:72–79. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yasui D, Miyano M, Cai S, Varga-Weisz P
and Kohwi-Shigematsu T: SATB1 targets chromatin remodelling to
regulate genes over long distances. Nature. 419:641–645. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han HJ, Russo J, Kohwi Y and
Kohwi-Shigematsu T: SATB1 reprogrammes gene expression to promote
breast tumour growth and metastasis. Nature. 452:187–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mao L, Yang C, Wang J, Li W, Wen R, Chen J
and Zheng J: SATB1 is overexpressed in metastatic prostate cancer
and promotes prostate cancer cell growth and invasion. J Transl
Med. 11:1112013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shukla S, Sharma H, Abbas A, MacLennan GT,
Fu P, Danielpour D and Gupta S: Upregulation of SATB1 is associated
with prostate cancer aggressiveness and disease progression. PLoS
One. 8:e535272013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mao LJ, Zhang J, Liu N, Fan L, Yang DR,
Xue BX, Shan YX and Zheng JN: Oncolytic virus carrying shRNA
targeting SATB1 inhibits prostate cancer growth and metastasis.
Tumour Biol. 36:9073–9081. 2015. View Article : Google Scholar : PubMed/NCBI
|