Introduction
According to the National Cancer Institute, breast
cancer was the most frequently diagnosed malignancy in 2015
(1). It is widely accepted that
breast cancer develops when a series of gene disorders occur
(2). It was demonstrated that breast
cancer negative for murine double minute gene 2 and wild-type
p53-activated fragment 1 expression, irrespective of p53 status,
exhibited an increased response rate to docetaxel but no response
to methotrexate and 5-fluorouracil, compared with breast cancer
positive for murine double minute gene 2 and wild-type
p53-activated fragment 1 expression (3). Immunohistochemical data has revealed
that p53 mutation is the most common genetic alteration detected in
primary breast cancer (4). Ertel
et al (5) reported that
retinoblastoma-loss signature expression is associated with poor
outcome in breast cancer, but predicts improved response to
chemotherapy based on data in oestrogen receptor (ER)-negative
populations (5). Additionally, the
Akt pathway is involved in the regulation of growth and migration
of breast cancer (6). Oncogene Sam68
upregulation is associated with, and its downregulation inhibits,
proliferation and tumorigenicity of breast cancer cells (7). Even so, a considerable number of
patients succumb to breast cancer every year. New regulating
factors that are targets for breast cancer therapy are urgently
required.
KIF11, distributed throughout the cytoplasm
(8), is a mitotic kinesin that plays
a crucial role in the formation of bipolar mitotic spindles by
hydrolysing ATP to push apart anti-parallel microtubules (9,10).
Previous studies reported that five mutations caused a broader
spectrum of ocular disease, including retinal detachment (11–13). It
has been hypothesized that KIF11 is involved in the progression of
numerous diseases, including lymphedema (14), Alzheimer's disease (15), type 2 diabetes (16) and xeroderma pigmentosum (17). Wakana et al (18) revealed that disrupting the function of
KIF11 in HeLa cells inhibited the secretion of pancreatic
adenocarcinoma upregulated factor. KIF11 overexpression is
associated with the poor differentiation of bladder cancer, and is
an independent prognostic factor for predicting early intravesical
recurrence in patients with non-muscle invasive bladder carcinoma
(19). Dimethylenastron prevents the
growth of pancreatic and lung cancer cells by halting mitotic
progression and triggering apoptosis (9). Small molecule inhibitors of kinesin-5,
which were developed as potential anti-cancer drugs, arrest cell
cycle progression in mitosis and promote apoptosis of cancer cells
(20). Additionally, the KIF11
inhibitor ARRY-520 may represent an alternative to paclitaxel in
this subgroup of epithelial ovarian cancer patients (21–23).
Overall, KIF11 was indicated to be involved in the progression and
therapy of several types of cancer, including prostate cancer,
colorectal cancer and gastric cancer (24–27).
However, the expression state and effect of KIF11 on breast cancer
remains unclear. As KIF11 plays an essential role in mitosis and is
an interesting drug target against cancer, it is worthwhile to
validate its role in breast cancer.
In the present study, it was revealed that the
expression of KIF11 was overexpressed in human breast cancer cells
and breast cancer tissues. Statistical analysis demonstrated a
significant association between the upregulation of KIF11
expression and the progression of breast cancer. Multivariate
analysis revealed that KIF11 upregulation may be an independent
prognostic indicator for the survival of patients with breast
cancer. Furthermore, silencing KIF11 with specific RNA interference
(RNAi) inhibited the cell growth rate in vitro and in
vivo. The present findings suggest that KIF11 serves an
important function in the proliferation and tumorigenesis of human
breast cancer, indicating that KIF11 may represent a valuable
target for human breast cancer treatment.
Materials and methods
Cell lines and tissues
Primary normal breast epithelial cells (NBECs) were
isolated from the mammoplasty material of two 32-year-old women at
the Department of Plastic Surgery, Shenzhen Longgang Maternal and
Child Health Hospital (Shenzhen, China) and cultured in the
Keratinocyte serum-free medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) (27,28).
Breast cancer MDA-MB-453, T47D, MCF-7, ZR-75-30, MDA-MB-231 and
BT-549 cell lines were cultured in DMEM medium (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(HyClone, Logan, Utah). Fresh tissues included six paired breast
cancer tissues and adjacent non-tumour tissues obtained from
individuals diagnosed with breast cancer at Shenzhen Longgang
Maternal and Child Health Hospital (Shenzhen, China) between March
2013 and June 2014.
A total of 268 paraffin-embedded, archived breast
cancer tissues were also collected, including 37 cases of low
histological grade, 111 cases of intermediate histological grade
and 120 cases of high histological grade. Samples were
histopathologically and clinically diagnosed at the Shenzhen
Longgang Maternal and Child Health Hospital between March 2003 and
December 2013, were also used in the current study. Patient consent
and approval from the Institutional Research Ethics Committee were
obtained prior to the use of these clinical specimens for research
purposes.
RNA extraction, reverse transcription
(RT) and quantitative polymerase chain reaction (qPCR)
Total RNA from cultured cells was extracted using
the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as
the manufacturer instructed. cDNA was amplified using the cDNA
Synthesis kit manual (cat. no. 6130 Takara Biotechnology Co., Ltd.-
Dalian, China) according to the manufacturer's protocol, and
quantified using an ABI Prism 7500 Sequence Detection system
(Applied Biosystems, Foster City, CA) using the dye SYBR Green I
(Invitrogen; Thermo Fisher Scientific, Inc.). PCR cycling
conditions were 50°C for 2 min, followed by 95°C for 10 min and
then 40 cycles for 95°C for 15 sec and 60°C for 1 min. The primers
used were: KIF11 forward, 5′-TAT TGA ATG GGC GCT AGC TT-3 and
reverse, 5′-TCG TCT GCG AAG AAG AAA GA-3; and GAPDH forward, 5′-ACC
ACA GTC CAT GCC ATC AC-3 and reverse, 5′-TCC ACC ACC CTG TTG CTG
TA-3′. Expression data were normalized to that of the housekeeping
gene GAPDH to control the variability in expression levels and
calculated using the 2−ΔΔCq method (28).
Western blot analysis
Western blotting was performed according to standard
methods, as described previously (29), using anti-KIF11 (cat. no. sc-365593)
and anti-mouse horseradish peroxidase (HRP)-conjugated
immunoglobulin (Ig)G (cat. no. sc-516102) antibodies (all 1:800;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA). The membranes
were stripped and re-probed with an anti-β-actin mouse monoclonal
antibody (1:2,000; cat. no. A2228; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) as a loading control. The expression of
indicated proteins was determined using an enhanced
chemiluminescence kit (cat. no. 3622ES60; Pierce, Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. ImageJ
1.48 software (National Institutes of Health, Bethesda, MD, USA)
was used to perform densitometry analysis. All detections were
performed in triplicate.
Plasmids and retroviral infection
For the depletion of KIF11, two human siRNA
sequences were cloned into pSuper-retro-puro (provided by Professor
Le Yang, Nanyang Medical College, Singapore) (30) to generate pSuper-retro-KIF11-RNAi#1
and #2, and the sequences were as follows: RNAi#1,
5′-UAUGGUGUUUGGAGCAUCUACUAAA-3′; and RNAi#2,
5′-CAGUACACAACAAGGAUGAAGUCUA-3′ (Invitrogen; Thermo Fisher
Scientific, Inc.). Retroviral production and infection were
performed as previously described (7).
Immunohistochemistry (IHC)
The IHC procedure and the KIF11 expression scores in
the 268 paraffin-embedded breast cancer samples, including 37 cases
of low histological grade, 111 cases of intermediate histological
grade and 120 cases of high histological grade, were performed as
previously described (31).
Anti-KIF11 and anti-mouse HRP-linked IgG (1:150; Santa Cruz
Biotechnology, Inc.) antibodies were used in this assay. Staining
for protein expression in tumor and normal tissues was quantitative
analyzed using the AxioVision Rel.4.6 computerized image analysis
system assisted with the automatic measurement program (Zeiss AG,
Oberkochen, Germany). Ten representative staining fields of each
section were analyzed to verify the mean optical density (MOD), and
the MOD data were statistically analyzed using a t-test to compare
the average MOD difference between different groups of tissues.
MTT assay
MCF-7-vector, MCF-7-KIF11 RNAi-1, MCF-7-KIF11
RNAi-2, MDA-MB-231-vector, MDA-MB-231-KIF11 RNAi-1 and
MDA-MB-231-KIF11 RNAi-2 cells were seeded on 96-well plates
(0.2×104 cells/well). At each time point, cells were
stained with MTT dye (0.5 mg/ml Sigma-Aldrich; Merck KGaA) for 4 h
at 37°C, followed by the removal of the culture medium and addition
of 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA). The
absorbance was measured at 570 nm, with 655 nm used as the
reference wavelength. All experiments were performed in
triplicate.
Anchorage-independent growth ability
assay
A total of 500 MCF-7-vector, MCF-7-KIF11 RNAi-1,
MCF-7-KIF11 RNAi-2, MDA-MB-231-vector, MDA-MB-231-KIF11 RNAi-1 and
MDA-MB-231-KIF11 RNAi-2 cells were trypsinized and suspended in 2
ml complete medium plus 0.3% agar (Sigma-Aldrich; Merck KGaA).
After 10 days, viable colonies that contained >50 cells or were
larger than 0.5 mm were counted. All experiments were performed in
triplicates.
Colony formation assays
MCF-7-vector, MCF-7-KIF11 RNAi-1, MCF-7-KIF11
RNAi-2, MDA-MB-231-vector, MDA-MB-231-KIF11 RNAi-1 and
MDA-MB-231-KIF11 RNAi-2 cells were plated on 60-mm plates
(0.5×103 cells per plate) and cultured at 37°C for 10
days. The colonies were stained with 1% crystal violet
(Sigma-Aldrich; Merck KGaA) for 30 sec following fixation with 10%
formaldehyde for 5 min at room temperature.
Xenograft tumour model
Each mouse was subcutaneously injected in
situ with MDA-MB-231-vector cells (5×106 cells) on
the left flank and with MDA-MB-231-KIF11 cells (5×106
cells) on the right flank. Tumours were examined every five days.
The length (L) and width (W) were measured using calipers, and
tumour volumes were calculated using the following equation: Tumour
volume (cm3)=(LxW2)/2. On day 35, the animals
were euthanized, and the tumours were excised and weighed. The
Institutional Animal Care and Use Committee of Shenzhen Longgang
Maternal and Child Health Hospital approved all experimental
procedures.
Statistical analysis
All statistical analyses were performed using SPSS
13.0 statistical software (SPSS, Inc., Chicago, IL, USA). The
association between KIF11 expression and clinicopathological
characteristics was analyzed using the χ2 test.
Bivariate correlations between study variables were calculated
using Spearman's rank correlation coefficients. Survival curves
were plotted using the Kaplan-Meier method and compared using the
log-rank test. Survival data were evaluated using univariate and
multivariate Cox regression analyses. P<0.05 was considered to
indicate a statistically significant difference.
Results
KIF11 is upregulated in breast cancer
cell lines
To assess the expression of KIF11, normal breast
epithelial cells (NBEC) from two different patients without breast
cancer were obtained and cultured. Western blot analysis and
RT-qPCR analysis revealed that KIF11 expression was extremely
difficult to detect in NBECs. Breast cancer cell lines were used to
detect the expression of KIF11 protein and mRNA in malignant cells.
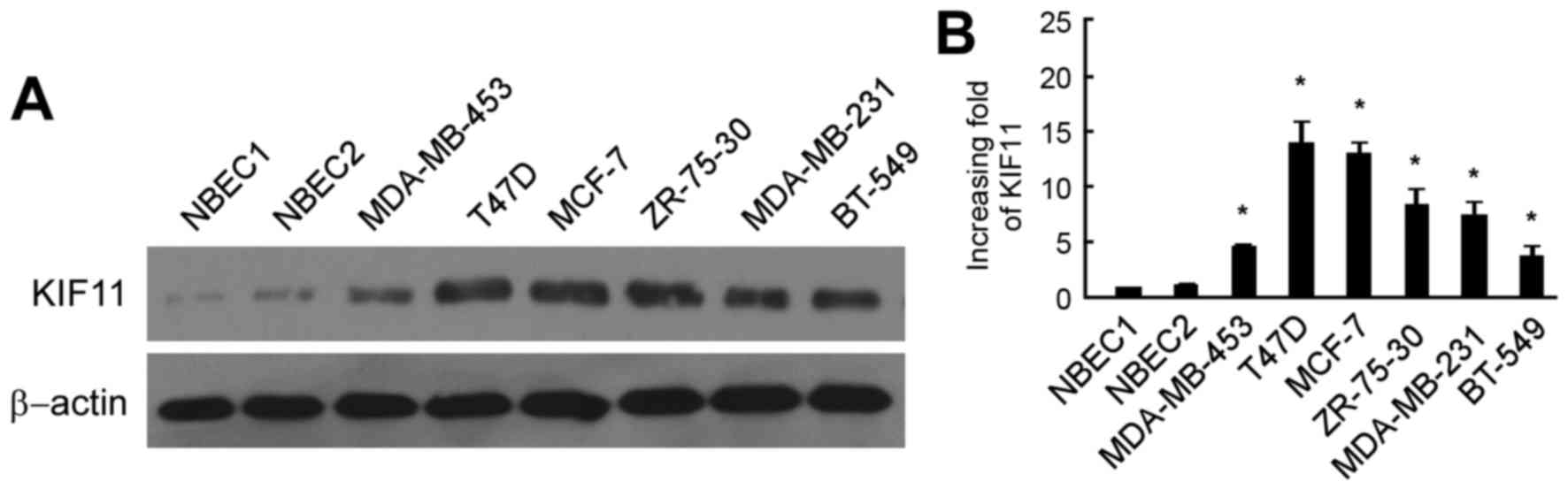
As shown in Fig. 1A, the protein
level of KIF11 was markedly upregulated in six breast cancer cells
lines, consisting of the MDA-MB-453, T47D, MCF-7, ZR-75-30,
MDA-MB-231 and BT-549 cell lines, in comparison to those in NBEC1
and NBEC2. RT-qPCR analysis also revealed similar results of mRNA
expression in the breast cancer cell lines MDA-MB-453, T47D, MCF-7,
ZR-75-30, MDA-MB-231 and BT-549, with a 4.7–14.9 fold increase
compared with that in the NBEC1 cells (Fig. 1B).
KIF11 is upregulated in paired fresh
tissues of breast cancer
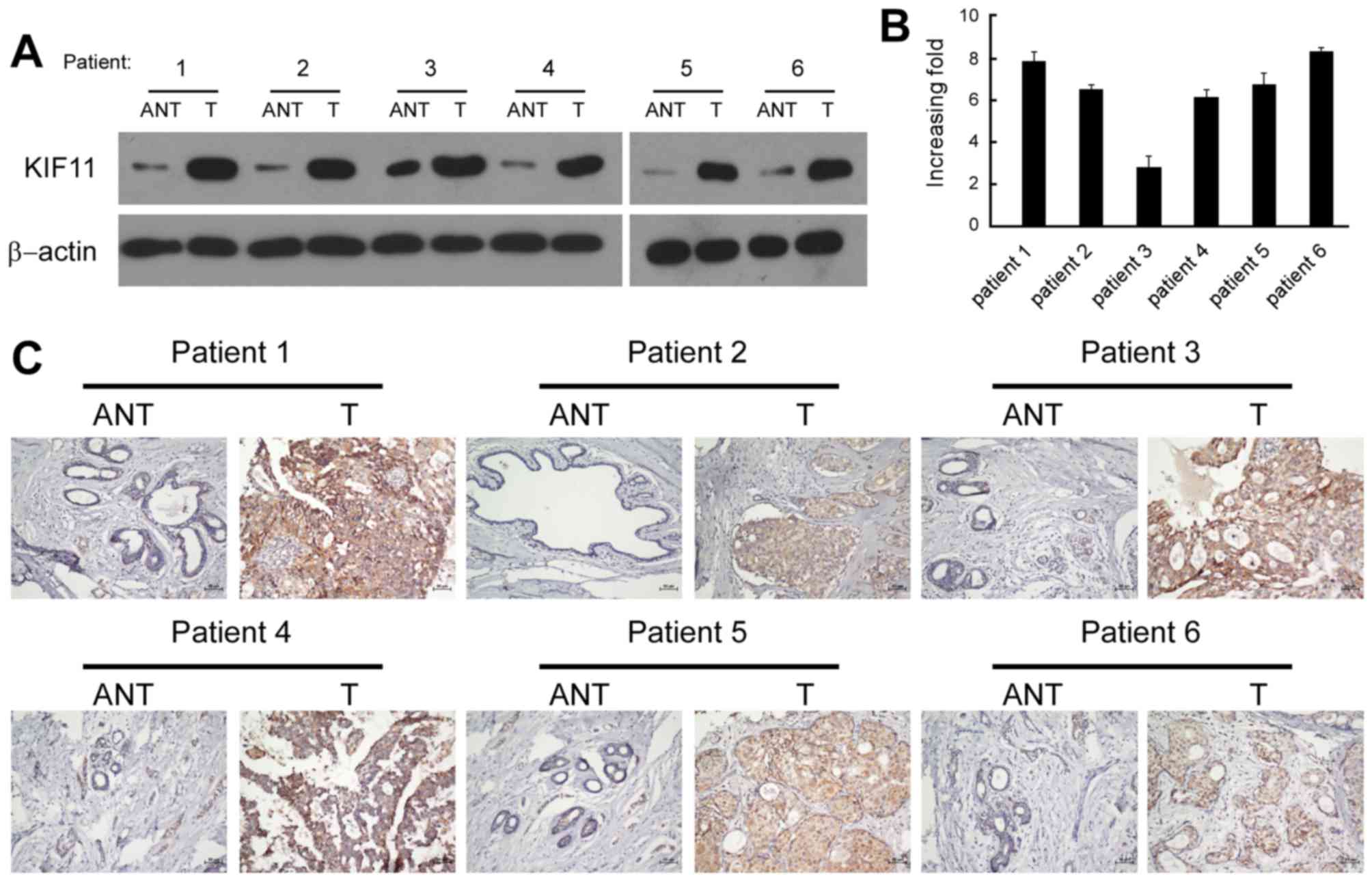
Six pairs of matched adjacent non-tumorous breast
tissue (ANT) and breast tumour tissue samples were used for
screening the expression of KIF11. Western blot and RT-qPCR
analysis revealed that the protein and mRNA levels of KIF11 were
significantly upregulated in the human primary breast tumour
tissues, with a ≥2.5-fold increase compared with each paired ANT
(Fig. 2A and B). The in situ
expression of KIF11 in the aforementioned six pairs of breast
tissues was examined by immunohistochemical staining. The
representative brown colour in Fig.
2C indicated the expression of the KIF11 protein, confirming
the upregulation of KIF11 in breast cancer. However, the IHC signal
of KIF11 was undetectable or only marginally detectable in the
ANTs. Overall, the present results indicated that KIF11 expression
was upregulated in breast cancer cell lines and breast cancer
tissues.
Upregulation of KIF11 is associated
with clinicopathological characteristics of breast cancer and
patient survival
To validate the universality and importance of the
upregulation of KIF11 in breast cancer, 268 paraffin-embedded,
archived breast cancer tissues were also collected, including 37
cases of low histological grade, 111 cases of intermediate
histological grade and 120 cases of high histological grade. These
samples were stained with KIF11 antibody, scored by a recognized
standard and summarized in Table I.
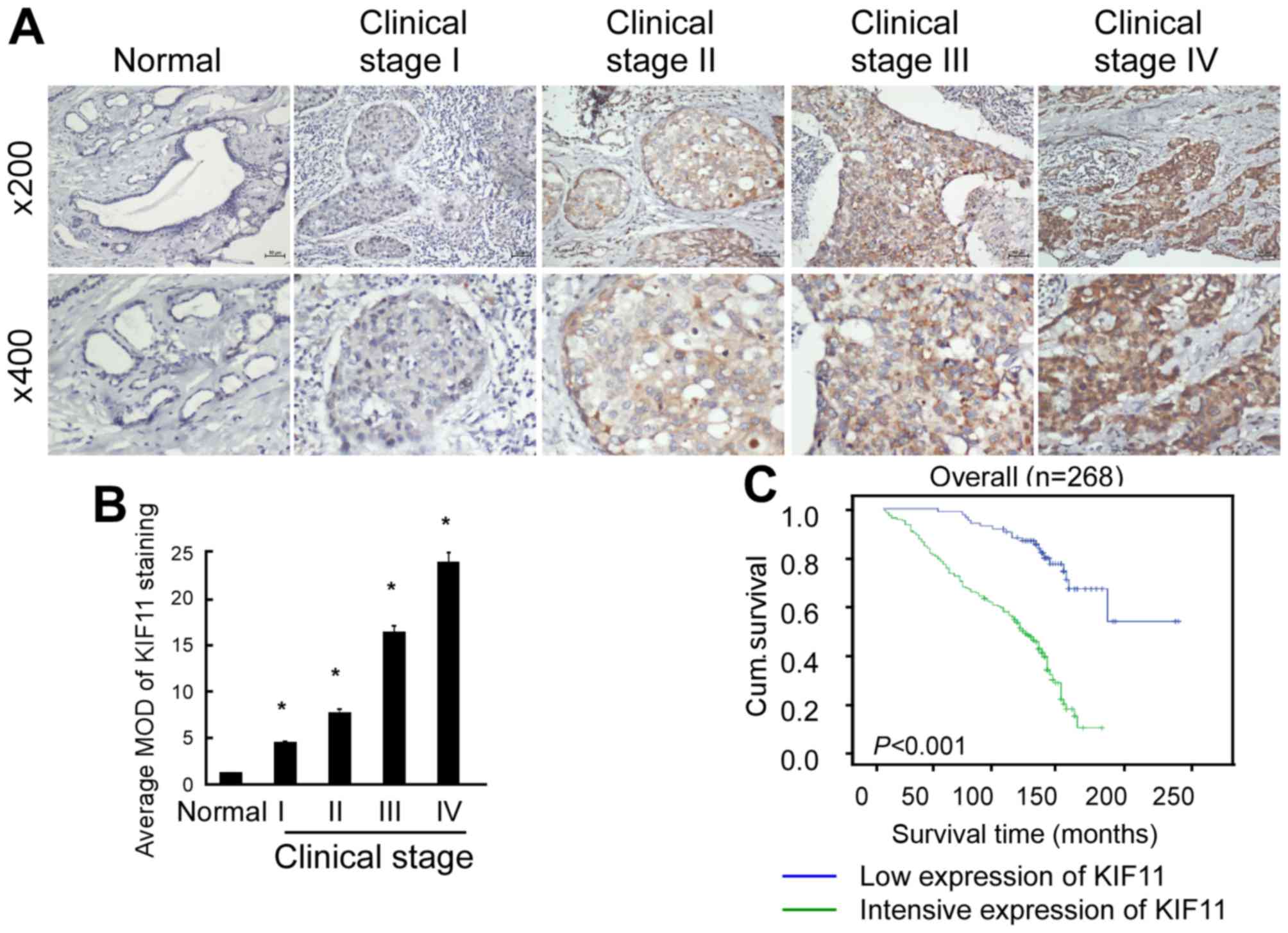
As shown in Fig. 3A, KIF11 protein
expression was detected in 256 of the 268 (95.5%) tested cases,
which was expressed at lower levels in early stages (stage I–II)
and more highly expressed in later stages (stage III–IV).
Quantitative analysis indicated that the average mean optical
densities (MODs) of KIF11 staining were markedly increased in
breast tumours compared with the MODs of normal breast tissues
(P<0.001; Fig. 3B). Overall, the
present results indicate that overexpression of KIF11 is a common
feature of breast cancer.
 | Table I.Association between KIF11 expression
and clinicopathological characteristics of breast cancer
patients. |
Table I.
Association between KIF11 expression
and clinicopathological characteristics of breast cancer
patients.
|
|
| KIF11 expression,
n |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Total, n | Low | High | P-value |
|---|
| Age |
|
|
| 0.685 |
| <45
years | 101 | 30 | 71 |
|
| ≥45
years | 167 | 54 | 113 |
|
| Clinical stage |
|
|
| <0.001 |
| I | 25 | 24 |
1 |
|
| II | 126 | 43 | 83 |
|
|
III | 79 | 11 | 68 |
|
| IV | 31 |
3 | 28 |
|
| T
classification |
|
|
| <0.001 |
| T1 | 44 | 29 | 15 |
|
| T2 | 141 | 44 | 97 |
|
| T3 | 50 |
8 | 42 |
|
| T4 | 26 |
0 | 26 |
|
| N
classification |
|
|
| <0.001 |
| N0 | 106 | 56 | 50 |
|
| N1 | 101 | 15 | 86 |
|
| N2 | 43 |
8 | 35 |
|
| N3 | 11 |
2 |
9 |
|
| Metastasis |
|
|
| 0.042 |
| No | 246 | 80 | 166 |
|
|
Yes | 15 |
1 | 14 |
|
| Histological
grade |
|
|
| <0.001 |
|
Low | 37 | 22 | 15 |
|
|
Intermediate | 111 | 48 | 63 |
|
|
High | 120 | 14 | 106 |
|
| ER |
|
|
| 0.088 |
|
Negative | 32 | 91 | 123 |
|
|
Positive | 52 | 93 | 145 |
|
| PR |
|
|
| 0.107 |
|
Negative | 107 | 28 | 79 |
|
|
Positive | 156 | 56 | 100 |
|
| erbB-2 |
|
|
| 1.000 |
|
Negative | 38 | 10 | 28 |
|
|
Positive | 115 | 30 | 84 |
|
Furthermore, the IHC score and classification (low
expression of KIF11; intensive expression of KIF11) of the
expression of KIF11 in the IHC assays were statistical analysed to
evaluate the association between KIF11 and the clinicopathological
characteristics of breast cancer. As shown in Table I, there was a strong association
between the expression of KIF11 and clinical stage (P<0.001), T
classification (P<0.001), N classification (P<0.001) and M
classification (P=0.042). However, the expression of KIF11 is not
associated with ER, PR or ErbB-2 expression.
Additionally, the effects of clinicopathological
characteristics and the expression of KIF11 protein on survival
were analysed using Kaplan-Meier analysis and the log-rank test. As
shown in Fig. 3C, the survival time
was evidently longer in the patients with low expression of KIF11
(P<0.001). Statistical analysis presented in Table II revealed an inverse association
between KIF11 level and patient survival (P=0.005). Furthermore,
log-rank test and Kaplan-Meier analysis were also applied to
calculate the effect of KIF11 expression and histological staging
of breast cancer on survival in more detail. The log-rank test
showed that the expression level of KIF11 protein in breast cancer
was significantly associated with the survival time of patients
(P<0.001). In particular, the mean survival time of patients
with high expression of the KIF11 protein was only 92.42 months,
whereas the mean survival time of those with low levels of KIF11
expression was 127.49 months. As shown in Fig. 2C, the cumulative survival rate was
significantly increased in the low KIF11 expression group compared
with the high KIF11 expression group. Multivariate survival
analysis shown in Table III
indicated that the KIF11 expression level was an independent
prognostic factor for the assessment of patient outcomes. This
finding suggested that KIF11 acted as a prognostic factor, which
may be useful to predict cancer evolution and provide appropriate
treatments for breast cancer patients.
 | Table II.Clinical pathological parameters and
expression of KIF11 for prognosis of 268 patients with breast
cancer by univariate survival analysis. |
Table II.
Clinical pathological parameters and
expression of KIF11 for prognosis of 268 patients with breast
cancer by univariate survival analysis.
|
Characteristics | Total, n | Mean survival time,
months | Median survival
time, months | P-value |
|---|
| Age |
|
|
| 0.219 |
| <45
years | 101 | 104.78 | 116 |
|
| ≥45
years | 167 | 102.59 | 118 |
|
| Clinical stage |
|
|
| <0.001 |
| I | 25 | 123.68 | 122 |
|
| II | 126 | 113.13 | 120 |
|
|
III | 79 |
90.53 | 106 |
|
| IV | 31 |
87.1 | 102 |
|
| T
classification |
|
|
| <0.001 |
| T1 | 44 | 109.16 | 120 |
|
| T2 | 141 | 113.85 | 120 |
|
| T3 | 50 |
94.64 | 108 |
|
| T4 | 26 |
61.92 | 46 |
|
| N
classification |
|
|
| <0.001 |
| N0 | 106 | 116.78 | 120 |
|
| N1 | 101 | 103.42 | 118 |
|
| N2 | 43 |
76.37 | 82 |
|
| N3 | 11 |
99.09 | 122 |
|
| Metastasis |
|
|
| <0.001 |
| No | 246 | 107.07 | 120 |
|
|
Yes | 15 |
57.2 | 40 |
|
| Histological
grade |
|
|
| <0.001 |
|
Low | 37 | 152.11 | 148 |
|
|
Intermediate | 111 | 125.06 | 124 |
|
|
High | 120 |
68.38 | 66 |
|
| KIF11
expression |
|
|
| <0.001 |
|
Low | 84 | 127.49 | 124 |
|
|
High | 181 |
92.42 | 107 |
|
 | Table III.Multivariate analysis of overall
survival (Cox regression model). |
Table III.
Multivariate analysis of overall
survival (Cox regression model).
| Variable | Relative risk | 95% confidence
interval | P-value |
|---|
| KIF11 | 3.177 | 1.684–5.991 | <0.001 |
| T
classification | 0.758 | 0.423–1.359 | 0.01 |
| N
classification | 1.233 | 0.502–3.029 | 0.005 |
| Metastasis | 0.215 | 0.111–0.417 | <0.001 |
| Histological
grade | 0.03 | 0.014–0.062 | <0.001 |
Downregulation of endogenous KIF11
inhibited the proliferation of breast cancer cells
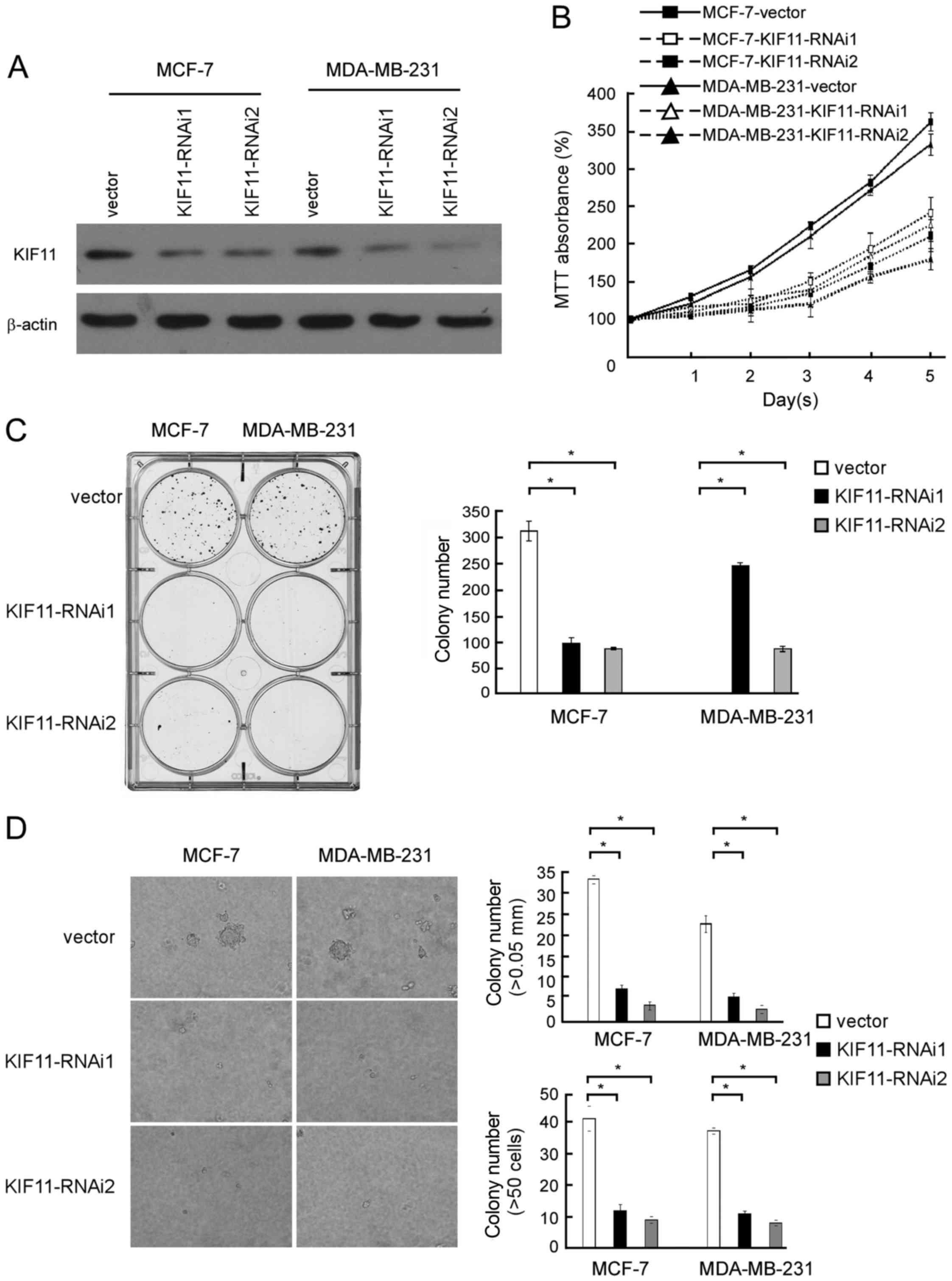
To investigate the biological role of KIF11
expression in the development and progression of breast cancer, two
specific KIF11-short hairpin (sh)RNAs were transfected into breast
cancer MCF-7 and MDA-MB-231 cell lines to further investigate the
effect of KIF11 in promoting proliferation of breast cancer
(Fig. 4A). MTT assay revealed that
downregulation of KIF11 significantly slowed down the proliferation
of breast cancer MCF-7 and MDA-MB-231 cells, with ~1.5-fold fewer
cells than the control by day 5 subsequent to plating (185 vs. 365%
MTT absorbance in MCF-7 cells and 155 vs. 337% MTT absorbance in
MDA-MB-231 cells; Fig. 4B). As shown
in Fig. 4C, the colony number was
also significantly decreased in the KIF11-transfected cells. These
experiments show that inhibition of KIF11 markedly reduced the
growth rate of MCF-7 and MDA-MB-231 cells compared with the growth
rate of vector-transfected cells. As shown in Fig. 4D, decreased colony number and colony
size in KIF11-shRNA-transfected breast cancer cells was identified
and indicated the inhibition effect of downregulation of KIF11 on
anchorage-independent growth ability (P<0.05). These results
further supported the hypothesis that KIF11 serves important
functions in the proliferation and tumorigenicity of breast cancer
cells.
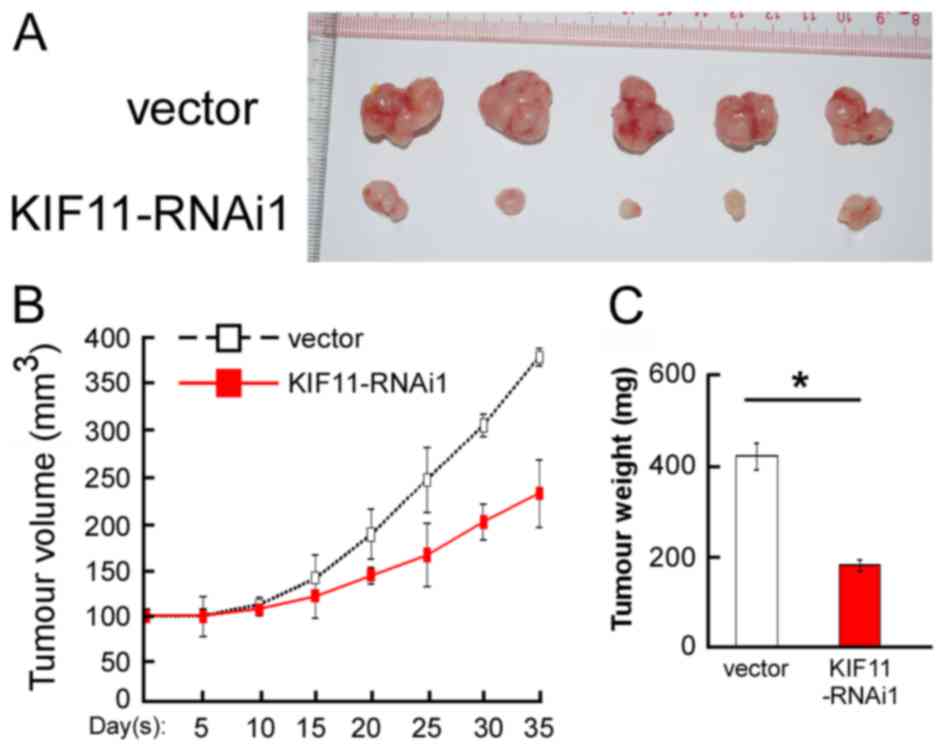
In vivo assay reveals the inhibition
role of KIF11-RNAi on tumorigenicity
To validate the aforementioned results obtained from
the in vitro cell proliferation assays, the present study
performed in vivo assays to evaluate the tumorigenic effect
of KIF11 in BALB/C nude mice using the MDA-MB-231 cell line. As
shown in Fig. 5A,
KIF11-RNAi#1-transfected cells showed an anti-proliferation
tendency in nude mice. Decreased tumour volume and tumour weight
generated from KIF11-RNAi#1-transfected cells were observed
compared with the vector infected group, as indicated in Fig. 5B and C. Overall, the present results
demonstrated that KIF11 has an important role in the tumorigenicity
of nude mice.
Discussion
The key finding of the present study is that the
progression of human breast cancer is associated with the
upregulation of KIF11. Furthermore, in vitro and in
vivo assays both demonstrated that the promoting effect of
KIF11 on breast cancer cells and may indicate a novel predictive
marker for the clinical outcome of the disease.
Previous studies have associated KIF11 expression
with cancer development and progression. Upregulation of the KIF11
protein and mRNA levels was reported in prostate cancer PC3 cells.
In the PC3 and LNCaP cell lines, deceased mRNA and protein levels
of KIF11 inhibited cell growth, induced G2/M phase arrest and
increased the apoptotic sub-G1 fraction. In vivo, decreased
KIF11 significantly reduced both LNCaP and PC-3 tumour growth
(32). Other studies may be utilized
to clarify the mechanisms of KIF11 in cancer progression. It was
reported that, in CD4-positive T-lymphocytes, Tat interacts with
KIF11 and allosterically modulates the ATPase activity of KIF11 by
affecting ADP release from the active centre of the enzyme. This
action of Tat impairs the formation of the mitotic spindle and
activates the spindle checkpoint, thereby blocking cell cycle
progression at mitosis and leading to apoptosis (33). Sun et al (34) revealed that the expression of KIF11 in
renal cell carcinoma was significantly associated with tumour
nuclear grade and stage, as well as tumour size. In univariate
analysis, KIF11 overexpression showed a statistically significant
unfavourable effect on recurrence-free survival (34). It was identified by Bartoli et
al (35) that, during interphase,
KIF11 is associated with ribosomes and is required for optimal
nascent polypeptide synthesis. When KIF11 was inhibited, ribosomes
no longer bound to microtubules in vitro, ribosome transit
rates slowed, and polysomes accumulated in intact cells, suggesting
defects in elongation or termination during polypeptide synthesis.
Furthermore, cycle-dependent kinase 1 (CDK1) and CDK2
phosphorylated KIF11 at Thr927, which supports the association
between KIF11 and cell-cycle regulation (36). Additionally, nucleophosmin/B23, an
abundant nucleolar protein, has multiple roles in cell growth and
proliferation. Both in vivo and in vitro studies have
demonstrated that B23 acts as an upstream regulator of KIF11 in
promoting microtubule polymerization. Additionally, it was further
demonstrated that B23 regulates microtubule dynamics by directly
inhibiting ATPase activity (37). The
present study found that KIF11 was frequently upregulated in breast
cancer and the positive association between the progression of
breast cancer and the expression of KIF11. In the more detailed
survival analysis of the present study, multivariate analyses have
shown that high expression of KIF11 is a predictor of poor
prognosis for breast cancer patients. Nevertheless, the mechanism
of the regulation of KIF protein in the progression of breast
cancer requires additional study.
Numerous studies have indicated that KIF11 is a
feasible drug target. It was reported that KIF11 has a critical
role in mitosis as it mediates centrosome separation and bipolar
spindle assembly and maintenance (38,39).
Knockdown of KIF11 by siRNA and SB-715992, another term for the
small-molecule inhibitor ispinesib, both induced G2 arrest.
Furthermore, growth of head and neck squamous cell carcinoma cells
engrafted in immunodeficient mice was significantly inhibited after
ispinesib treatment (40). Marra
et al (41) found that
KIF-specific siRNA markedly reduced outgrowth of subcutaneous
melanoma and ovarian cancer lesions. The ispinesib analogue 1, a
well characterized and potentially specific small molecule
inhibitor of KIF11, showed an anti-proliferative effect on
glioblastoma multiforme cell lines by blocking cell cycle
progression in the G2/M phase and increased caspase 3/7-induced
apoptosis in U87MG cells (42). In
addition, the kinesin spindle protein (KSP) inhibitor ARRY-520 may
be an alternative to paclitaxel in a Type I ovarian cancer patients
(23). Inhibition of KSP by ARRY-520
induces cell cycle block and cell death via the mitochondrial
pathway in AML cells (22).
Comparison between KSP inhibitor-induced apoptosis in matched cell
lines containing functional p53 and cells containing deficient p53
revealed that inhibition of KSP induces apoptosis independently of
p53, and that p53 is dispensable for spindle checkpoint function.
Thus, KIF11 inhibitors should be active in p53-deficient tumours
(43). However, it is rare for
studies to focus on the inhibition of KIF11 in breast cancer. In
the present study, it was found that downregulation of endogenous
KIF11 inhibited the proliferation of breast cancer cells in
vitro and in vivo, indicating that KIF11 may be a
valuable target for human breast cancer treatment.
In summary, the present study suggests that KIF11
overexpression is a common feature in breast cancer and may be a
potential target as a therapeutic strategy for breast cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81201568).
References
|
1
|
Stuckey AR and Onstad MA: Hereditary
breast cancer: An update on risk assessment and genetic testing in
2015. Am J Obstet Gynecol. 213:161–165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sutherland RL, Hamilton JA, Sweeney KJ,
Watts CK and Musgrove EA: Expression and regulation of cyclin genes
in breast cancer. Acta Oncol. 34:651–656. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sjöström J, Blomqvist C, Heikkilä P,
Boguslawski KV, Räisänen-Sokolowski A, Bengtsson NO, Mjaaland I,
Malmström P, Ostenstadt B, Bergh J, et al: Predictive value of p53,
mdm-2, p21, and mib-1 for chemotherapy response in advanced breast
cancer. Clin Cancer Res. 6:3103–3110. 2000.PubMed/NCBI
|
|
4
|
Bartek J, Iggo R, Gannon J and Lane DP:
Genetic and immunochemical analysis of mutant p53 in human breast
cancer cell lines. Oncogene. 5:893–899. 1990.PubMed/NCBI
|
|
5
|
Ertel A, Dean JL, Rui H, Liu C, Witkiewicz
AK, Knudsen KE and Knudsen ES: RB-pathway disruption in breast
cancer: Differential association with disease subtypes,
disease-specific prognosis and therapeutic response. Cell Cycle.
9:4153–4163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tomas NM, Masur K, Piecha JC, Niggemann B
and Zänker KS: Akt and phospholipase Cγ are involved in the
regulation of growth and migration of MDA-MB-468 breast cancer and
SW480 colon cancer cells when cultured with diabetogenic levels of
glucose and insulin. BMC Res Notes. 5:2142012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song L, Wang L, Li Y, Xiong H, Wu J, Li J
and Li M: Sam68 up-regulation correlates with, and its
down-regulation inhibits, proliferation and tumourigenicity of
breast cancer cells. J Pathol. 222:227–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iwakiri Y, Kamakura S, Hayase J and
Sumimoto H: Interaction of NuMA protein with the kinesin Eg5: It's
possible role in bipolar spindle assembly and chromosome alignment.
Biochem J. 451:195–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun L, Sun X, Xie S, Yu H and Zhong D:
Significant decrease of ADP release rate underlies the potent
activity of dimethylenastron to inhibit mitotic kinesin Eg5 and
cancer cell proliferation. Biochem Biophys Res Commun. 447:465–470.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McGrath MJ, Kuo IF, Hayashi S and Takada
S: Adenosine triphosphate hydrolysis mechanism in kinesin studied
by combined quantum-mechanical/molecular-mechanical metadynamics
simulations. J Am Chem Soc. 135:8908–8919. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Robitaille JM, Gillett RM, LeBlanc MA,
Gaston D, Nightingale M, Mackley MP, Parkash S, Hathaway J, Thomas
A, Ells A, et al: Phenotypic overlap between familial exudative
vitreoretinopathy and microcephaly, lymphedema, and chorioretinal
dysplasia caused by KIF11 mutations. JAMA Ophthalmol.
132:1393–1399. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hazan F, Ostergaard P, Ozturk T, Kantekin
E, Atlihan F, Jeffery S and Ozkinay F: A novel KIF11 mutation in a
Turkish patient with microcephaly, lymphedema, and chorioretinal
dysplasia from a consanguineous family. Am J Med Genet A.
158A:1–1689. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Riedl J, Voβmerbäumer U, Stoffelns B and
Elflein H: Total retinal detachment caused by a KIF11 mutation. Eur
J Ophthalmol. May 24–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jones GE, Ostergaard P, Moore AT, Connell
FC, Williams D, Quarrell O, Brady AF, Spier I, Hazan F, Moldovan O,
et al: Microcephaly with or without chorioretinopathy, lymphoedema,
or mental retardation (MCLMR): Review of phenotype associated with
KIF11 mutations. Eur J Hum Genet. 22:881–887. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bullock JM, Medway C, Cortina-Borja M,
Turton JC, Prince JA, Ibrahim-Verbaas CA, Schuur M, Breteler MM,
van Duijn CM, Kehoe PG, et al: Discovery by the Epistasis Project
of an epistatic interaction between the GSTM3 gene and the
HHEX/IDE/KIF11 locus in the risk of Alzheimer's disease. Neurobiol
Aging. 34:1309.e1–e7. 2013. View Article : Google Scholar
|
|
16
|
Qian Y, Lu F, Dong M, Lin Y, Li H, Chen J,
Shen C, Jin G, Hu Z and Shen H: Genetic variants of IDE-KIF11-HHEX
at 10q23.33 associated with type 2 diabetes risk: A fine-mapping
study in Chinese population. PLoS One. 7:e350602012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tan LJ, Saijo M, Kuraoka I, Narita T,
Takahata C, Iwai S and Tanaka K: Xeroderma pigmentosum group F
protein binds to Eg5 and is required for proper mitosis:
Implications for XP-F and XFE. Genes Cells. 17:173–185. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wakana Y, Villeneuve J, van Galen J,
Cruz-Garcia D, Tagaya M and Malhotra V: Kinesin-5/Eg5 is important
for transport of CARTS from the trans-Golgi network to the cell
surface. J Cell Biol. 202:241–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ding S, Xing N, Lu J, Zhang H, Nishizawa
K, Liu S, Yuan X, Qin Y, Liu Y, Ogawa O, et al: Overexpression of
Eg5 predicts unfavorable prognosis in non-muscle invasive bladder
urothelial carcinoma. Int J Urol. 18:432–438. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tsui M, Xie T, Orth JD, Carpenter AE,
Rudnicki S, Kim S, Shamu CE and Mitchison TJ: An intermittent live
cell imaging screen for siRNA enhancers and suppressors of a
kinesin-5 inhibitor. PLoS One. 4:e73392009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Woessner R, Tunquist B, Lemieux C,
Chlipala E, Jackinsky S, Dewolf W Jr, Voegtli W, Cox A, Rana S, Lee
P and Walker D: ARRY-520, a novel KSP inhibitor with potent
activity in hematological and taxane-resistant tumor models.
Anticancer Res. 29:4373–4380. 2009.PubMed/NCBI
|
|
22
|
Carter BZ, Mak DH, Woessner R, Gross S,
Schober WD, Estrov Z, Kantarjian H and Andreeff M: Inhibition of
KSP by ARRY-520 induces cell cycle block and cell death via the
mitochondrial pathway in AML cells. Leukemia. 23:1755–1762. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim KH, Xie Y, Tytler EM, Woessner R, Mor
G and Alvero AB: KSP inhibitor ARRY-520 as a substitute for
Paclitaxel in Type I ovarian cancer cells. J Transl Med. 7:632009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Asbaghi Y, Thompson LL, Lichtensztejn Z
and McManus KJ: KIF11 silencing and inhibition induces chromosome
instability that may contribute to cancer. Genes Chromosomes
Cancer. 56:668–680. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schneider MA, Christopoulos P, Muley T,
Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller
NS, Theis F, et al: AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five
specific mitosis-associated genes correlate with poor prognosis for
non-small cell lung cancer patients. Int J Oncol. 50:365–372. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Imai T, Oue N, Sentani K, Sakamoto N,
Uraoka N, Egi H, Hinoi T, Ohdan H, Yoshida K and Yasui W: KIF11 is
required for spheroid formation by oesophageal and colorectal
cancer cells. Anticancer Res. 37:47–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Imai T, Oue N, Nishioka M, Mukai S, Oshima
T, Sakamoto N, Sentani K, Matsusaki K, Yoshida K and Yasui W:
Overexpression of KIF11 in gastric cancer with intestinal mucin
phenotype. Pathobiology. 84:16–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li J, Zhang N, Song LB, Liao WT, Jiang LL,
Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS and Li M: Astrocyte
elevated gene-1 is a novel prognostic marker for breast cancer
progression and overall patient survival. Clin Cancer Res.
14:3319–3326. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li J, Yang L, Song L, Xiong H, Wang L, Yan
X, Yuan J, Wu J and Li M: Astrocyte elevated gene-1 is a
proliferation promoter in breast cancer via suppressing
transcriptional factor FOXO1. Oncogene. 28:3188–3196. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Z, Li J, Zheng H, Yu C, Chen J, Liu
Z, Li M, Zeng M, Zhou F and Song L: Expression and cytoplasmic
localization of SAM68 is a significant and independent prognostic
marker for renal cell carcinoma. Cancer Epidemiol Biomarkers Prev.
18:2685–2693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hayashi N, Koller E, Fazli L and Gleave
ME: Effects of Eg5 knockdown on human prostate cancer xenograft
growth and chemosensitivity. Prostate. 68:1283–1295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu M, Li D, Sun L, Chen J, Sun X, Zhang
L, Huo L and Zhou J: Modulation of Eg5 activity contributes to
mitotic spindle checkpoint activation and Tat-mediated apoptosis in
CD4-positive T-lymphocytes. J Pathol. 233:138–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun D, Lu J, Ding K, Bi D, Niu Z, Cao Q,
Zhang J and Ding S: The expression of Eg5 predicts a poor outcome
for patients with renal cell carcinoma. Med Oncol. 30:4762013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bartoli KM, Jakovljevic J, Woolford JL Jr
and Saunders WS: Kinesin molecular motor Eg5 functions during
polypeptide synthesis. Mol Biol Cell. 22:3420–3430. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Smith E, Hégarat N, Vesely C, Roseboom I,
Larch C, Streicher H, Straatman K, Flynn H, Skehel M, Hirota T, et
al: Differential control of Eg5-dependent centrosome separation by
Plk1 and Cdk1. EMBO J. 30:2233–2245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang G, Gao X, Huang Y, Yao Z, Shi Q and
Wu M: Nucleophosmin/B23 inhibits Eg5-mediated microtubule
depolymerization by inactivating its ATPase activity. J Biol Chem.
285:19060–19067. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cochran JC, Sontag CA, Maliga Z, Kapoor
TM, Correia JJ and Gilbert SP: Mechanistic analysis of the mitotic
kinesin Eg5. J Biol Chem. 279:38861–38870. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rosenfeld SS, Xing J, Jefferson GM and
King PH: Docking and rolling, a model of how the mitotic motor Eg5
works. J Biol Chem. 280:35684–35695. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martens-de Kemp SR, Nagel R, Stigter-van
Walsum M, Van der Meulen IH, van Beusechem VW, Braakhuis BJ and
Brakenhoff RH: Functional genetic screens identify genes essential
for tumor cell survival in head and neck and lung cancer. Clin
Cancer Res. 19:1994–2003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Marra E, Palombo F, Ciliberto G and
Aurisicchio L: Kinesin spindle protein SiRNA slows tumor
progression. J Cell Physiol. 228:58–64. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Valensin S, Ghiron C, Lamanna C, Kremer A,
Rossi M, Ferruzzi P, Nievo M and Bakker A: KIF11 inhibition for
glioblastoma treatment: Reason to hope or a struggle with the
brain? BMC Cancer. 9:1962009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tao W, South VJ, Diehl RE, Davide JP,
Sepp-Lorenzino L, Fraley ME, Arrington KL and Lobell RB: An
inhibitor of the kinesin spindle protein activates the intrinsic
apoptotic pathway independently of p53 and de novo protein
synthesis. Mol Cell Biol. 27:689–698. 2007. View Article : Google Scholar : PubMed/NCBI
|