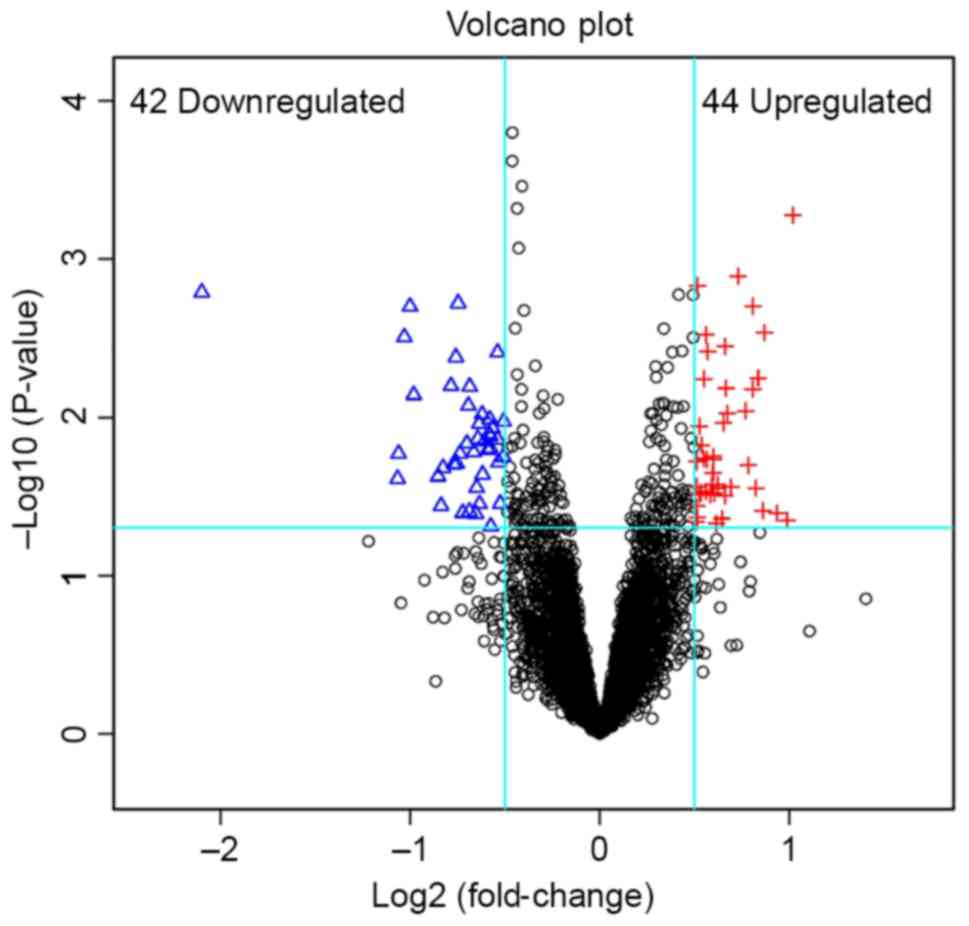
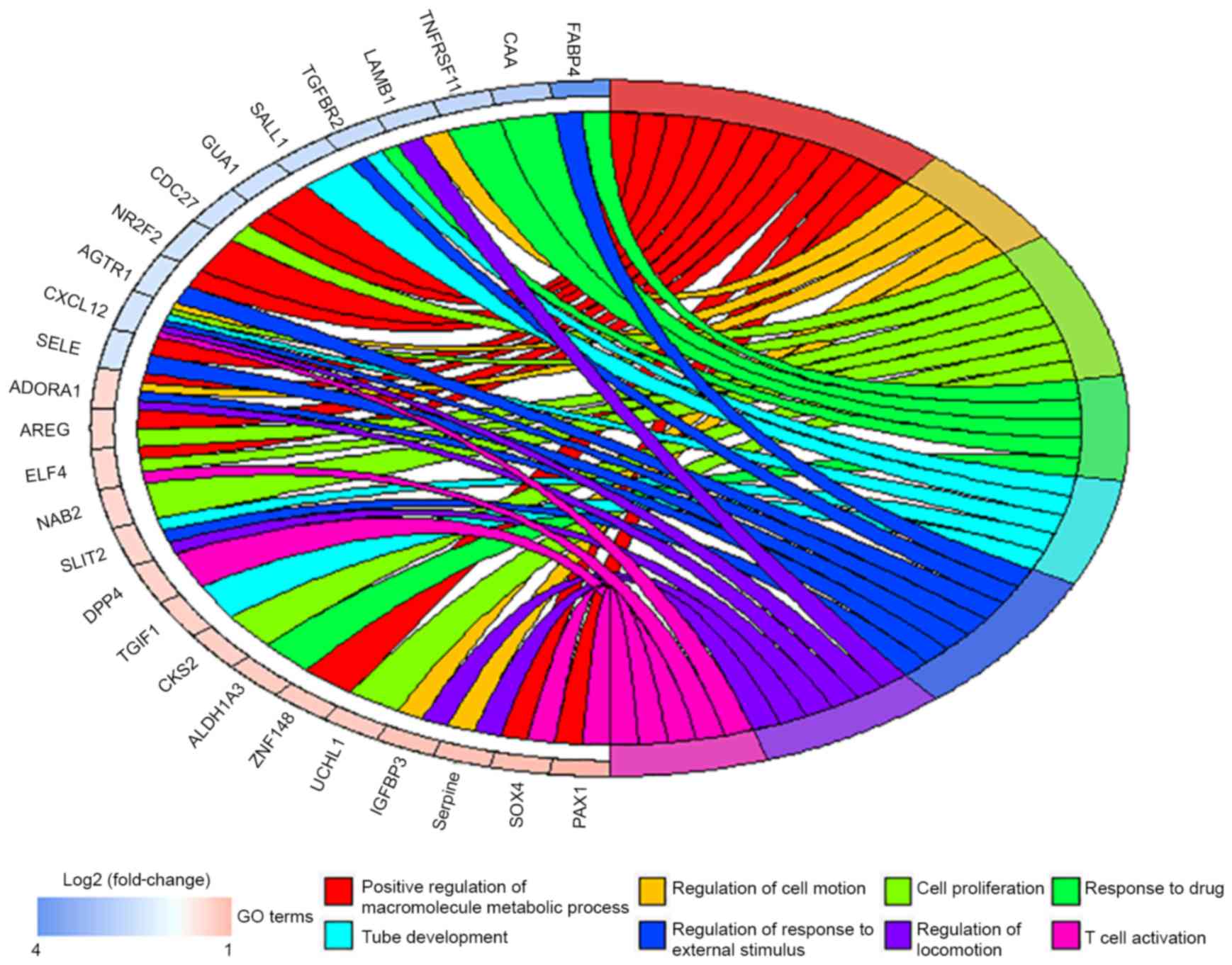
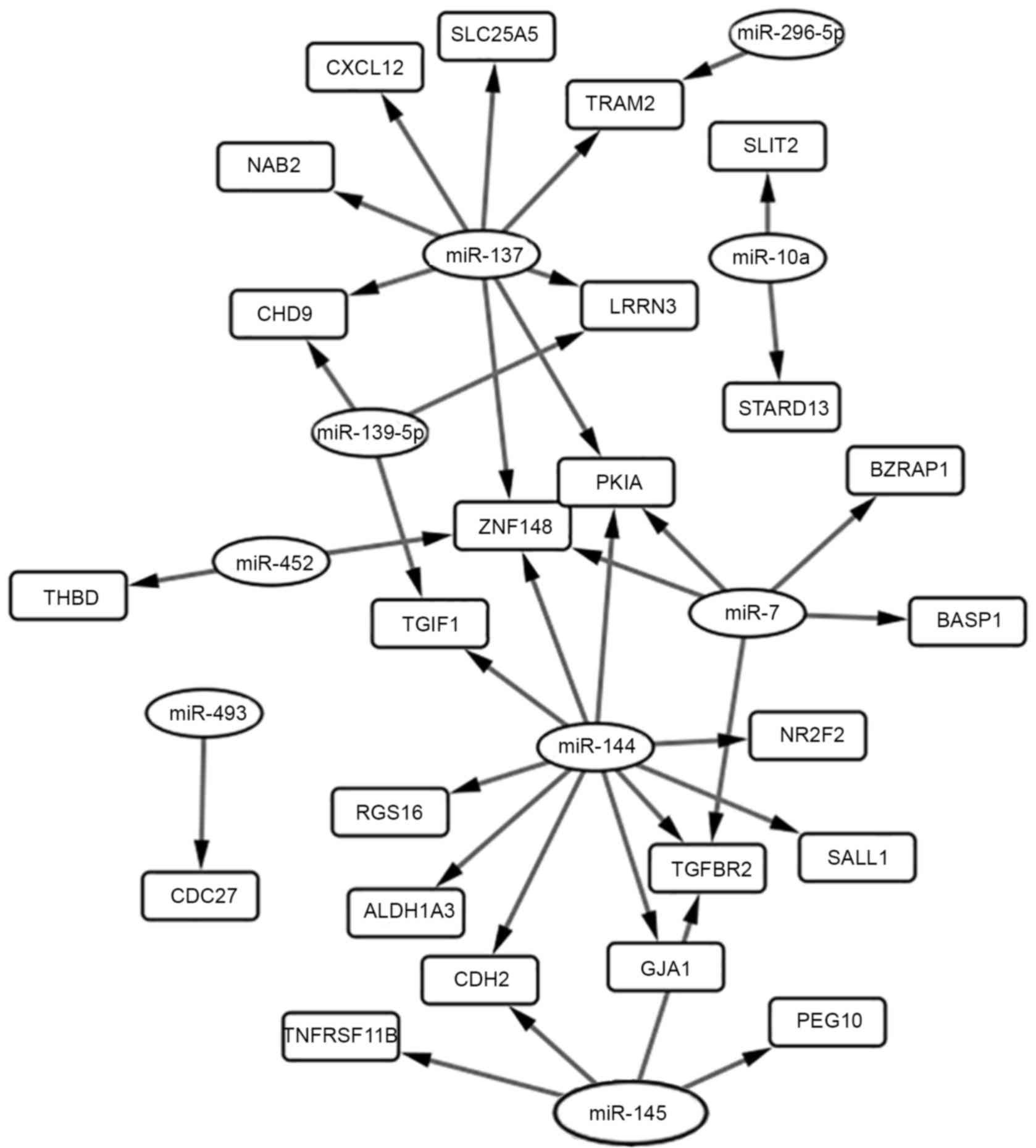
|
1
|
Kondo T, Ezzat S and Asa SL: Pathogenetic
mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer.
6:292–306. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Enewold L, Zhu K, Ron E, Marrogi AJ,
Stojadinovic A, Peoples GE and Devesa SS: Rising thyroid cancer
incidence in the United States by demographic and tumor
characteristics, 1980–2005. Cancer Epidemiol Biomarkers Prev.
18:784–791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McHenry CR and Phitayakorn R: Follicular
adenoma and carcinoma of the thyroid gland. Oncologist. 16:585–593.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Francis GL, Waguespack SG, Bauer AJ,
Angelos P, Benvenga S, Cerutti JM, Dinauer CA, Hamilton J, Hay ID,
Luster M, et al: Management guidelines for children with thyroid
nodules and differentiated thyroid cancer. Thyroid. 25:716–759.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu C, Chen WP and Wang XH: MicroRNA in
osteoarthritis. J Int Med Res. 39:1–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang C, Wan S, Yang T, Niu D, Zhang A,
Yang C, Cai J, Wu J, Song J, Zhang CY, et al: Increased serum
microRNAs are closely associated with the presence of microvascular
complications in type 2 diabetes mellitus. Sci Rep. 6:200322016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chiang HR, Schoenfeld LW, Ruby JG, Auyeung
VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al:
Mammalian microRNAs: Experimental evaluation of novel and
previously annotated genes. Genes Dev. 24:992–1009. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu
Y, Chen Y, Xu L, Zen K, Zhang C and Shen H: Serum microRNA
signatures identified in a genome-wide serum microRNA expression
profiling predict survival of non-small-cell lung cancer. J Clin
Oncol. 28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Finley DJ, Zhu B, Barden CB and Fahey TJ
III: Discrimination of benign and malignant thyroid nodules by
molecular profiling. Ann Surg. 240:425–437. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barden CB, Shister KW, Zhu B, Guiter G,
Greenblatt DY, Zeiger MA and Fahey TJ III: Classification of
follicular thyroid tumors by molecular signature: Results of gene
profiling. Clin Cancer Res. 9:1792–1800. 2003.PubMed/NCBI
|
|
17
|
Aldred MA, Huang Y, Liyanarachchi S,
Pellegata NS, Gimm O, Jhiang S, Davuluri RV, de la Chapelle A and
Eng C: Papillary and follicular thyroid carcinomas show distinctly
different microarray expression profiles and can be distinguished
by a minimum of five genes. J Clin Oncol. 22:3531–3539. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
de la Chapelle A and Jazdzewski K:
MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 96:3326–3336.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stokowy T, Wojtaś B, Krajewska J,
Stobiecka E, Dralle H, Musholt T, Hauptmann S, Lange D, Hegedüs L,
Jarząb B, et al: A two miRNA classifier differentiates follicular
thyroid carcinomas from follicular thyroid adenomas. Mol Cell
Endocrinol. 399:43–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ak G, Tomaszek SC, Kosari F, Metintas M,
Jett JR, Metintas S, Yildirim H, Dundar E, Dong J, Aubry MC, Wigle
DA and Thomas CF Jr: MicroRNA and mRNA features of malignant
pleural mesothelioma and benign asbestos-related pleural effusion.
Biomed Res Int. 635748:2015.
|
|
22
|
Liu MY, Zhang H, Hu YJ, Chen YW and Zhao
XN: Identification of key genes associated with cervical cancer by
comprehensive analysis of transcriptome microarray and methylation
microarray. Oncol Lett. 12:473–478. 2016.PubMed/NCBI
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:(Database issue). D105–D110.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu PF, Jiang WH, Han YT, He LF, Zhang HL
and Ren H: Integrated microRNA-mRNA analysis of pancreatic ductal
adenocarcinoma. Genet Mol Res. 14:10288–10297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yeung N, Cline MS, Kuchinsky A, Smoot ME
and Bader GD: Exploring biological networks with Cytoscape
software. Curr Protoc Bioinformatics Chapter. 8:Unit 8.13. 2008.
View Article : Google Scholar
|
|
28
|
Stokowy T, Wojtas B, Jarzab B, Krohn K,
Fredman D, Dralle H, Musholt T, Hauptmann S, Lange D, Hegedüs L, et
al: Two-miRNA classifiers differentiate mutation-negative
follicular thyroid carcinomas and follicular thyroid adenomas in
fine needle aspirations with high specificity. Endocrine.
54:440–447. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cerutti JM, Delcelo R, Amadei MJ,
Nakabashi C, Maciel RM, Peterson B, Shoemaker J and Riggins GJ: A
preoperative diagnostic test that distinguishes benign from
malignant thyroid carcinoma based on gene expression. J Clin
Invest. 113:1234–1242. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Umbricht CB, Conrad GT, Clark DP, Westra
WH, Smith DC, Zahurak M, Saji M, Smallridge RC, Goodman S and
Zeiger MA: Human telomerase reverse transcriptase gene expression
and the surgical management of suspicious thyroid tumors. Clin
Cancer Res. 10:5762–5768. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weber F, Shen L, Aldred MA, Morrison CD,
Frilling A, Saji M, Schuppert F, Broelsch CE, Ringel MD and Eng C:
Genetic classification of benign and malignant thyroid follicular
neoplasia based on a three-gene combination. J Clin Endocrinol
Metab. 90:2512–2521. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Weber F, Teresi RE, Broelsch CE, Frilling
A and Eng C: A limited set of human MicroRNA is deregulated in
follicular thyroid carcinoma. J Clin Endocrinol Metab.
91:3584–3591. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He H, Jazdzewski K, Li W, Liyanarachchi S,
Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al:
The role of microRNA genes in papillary thyroid carcinoma. Proc
Natl Acad Sci USA. 102:pp. 19075–19080. 2005, View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma.
Hepatology. 55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang L, Liu X, Chen Z, Jin Y, Heidbreder
CE, Kolokythas A, Wang A, Dai Y and Zhou X: MicroRNA-7 targets
IGF1R (insulin-like growth factor 1 receptor) in tongue squamous
cell carcinoma cells. Biochem J. 432:199–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Reddy SD, Ohshiro K, Rayala SK and Kumar
R: MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase
1 and regulates its functions. Cancer Res. 68:8195–8200. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fang L, Ellims AH, Moore XL, White DA,
Taylor AJ, Chin-Dusting J and Dart AM: Circulating microRNAs as
biomarkers for diffuse myocardial fibrosis in patients with
hypertrophic cardiomyopathy. J Transl Med. 13:3142015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lopez-Bertoni H, Lal B, Michelson N,
Guerrero-Cázares H, Quiñones-Hinojosa A, Li Y and Laterra J:
Epigenetic modulation of a miR-296-5p:HMGA1 axis regulates Sox2
expression and glioblastoma stem cells. Oncogene. 35:4903–4913.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Santarpia L, Calin GA, Adam L, Ye L, Fusco
A, Giunti S, Thaller C, Paladini L, Zhang X, Jimenez C, et al: A
miRNA signature associated with human metastatic medullary thyroid
carcinoma. Endocr Relat Cancer. 20:809–823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hudson J, Duncavage E, Tamburrino A,
Salerno P, Xi L, Raffeld M, Moley J and Chernock RD: Overexpression
of miR-10a and miR-375 and downregulation of YAP1 in medullary
thyroid carcinoma. Exp Mol Pathol. 95:62–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guan H, Liang W, Xie Z, Li H, Liu J, Liu
L, Xiu L and Li Y: Down-regulation of miR-144 promotes thyroid
cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine.
48:566–574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shomron N: MicroRNAs and pharmacogenomics.
Pharmacogenomics. 11:629–632. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang HD, Jiang LH, Sun DW, Li J and Tang
JH: MiR-139-5p: Promising biomarker for cancer. Tumour Biol.
36:1355–1365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kent OA, McCall MN, Cornish TC and
Halushka MK: Lessons from miR-143/145: The importance of cell-type
localization of miRNAs. Nucleic Acids Res. 42:7528–7538. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kobayashi K, Imazu Y, Kawabata M and
Shohmori T: Effect of long-term storage on monoamine metabolite
levels in human cerebrospinal fluid. Acta Med Okayama. 41:179–181.
1987.PubMed/NCBI
|
|
49
|
Jia X, Li N, Peng C, Deng Y, Wang J, Deng
M, Lu M, Yin J, Zheng G, Liu H and He Z: miR-493 mediated DKK1
down-regulation confers proliferation, invasion and
chemo-resistance in gastric cancer cells. Oncotarget. 7:7044–7054.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tommerup N and Vissing H: Isolation and
fine mapping of 16 novel human zinc finger-encoding cDNAs identify
putative candidate genes for developmental and malignant disorders.
Genomics. 27:259–264. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
de Celis JF and Barrio R: Regulation and
function of Spalt proteins during animal development. Int J Dev
Biol. 53:1385–1398. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
May CD, Sphyris N, Evans KW, Werden SJ,
Guo W and Mani SA: Epithelial-mesenchymal transition and cancer
stem cells: A dangerously dynamic duo in breast cancer progression.
Breast Cancer Res. 13:2022011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Link LA, Howley BV, Hussey GS and Howe PH:
PCBP1/HNRNP E1 protects chromosomal integrity by translational
regulation of CDC27. Mol Cancer Res. 14:634–646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Thornton BR, Ng TM, Matyskiela ME, Carroll
CW, Morgan DO and Toczyski DP: An architectural map of the
anaphase-promoting complex. Genes Dev. 20:449–460. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lee SJ and Langhans SA: Anaphase-promoting
complex/cyclosome protein Cdc27 is a target for curcumin-induced
cell cycle arrest and apoptosis. BMC Cancer. 12:442012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Browning BL and Browning SR: Haplotypic
analysis of wellcome trust case control consortium data. Hum Genet.
123:273–280. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Huggins GS, Bacani CJ, Boltax J, Aikawa R
and Leiden JM: Friend of GATA 2 physically interacts with chicken
ovalbumin upstream promoter-TF2 (COUP-TF2) and COUP-TF3 and
represses COUP-TF2-dependent activation of the atrial natriuretic
factor promoter. J Biol Chem. 276:28029–28036. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Song HM, Luo Y, Li DF, Wei CK, Hua KY,
Song JL, Xu H, Maskey N and Fang L: MicroRNA-96 plays an oncogenic
role by targeting FOXO1 and regulating AKT/FOXO1/Bim pathway in
papillary thyroid carcinoma cells. Int J Clin Exp Pathol.
8:9889–9900. 2015.PubMed/NCBI
|
|
59
|
Chruścik A and Lam AK: Clinical
pathological impacts of microRNAs in papillary thyroid carcinoma: A
crucial review. Exp Mol Pathol. 99:393–398. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Nicholson KM and Anderson NG: The protein
kinase B/Akt signalling pathway in human malignancy. Cell Signal.
14:381–395. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xue G and Hemmings BA: PKB/Akt-dependent
regulation of cell motility. J Natl Cancer Inst. 105:393–404. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Osaki M, Oshimura M and Ito H: PI3K-Akt
pathway: Its functions and alterations in human cancer. Apoptosis.
9:667–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Manzella L, Stella S, Pennisi MS, Tirrò E,
Massimino M, Romano C, Puma A, Tavarelli M and Vigneri P: New
insights in thyroid cancer and p53 family proteins. Int J Mol Sci.
18:pii: E1325. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhu X, Zhao L, Park JW, Willingham MC and
Cheng SY: Synergistic signaling of KRAS and thyroid hormone
receptor β mutants promotes undifferentiated thyroid cancer through
MYC up-regulation. Neoplasia. 16:757–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kataki A, Sotirianakos S, Memos N,
Karayiannis M, Messaris E, Leandros E, Manouras A and Androulakis
G: P53 and C-FOS overexpression in patients with thyroid cancer: An
immunohistochemical study. Neoplasma. 50:26–30. 2003.PubMed/NCBI
|
|
66
|
Aliyev A, Soundararajan S, Bucak E, Gupta
M, Hatipoglu B, Nasr C, Siperstein A and Berber E: The utility of
peripheral thyrotropin receptor mRNA in the management of
differentiated thyroid cancer. Surgery. 158:1089–1094. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Huang FJ, Zhou XY, Ye L, Fei XC, Wang S,
Wang W and Ning G: Follicular thyroid carcinoma but not adenoma
recruits tumor-associated macrophages by releasing CCL15. BMC
Cancer. 16:982016. View Article : Google Scholar : PubMed/NCBI
|