Introduction
Lung cancer is the most common cause of cancer
mortality in the world, with non-small cell lung cancer (NSCLC) its
most prevalent form (1). Despite
recent development of oncogene-targeting therapy and immunotherapy,
the prognosis for NSCLC at advanced stages remains poor (2,3). Either
chemically-induced or genetically-engineered mouse models, as well
as patient-derived xenograft models, have provided preclinical
models to understand NSCLC and to test novel therapeutic approaches
(4). Nonetheless, more efficient
methods to culture, expand and transform lung epithelial (LE) cells
are required.
The linings of organs are constructed from
epithelial tissues, which are the origin of most of solid
malignancies. Epithelial cells are connected to each other and/or
with the basement membrane, and these cell-cell or cell-matrix
interactions cannot be completely recapitulated in 2D culture,
rendering analysis difficult using non-transformed epithelial cells
in vitro and ex vivo. But recent advances in
three-dimensional (3D) culture techniques have enabled in
vitro expansion, genetic or environmental manipulation and
real-time observations of epithelial cells from numerous human and
rodent organs (5,6).
In this study, we applied a 3D culture technique to
isolate epithelial cells from a mixed population of mouse lung
cells. Almost all isolated cells were positive for EpCAM, an
epithelial surface-marker. LE cells could be passaged and expanded
for several months and were easily transformed by genetic
manipulation.
Materials and methods
Materials
Human EGFR cDNA encoding the exon 19-deletion mutant
(EGFRex19del) was generated by a polymerase chain
reaction-based method (7) and
subcloned into a retroviral vector, pMXs-Puro-3×HA, as described
previously (7). The plasmid
pBabe-puro Kras-V12 was a gift from Dr C. Counter (8). The plasmid pBabe-neo largeT cDNA was a
gift from Dr R. Weinberg (9).
Matrigel was obtained from BD Biosciences (Franklin Lakes, NJ,
USA). Murine EGF (mEGF) was purchased from PeproTech Inc. (Rocky
Hill, NJ, USA). Erlotinib was purchased from Wako Pure Chemical
Industries (Osaka, Japan). Trametinib and Captisol were purchased
from ChemScene (Monmouth Junction, NJ, USA). Y-27632 was purchased
from Calbiochem (Darmstadt, Germany). TrypLE Express from Thermo
Fisher Scientific Inc. (Waltham, MA, USA).
Isolation and 3D culture of mouse LE
cells
C57BL/6 mice were used at 6–9 weeks of age.
Isolation, 3D culture and passage were performed similarly to
methods previously described for colon epithelial cells, except
that R-Spondin, Noggin and Jagged-1 were not added to culture media
(10). In various analyses, mEGF and
Y-27632 were added to the medium both in 3D and 2D culture, both at
final concentrations of 50 nM. To passage cells in 2D culture,
cells were washed with PBS, treated with Trypl Express reagent,
which is a mixture of protease and collagenase, and then detached
from the dish using a cell-scraper. Time-lapse imaging was
performed using an Incucyte cell-analyzer (Essen Bioscience, Ann
Arbor, MI, USA).
Caspase activity assay
Cellular caspase activity was measured using the
Caspase-Glo 3/7 Assay kit according to the manufacturer's
recommendation (Promega, Madison, WI, USA). The values were
normalized to cell numbers. The results shown in Fig. 2C is results from analyses of cells 2 h
after the re-plating upon passage.
Infection of LE cells with
retrovirus
LE cells were transduced with retroviral vectors
harboring KrasG12V, EGFRex19del, or SV40
Large-T plus selection marker (Puror for
KrasG12V and EGFRex19del, and Neor
for SV40 Large-T construct). Ecotropic viruses were packaged using
PLAT-E cells (a gift from Dr T. Kitamura, Tokyo University)
(11) and Fugene 6 transfection
reagent. Infection of LE cells in 3D culture with retroviruses was
performed as described (12).
Animal experiments
All animal experiments were performed after approval
of Miyagi Cancer Center Research Institute Animal Care and Use
committee. In allograft experiments, 4×105 of
LE-LT/KrasG12V cells were injected sc into the
dorsal flank of nude mice. LE-LT/EGFRex19del cells were
injected as 1:1 mixtures with Matrigel at 2×106 cells
per injection site.
Immunostaining analyses and flow
cytometry
Immunohistochemical analyses were performed using
reagents from Roche Ventana systems (Basel, Switzerland).
Antibodies against thyroid transcription factor 1 (TTF1) (SP141)
and Ki-67 (30–9) were from Roche. Anti-cytokeratin 14 (CK14) goat
antibody was purchased from Santa Cruz Biotechnology (Dallas, TX,
USA). Anti-prosurfactant protein-C (SPC) antibody (ab40879) was
obtained from Abcam (Cambridge, UK). Anti-EpCAM monoclonal antibody
(2–17-F-1) was obtained from MBL (Nagoya, Japan). Frozen sections
were prepared for anti-EpCAM staining only, as the antibody did not
work on paraffin-embedded sections (data not shown). Flow cytometry
was performed with a FACS Canto-II flow cytometer (BD Bioscences
Inc.).
Statistics
We used Student's t-test (2-tailed) to compare two
groups. A P-value of <0.05 was considered significant. Data are
presented as means with the SD (Fig. 2B
and D) or SEM (all others).
Results
Isolation and characterization of
primary LE cells
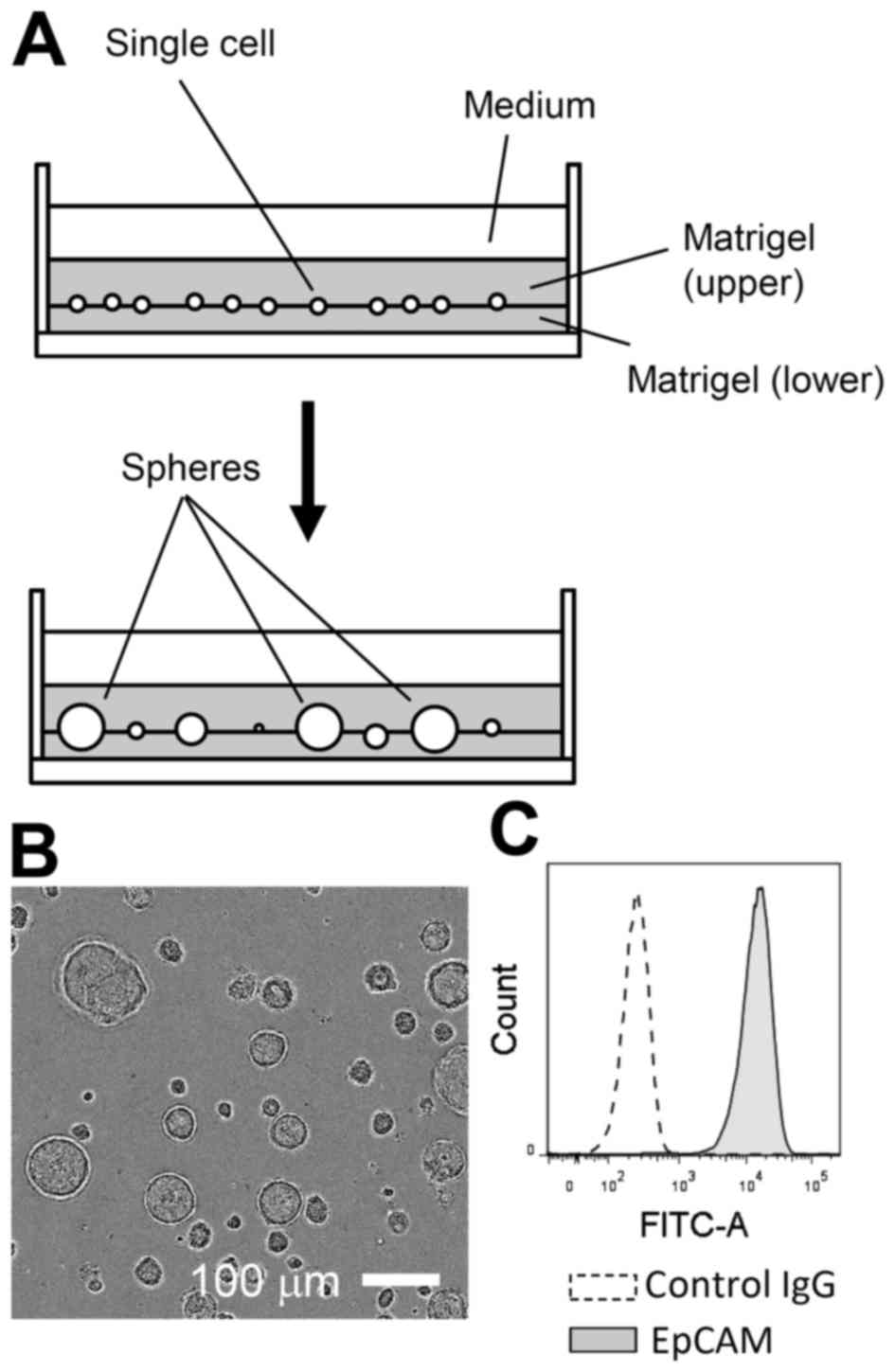
Primary LE cells were isolated from adult mice and
expanded in Matrigel-assisted 3D culture for 2 to 3 weeks in the
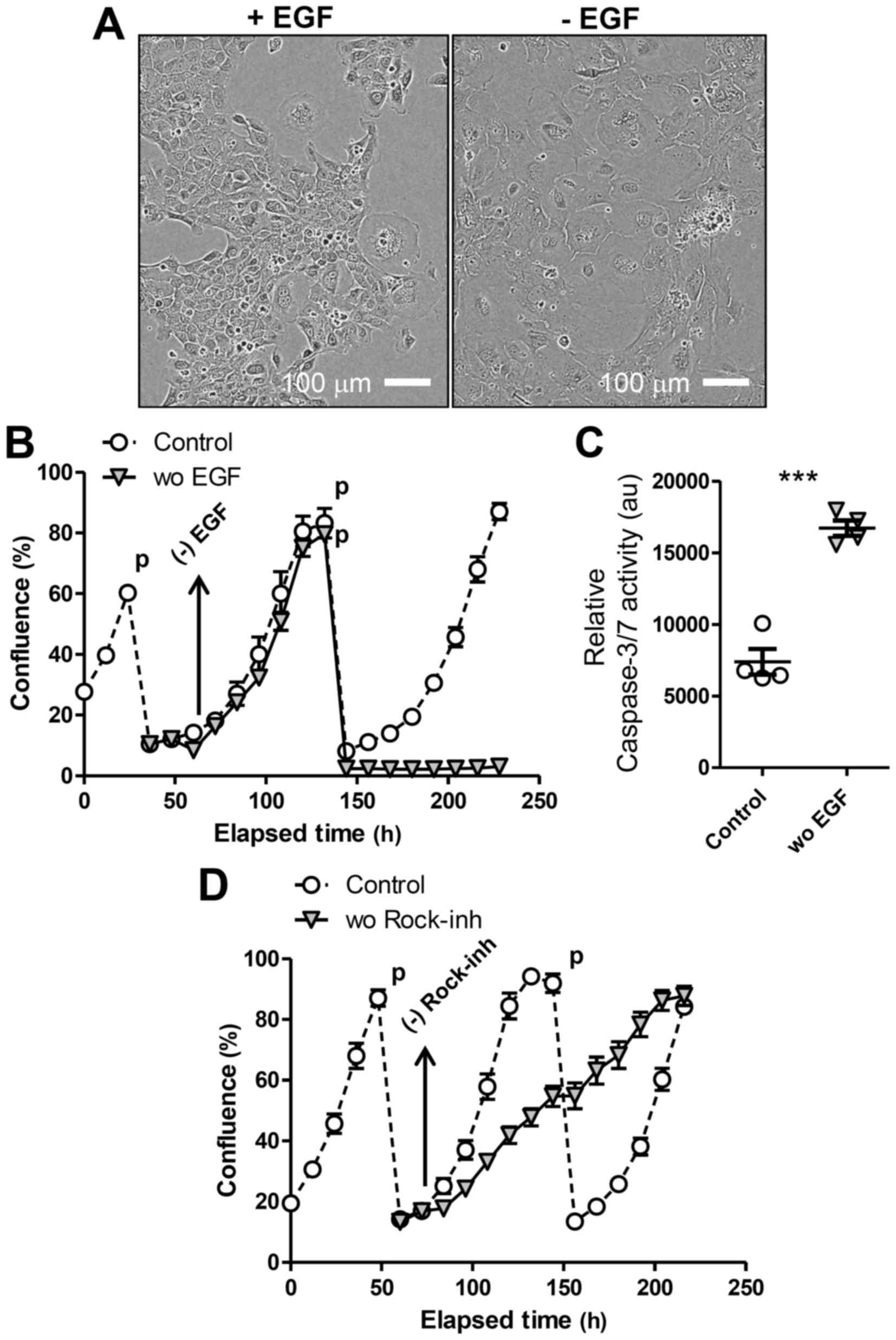
presence of EGF (Fig. 1A and B).
During this period, non-epithelial cells, such as fibroblasts, were
likely lost or selected against, as nearly all expanded cells were
positive for EpCAM, an established marker of the epithelial lineage
(Fig. 1C) (13). In 3D culture, LE cells could be
propagated, for at least two to three months, with passage once
weekly at an approximately 1:4 dilution.
To test EGF-dependence of LE cells, we transferred
cells to 2D culture, as Matrigel used for 3D culture contains
substantial amounts of EGF (14).
Then, we prepared single-cell suspensions from LE spheres formed in
3D culture and seeded them in tissue-culture plates in the presence
of EGF and the Rock-inhibitor Y-27632. Cells adapted to 2D culture
(Fig. 2A) and were passaged (1:5
dilution) several times, always replacing EGF and Y-27632 in the
media (‘Control’ in Fig. 2B and C).
We confirmed that LE cells passaged in 2D were still
EpCAM-positive. In addition, the 2D-cultured cells as monolayer
were converted into 3D-spheres again when re-transferred on
Matrigel. To evaluate the requirement for either of these factors,
we prepared replicates of culture when cells were passaged and then
withdrew EGF or Y-27632 from the media the next day. Upon removing
EGF, LE cells in 2D culture ceased proliferating and became more
flattened and enlarged (Fig. 2A),
exhibiting the morphology of senescent cells, and died after the
next passage (Fig. 2B). Upon the
passage, the LE cells deprived of EGF showed markedly higher
activity of caspase, suggesting apoptosis induction, compared to
control cells (Fig. 2C). We also
observed that the presence of both EGF and the Rock-inhibitor was
required for maximal LE cell proliferation in 2D culture (Fig. 2D), as Y-27632 withdrawal immediately
decreased cell proliferation. These results overall indicate that
LE cells expanded in 3D culture over a long period maintain their
requirement for EGF to proliferate.
LE cell transformation requires both
oncogene activation and tumor-suppressor loss
We next transformed LE cells by transduction with
SV40 Large-T and the active Kras-mutant KrasG12V, either
singly or together, using retroviral vectors harboring
Neor and Puror, respectively,
as selection marker. Transduced cells were then drug-selected and
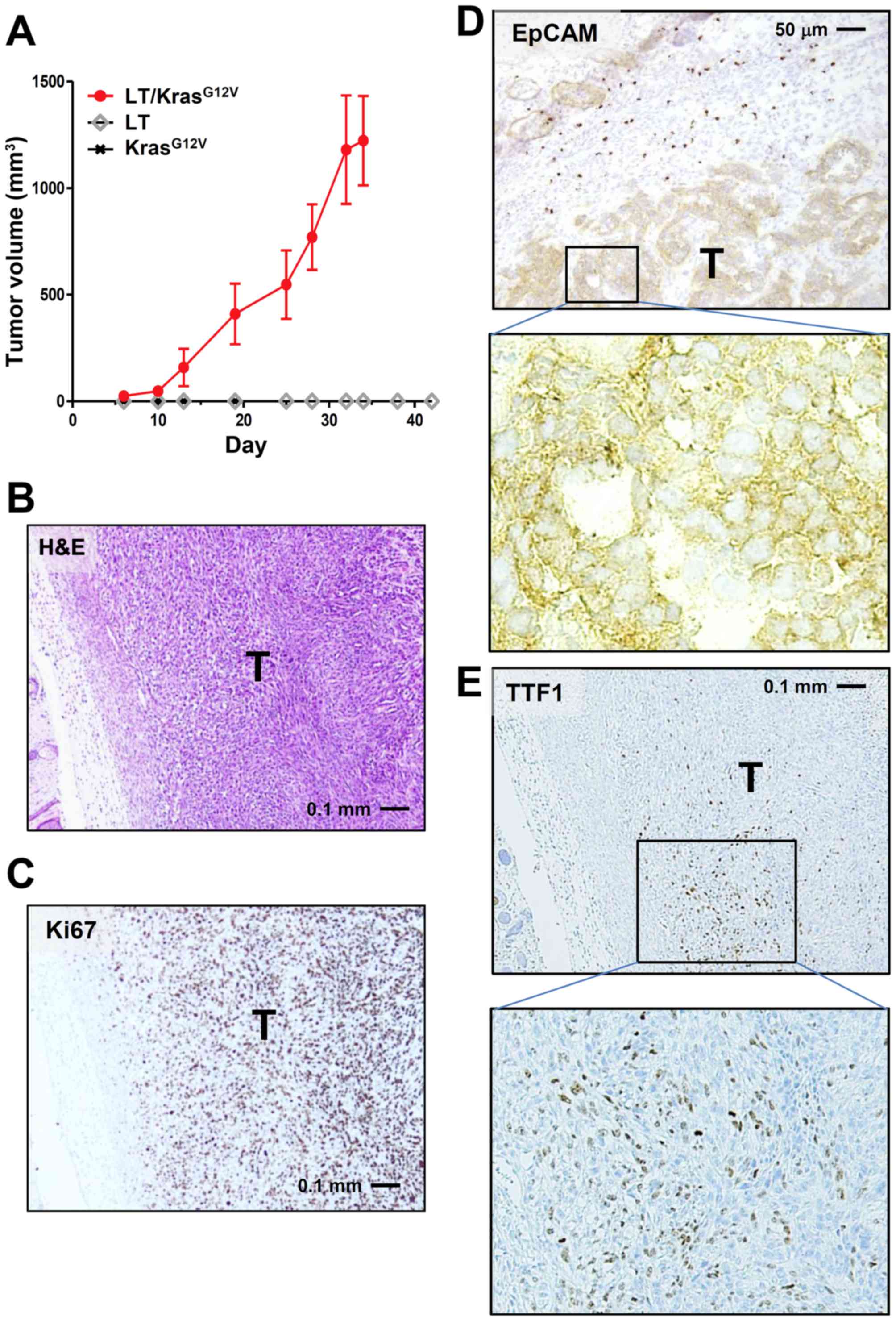
nude mice were inoculated with drug-resistant cells. Only LE cells
receiving both Large-T and KrasG12V
(LE-LT/KrasG12V cells) formed allograft tumors, while
cells harboring either Large-T or KrasG12V alone did not
(Fig. 3A and B). These results are
consistent with previous observations that cellular transformation
requires both oncogene activation and inactivation of tumor
suppressors such as p53 and pRb (15).
Immunostaining of LE-LT/KrasG12V tumor
showed that the Ki-67 index of the tumor was very high (~85%;
Fig. 3C) and that tumor cells arising
from LE cells were mostly EpCAM-positive (Fig. 3D). In addition, a part of tumor cells
were positive for TTF1, an established marker of pulmonary
adenocarcinoma (Fig. 3E). No staining
in tumors was observed for SP-C and CK14, respective markers of
lung adenocarcinoma and squamous-cell carcinoma (data not shown).
Together with H&E analysis (Fig.
3B), these results suggest that tumors derived from
LE-LT/KrasG12V cells are largely anaplastic (expressing
neither adenocarcinoma marker nor squamous-cell carcinoma marker)
and that a subpopulation of tumor cells tends to differentiate into
adenocarcinoma.
In addition to KrasG12V, we also
transformed LE cells with EGFRex19del, the
constitutively active mutant of EGFR, in combination with Large-T.
Like the LE-LT/KrasG12V cells, the
LE-LT/EGFRex19del cells formed tumors when transplanted
into nude mice (data not shown).
Oncogene addiction of transformed LE
cells
It is well known that tumorigenicity due to
KrasG12V activity requires activation of the classical
mitogen-activated protein kinase (MAPK) cascade, in which MAPK/ERK
kinase (MEK) phosphorylates and activates extracellular regulated
kinase (Erk) (16). Proliferation of
LE-LT/KrasG12V cells in vitro was independent of
exogenous EGF (data not shown), which activates the MEK-ERK
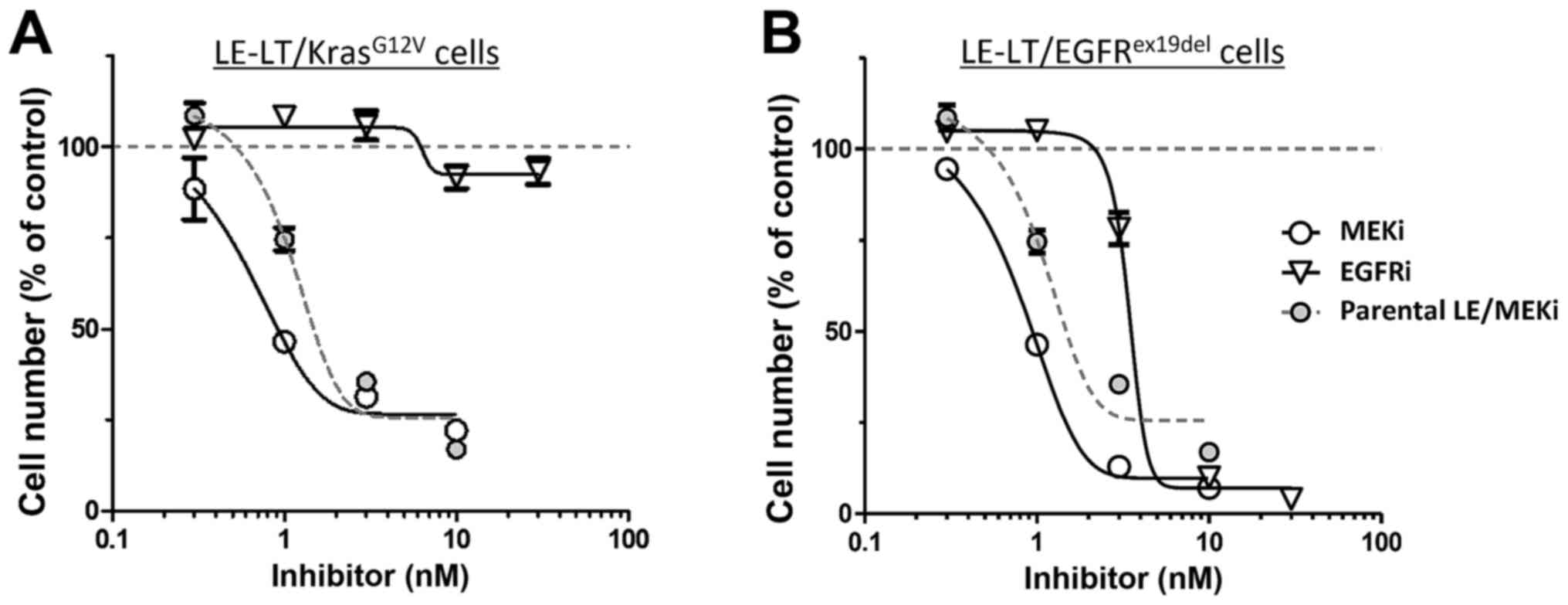
pathway. We then tested the sensitivity of
LE-LT/KrasG12V cells in vitro to trametinib, a
MEK1-inhibitor. As shown in Fig. 4A,
proliferation of LE-LT/KrasG12V cells markedly decreased
following trametinib treatment, whereas comparable treatment with
erlotinib, which potently inhibits EGFR kinase activity, had no
effect. In contrast, both trametinib and erlotinib treatment
inhibited LE-LT/EGFRex19del cell proliferation (Fig. 4B). These results are consistent with
the idea that Kras activates MAPK signaling downstream of EGFR,
making cells resistant to EGFR inhibition and that transformation
by EGFRex19del requires its kinase activity and
downstream MAPK signaling (17).
Overall, these results show that transformed LE cells are addicted
to oncogenic Kras- or EGFR-stimulated signaling.
Discussion
Use of 3D-culture has become a powerful tool for
development of tissue engineering methods for regenerative medicine
and to understand cancer biology (6,18).
Relevant to the latter, in colorectal cancer research, use of 3D
culture has revealed the identity of cancer cells of origin, the
function of Yap-dependent regenerative signaling in cancer
initiation, and how niche factor requirements are lost during colon
cancer progression (19,20). Given the utility of 3D culture in
expanding normal epithelial cells, we used this method here to
isolate, expand, and transform mouse LE cells.
While our method could be applied to numerous
experimental settings, one of particular interest is analysis of
cells from genetically-engineered mice in several ways. For
example, although several sophisticated mouse models of NSCLC have
been established in the last decade (4), it is time-consuming to obtain mice
carrying the multiple mutant alleles required for analyses, often
requiring a year or more. Our method yields either transformed LE
cells or NSCLC-like allograft tumors normally within 2 or 3 months,
respectively.
Pharmacological experiments using MEK and/or EGFR
inhibitors show that proliferation of transformed LE cells is
highly dependent on signaling by the transduced oncogene. Thus, our
system could be useful to identify novel chemo-preventatives and
therapeutics and to test their efficacy in vitro or in
transplantation models. Also, allograft models using syngeneic
strains will enable experiments in immune-competent mice, providing
a unique opportunity to explore novel immunotherapy approaches and
pre-clinical models for testing their efficacy (21).
Tumors arising from transformed mouse LE cells are
largely anaplastic. It may be possible to create more
differentiated tumors using different combination(s) of oncogenes
and tumor-suppressor inhibitions. Alternatively, more
differentiated tumors may be achieved using environmental cues, as
it is well known that specific culture conditions are critical to
promote induction of specific lineages from stem cells (22). To date, several cell types found in
the pulmonary system, such as alveolar type 2 cells, club cells,
and tracheobronchial basal cells, have been reported as NSCLC cells
of origin (13,23). Accordingly, detailed characterization
such as comprehensive marker and transcriptome analyses are needed
to accurately define identities of LE cells cultured long-term.
In summary, we established an ex vivo method
to recapitulate tumorigenesis of mouse LE cells. Our experimental
system provides a unique opportunity to study lung tumorigenesis
and to develop novel therapeutics against NSCLC. Characterization
of long-term expanded LE cells also has implications for
regenerative approaches to lung disease, such as chronic
obstructive pulmonary disease (24).
Acknowledgements
We would like to acknowledge Dr C. Counter and Dr R.
Weinberg for providing plasmid constructs. Thanks are also due to
members of the Pathology facility of Miyagi Cancer Center Hospital
for technical help in tissue analyses, and to Y. Chiba for
secretarial assistance. The present study was supported by grants
from JSPS KAKENHI grants (16K14621) to N. T., 26430130 to H. S.,
16K07187 to S. I., 15K14387 to I. S., and 16K10486 to K. M.), and
to the Takeda Foundation (N. T.), the Mochida Memorial Foundation
for Medical and Pharmaceutical Research (N. T.), the Kato Memorial
Bioscience Foundation (N. T.), the Uehara Memorial Foundation (N.
T.) and the Sagawa Foundation for Promotion of Cancer Research (N.
T.).
References
|
1
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sundar R, Cho BC, Brahmer JR and Soo RA:
Nivolumab in NSCLC: Latest evidence and clinical potential. Ther
Adv Med Oncol. 7:85–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
DuPage M, Dooley AL and Jacks T:
Conditional mouse lung cancer models using adenoviral or lentiviral
delivery of Cre recombinase. Nat Protoc. 4:1064–1072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shamir ER and Ewald AJ: Three-dimensional
organotypic culture: Experimental models of mammalian biology and
disease. Nat Rev Mol Cell Biol. 15:647–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clevers H: Modeling development and
disease with organoids. Cell. 165:1586–1597. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Watanuki Z, Kosai H, Osanai N, Ogama N,
Mochizuki M, Tamai K, Yamaguchi K, Satoh K, Fukuhara T, Maemondo M,
et al: Synergistic cytotoxicity of afatinib and cetuximab against
EGFR T790M involves Rab11-dependent EGFR recycling. Biochem Biophys
Res Commun. 455:269–276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lampson BL, Pershing NL, Prinz JA, Lacsina
JR, Marzluff WF, Nicchitta CV, MacAlpine DM and Counter CM: Rare
codons regulate KRas oncogenesis. Curr Biol. 23:70–75. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hahn WC, Dessain SK, Brooks MW, King JE,
Elenbaas B, Sabatini DM, DeCaprio JA and Weinberg RA: Enumeration
of the simian virus 40 early region elements necessary for human
cell transformation. Mol Cell Biol. 22:2111–2123. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Onuma K, Ochiai M, Orihashi K, Takahashi
M, Imai T, Nakagama H and Hippo Y: Genetic reconstitution of
tumorigenesis in primary intestinal cells. Proc Natl Acad Sci USA.
110:pp. 11127–11132. 2013, View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morita S, Kojima T and Kitamura T: Plat-E:
An efficient and stable system for transient packaging of
retroviruses. Gene Ther. 7:1063–1066. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tanuma N, Nomura M, Ikeda M, Kasugai I,
Tsubaki Y, Takagaki K, Kawamura T, Yamashita Y, Sato I, Sato M, et
al: Protein phosphatase Dusp26 associates with KIF3 motor and
promotes N-cadherin-mediated cell-cell adhesion. Oncogene.
28:752–761. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Asselin-Labat ML and Filby CE: Adult lung
stem cells and their contribution to lung tumourigenesis. Open
Biol. 2:1200942012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hughes CS, Postovit LM and Lajoie GA:
Matrigel: A complex protein mixture required for optimal growth of
cell culture. Proteomics. 10:1886–1890. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Katz M, Amit I and Yarden Y: Regulation of
MAPKs by growth factors and receptor tyrosine kinases. Biochim
Biophys Acta. 1773:1161–1176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sharifnia T, Rusu V, Piccioni F, Bagul M,
Imielinski M, Cherniack AD, Pedamallu CS, Wong B, Wilson FH,
Garraway LA, et al: Genetic modifiers of EGFR dependence in
non-small cell lung cancer. Proc Natl Acad Sci USA. 111:pp.
18661–18666. 2014, View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fatehullah A, Tan SH and Barker N:
Organoids as an in vitro model of human development and disease.
Nat Cell Biol. 18:246–254. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gregorieff A, Liu Y, Inanlou MR, Khomchuk
Y and Wrana JL: Yap-dependent reprogramming of Lgr5(+) stem cells
drives intestinal regeneration and cancer. Nature. 526:715–718.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fujii M, Shimokawa M, Date S, Takano A,
Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, et
al: A colorectal tumor organoid library demonstrates progressive
loss of niche factor requirements during tumorigenesis. Cell Stem
Cell. 18:827–838. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Akbay EA, Koyama S, Carretero J, Altabef
A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp
EM, Pugh TJ, et al: Activation of the PD-1 pathway contributes to
immune escape in EGFR-driven lung tumors. Cancer Discov.
3:1355–1363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang SX, Islam MN, O'Neill J, Hu Z, Yang
YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J
and Snoeck HW: Efficient generation of lung and airway epithelial
cells from human pluripotent stem cells. Nat Biotechnol. 32:84–91.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Desai TJ, Brownfield DG and Krasnow MA:
Alveolar progenitor and stem cells in lung development, renewal and
cancer. Nature. 507:190–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kotton DN and Morrisey EE: Lung
regeneration: Mechanisms, applications, and emerging stem cell
populations. Nat Med. 20:822–832. 2014. View Article : Google Scholar : PubMed/NCBI
|