Introduction
Hepatocellular carcinoma (HCC) is the most common
type of liver cancer associated with a poor clinical outcome
(1). Cancer stem cells (CSCs) have
been identified in a number of solid tumors, including HCC
(2–4).
CSCs may lead to recurrence, drug-resistance and tumor formation in
HCC (5). Cluster of differentiation
(CD)133 is a membrane-bound pentaspan glycoprotein that has been
identified as a surface biomarker in a variety of cancer types,
including breast, colon, prostate, pancreatic, lung and liver
carcinoma (2,3,6–11). Although its function remains unclear,
directly targeting CD133 has been demonstrated to be a potentially
effective strategy for eliminating CSCs (11,12). Thus,
drug screens that target CSC markers may improve the efficacy of
therapeutic strategies for the treatment of HCC.
Lidamycin (LDM), an anticancer antibiotic, has been
demonstrated to exhibit distinct antitumor effects in various types
of cancer, including liver, breast, pancreatic, colon, lung,
gastric and brain cancer, as well as in lymphoma and myeloma
(13). Furthermore, LDM is toxic to
multi-drug resistant HCC cells (14).
In our previous study, LDM was revealed to inhibit the expression
of epithelial cell adhesion module (EpCAM) and the population of
CSCs through regulating the glycogen synthase kinase
(GSK)3β/β-catenin signaling pathway (15). Thus, LDM has been demonstrated to be a
potentially effective strategy to target CSCs in HCC. However, the
effect of LDM on CD133 remains unknown.
In the present study, it was hypothesized that LDM
may suppress CD133 expression. To investigate this, the proportion
of CD133+ cells, the expression of CD133 protein and the
sphere formation of sorted CD133+ cells were evaluated
in vitro following LDM treatment. Subsequently, the tumor
volume of enriched CD133+ cells and CD133 expression was
measured in vivo following LDM treatment. Additionally, the
effect of LDM on the Notch signaling pathway was investigated.
Materials and methods
Cell culture
Human hepatocellular carcinoma Huh7 cells (American
Type Culture Collection, Manassas, VA, USA) were cultured in
Dulbecco's modified Eagle's medium, supplemented with 10% fetal
bovine serum and 1% penicillin-streptomycin (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C and 5%
CO2.
Chemicals
LDM was supplied by Professor Lian-fang Jin of the
Institute of Medicinal Biotechnology, Chinese Academy of Medical
Sciences and Peking Union Medical College (Beijing, China), with a
purity of >95.0%. LDM was prepared as previously described
(15). In brief, 1 µM LDM stock
solution in saline was stored at −80°C for in vitro
experiments, and diluted in saline at 5 µg/ml prior to intravenous
injection. N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine
t-butyl ester (DAPT; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
was prepared as a 20 mM stock solution in DMSO, and stored at
−20°C.
Flow cytometry analysis and
sorting
FcR blocking reagent (Miltenyi Biotec GmbH, Bergisch
Gladbach, Germany) was added to 5×105 cells suspended in
PBS and incubated at 4°C for 10 min. Subsequently, cells were
separately stained with phycoerythrin-conjugated anti-human CD133
or anti-human IgG isotype antibodies (R&D Systems, Inc.,
Minneapolis, MN, USA) for 30–40 min in 4°C. IgG isotype was used as
negative control. Ice-cold PBS was used as washing reagent. Flow
cytometry analysis was performed on Accuri™ C6 (BD Biosciences, San
Jose, CA, USA) using CFlow software (FCS3.0; BD Biosciences). Flow
cytometry sorting was conducted using a BD FACSAria™ I (BD
Biosciences).
Sphere formation assay
The sphere formation medium for HCC was created as
previously described (2,15). Sorted CD133+ cells in
sphere formation medium were cultured in ultra-low attachment
24-well plates (Corning Inc., Corning, NY, USA) at a density of
5,000 cells/well. Sphere formation medium with 0.03 or 0.3 nM LDM
was refreshed twice a week. After 7 days of culture, spheres were
counted and images were captured at a magnification of ×200 using a
light microscope (Eclipse TE2000-U; Nikon Corporation, Tokyo,
Japan). The size of the spheres was analyzed using ImageJ software
(version 1.51K; National Institutes of Health, Bethesda, MD,
USA).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total mRNA was extracted from the Huh7 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and purified using acid phenol-chloroform. A SuperScript™
RT-PCR kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used
according to the manufacturer's protocol. RT-qPCR was performed in
triplicate using SYBR® Green reagents (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and CFX96 qPCR system
(Bio-Rad Laboratories, Inc.) under the following conditions: 10 min
at 95°C, followed by 45 cycles of 15 sec at 95°C and 30 sec at
60°C. The Cq values, analyzed by CFX Manager (Bio-Rad Laboratories,
Inc.), were used for calculation of relative expression levels with
the 2−ΔΔCq method (16).
GAPDH was used as the reference gene; all primer sequences are
presented in Table I.
 | Table I.List of polymerase chain reaction
primers. |
Table I.
List of polymerase chain reaction
primers.
| Gene | Species | Forward primer | Reverse primer |
|---|
| Notch1 | Human |
5′-CACTGTGGGCGGGTCC-3′ |
5′-GTTGTATTGGTTCGGCACCAT-3′ |
| Hes1 | Human |
5′-GTCAAGCACCTCCGGAAC-3′ |
5′-CGTTCATGCACTCGCTGA-3′ |
| Hey1 | Human |
5′-TCTGAGCTGAGAAGGCTGGT-3′ |
5′-CGAAATCCCAAACTCCGATA-3′ |
| GAPDH | Human |
5′-TGAAGGTCGGTGTGAACGG-3′ |
5′-CGTGAGTGGAGTCATACTGGAA-3′ |
Western blot analysis
Whole cell lysates were prepared as described
previously (17). Briefly, cells were
washed with ice-cold PBS and lysed with radioimmunoprecipitation
assay lysis buffer (cat. no. 89900; Thermo Fisher Scientific, Inc.)
containing protease inhibitor (Sigma-Aldrich; Merck KGaA). Protein
concentrations were determined using a Bradford assay. Proteins
(~30 µg/lane) were separated by SDS-PAGE (10% gel). Proteins were
transferred onto a polyvinylidene difluoride membrane (Merck KGaA)
blocked with dried non-fat skimmed milk (5% in Tris-buffered saline
containing Tween-20) at room temperature for 2 h. The membranes
were subsequently probed with primary mouse anti-CD133 (1:200
dilution; cat. no. 130-090-422; Miltenyi Biotec GmbH), rabbit
anti-Notch intracellular domain (NICD; 1:500 dilution; cat. no.
4147; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit
anti-NOTCH1 (1:1,000 dilution; cat. no. 3447; Cell Signaling
Technology, Inc.), rabbit anti-Hes1 (1:1,000 dilution; cat. no.
11988; Cell Signaling Technology, Inc.) and mouse anti-β-actin
(1:3,000 dilution; cat. no. A3854; Sigma-Aldrich; Merck KGaA)
antibodies overnight at 4°C. The membranes were then incubated with
horseradish peroxidase (HRP)-linked anti-rabbit secondary antibody
(1:3,000 dilution; cat. no. 7074; Cell Signaling Technology, Inc.)
or HRP-linked anti-mouse secondary antibody (1:3,000 dilution; cat.
no. 7076; Cell Signaling Technology, Inc.) at room temperature for
2 h. Enhanced chemiluminescence (ECL) was performed using ECL
Western Blotting Substrate (cat. no. 32106; Pierce; Thermo Fisher
Scientific, Inc.) and the ChemiImager 5500 imaging system
(ProteinSimple, San Jose, CA, USA), according to the manufacturer's
protocol. The intensity of the bands was analyzed by densitometry
using the ChemiImager AlphaEaseFC™ software version 4.0
(ProteinSimple).
Animal care and ethical statement
Nude mice (n=18, 50:50 male/female, aged 5–6 weeks,
weighing 18–20 g) were obtained from the Chinese Academy of
Military Medical Sciences (Beijing, China). They were housed in a
room with a 12-h light/12-h dark cycle, ad libitum access to
food and water, an ambient temperature of 22°C. This study was
carried out in strict accordance with the recommendations in the
Guide for the Care and Use of Laboratory animals of the Chinese
Academy of Medical Science [permit number, SYXK (Jing) 2010–0013].
The experimental protocols were approved by the Ethics Committee of
the Institute of Medicinal Biotechnology at the Chinese Academy of
Medical Science. To minimize suffering, sodium pentobarbital
anesthesia (100 mg/kg i.p.) was administered prior to surgery. Mice
were sacrificed at 21 days after CO2 injection.
Tumorigenicity assay in nude mice
Following cell separation using a MACS kit (Miltenyi
Biotec GmbH), according to the manufacturer's protocol, enriched
CD133+ Huh7 cells (3×105 cells/mouse) were
subcutaneously injected into each nude mouse. The mice were
randomly separated into three groups (n=6): The control group was
intravenously injected with an equal volume of saline; the LDM
groups were intravenously injected with 0.025 or 0.05 mg/kg LDM.
All treatments were applied in a single injection, 24 h after cell
transplantation. Tumor volume was examined bi-weekly and the mice
were sacrificed 21 days after cell injection. Tumors were measured
with a Vernier caliper and the volume was calculated using the
following equation: V = 1/2[(width2) × length].
Statistical analysis
The flow cytometry data are presented as the
arithmetic mean. Other data are presented as the arithmetic mean ±
standard deviation, with all experiments repeated in triplicate.
The data for LDM regarding the NOTCH1, NICD, Hes1 and Hey1 genes
and protein levels were compared with the two independent-samples
t-test. One-way analysis of variance was used for comparisons of
the data when there were >2 groups, using SPSS software (version
21.0; IBM Corp., Armonk, NY, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
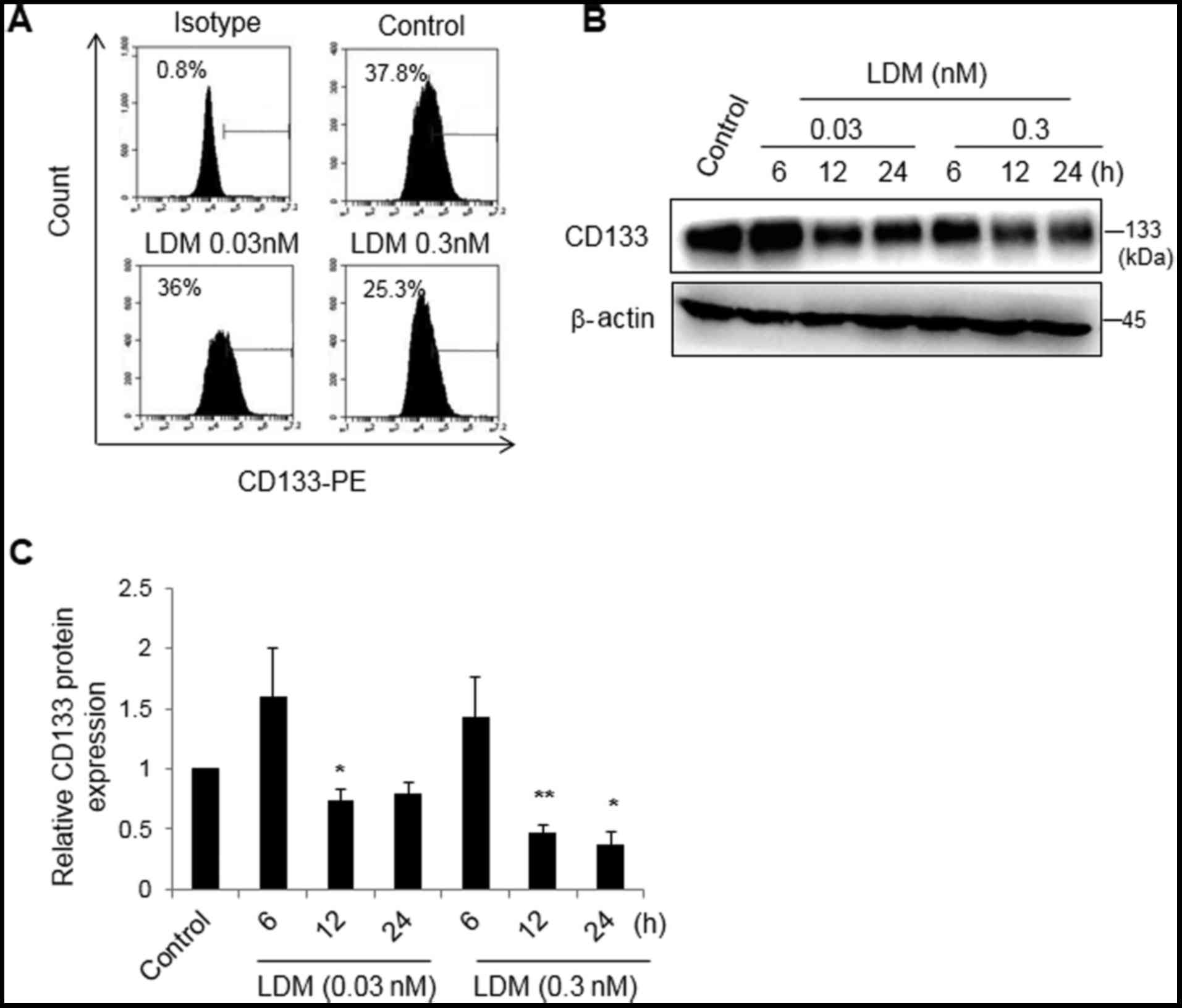
LDM decreases CD133 expression in
vitro
CD133 is considered to be a surface marker of CSCs
in HCC (2,4). As aforementioned, the IC50 of
LDM was 0.3±0.1 nM in the Huh7 cells treated for 48 h (data not
shown). Thus, 0.03 and 0.3 nM LDM was used in vitro. Using a
flow cytometry assay, it was revealed that 0.3 nM LDM decreased the
population of CD133+ cells (25.2±0.3%) compared with the
control group (37.8±0.4%) (P<0.05), while there was no marked
difference between 0.03 nM LDM (36.0±0.3%) and the control
(Fig. 1A). Following treatment with
0.03 or 0.3 nM LDM for 6, 12 or 24 h, proteins were collected for
western blot analysis. The relative densitometry measurements of
bands in 0.03 nM LDM for 12 h and 0.3 nM LDM for 12 and 24 h
(0.7±0.1, 0.5±0.1 and 0.3±0.1, respectively) were lower than that
of the control group. The results demonstrated that LDM
significantly decreased the expression of CD133 protein in a dose-
and time-dependent manner (P<0.05). At 6 h, there was no marked
difference between the LDM-treated groups and the control group
(Fig. 1B and C). The data suggested
that LDM may inhibit the formation of a population of
CD133+ cells and CD133 protein expression.
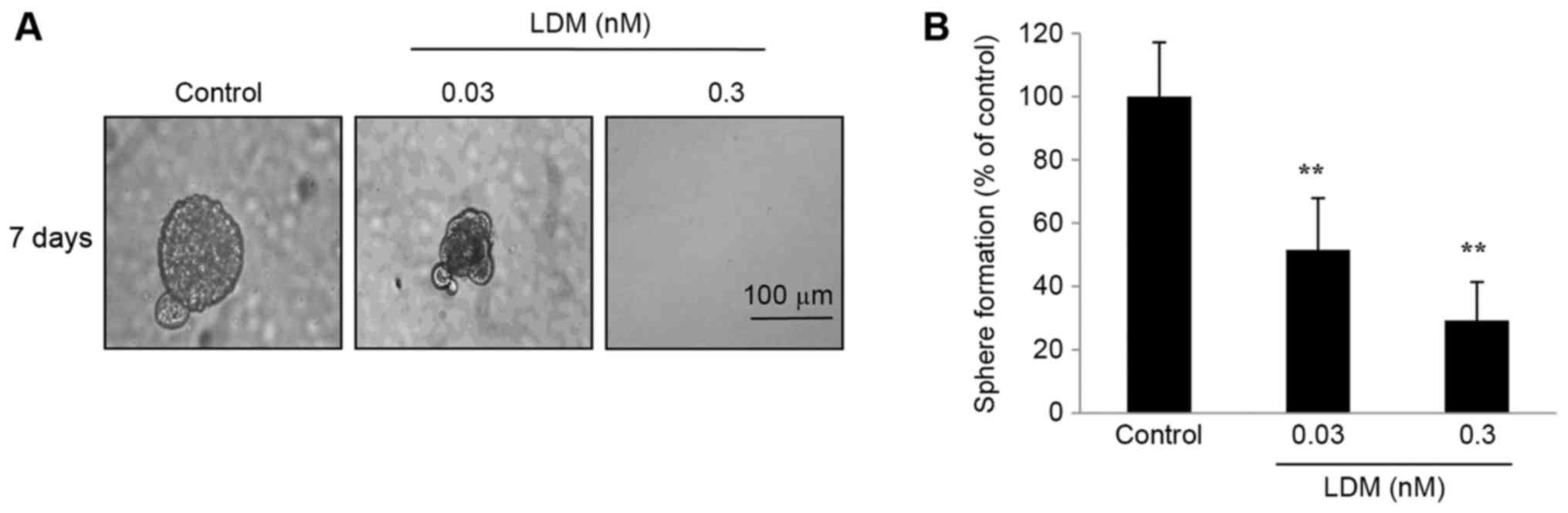
Sphere formation of CD133+
cells is inhibited by LDM
Under non-adherent conditions, sphere formation has
been demonstrated to resemble the characteristics of CSCs (18). CD133+ cells were separated
using fluorescence-activated cell sorting and sphere formation was
detected. The size of spheres formed from sorted CD133+
cells was significantly diminished following exposure to LDM for 7
days (Fig. 2A). Additionally, LDM
decreased the number of spheres compared with the control after 7
days (P<0.01; Fig. 2B). It was
notable that the proportion of inhibition of 0.03 nM LDM was 49%,
while 0.03 nM was 10-fold less than the IC50. The data
suggested that the cell spheres are highly sensitive to LDM in
conditional culture systems.
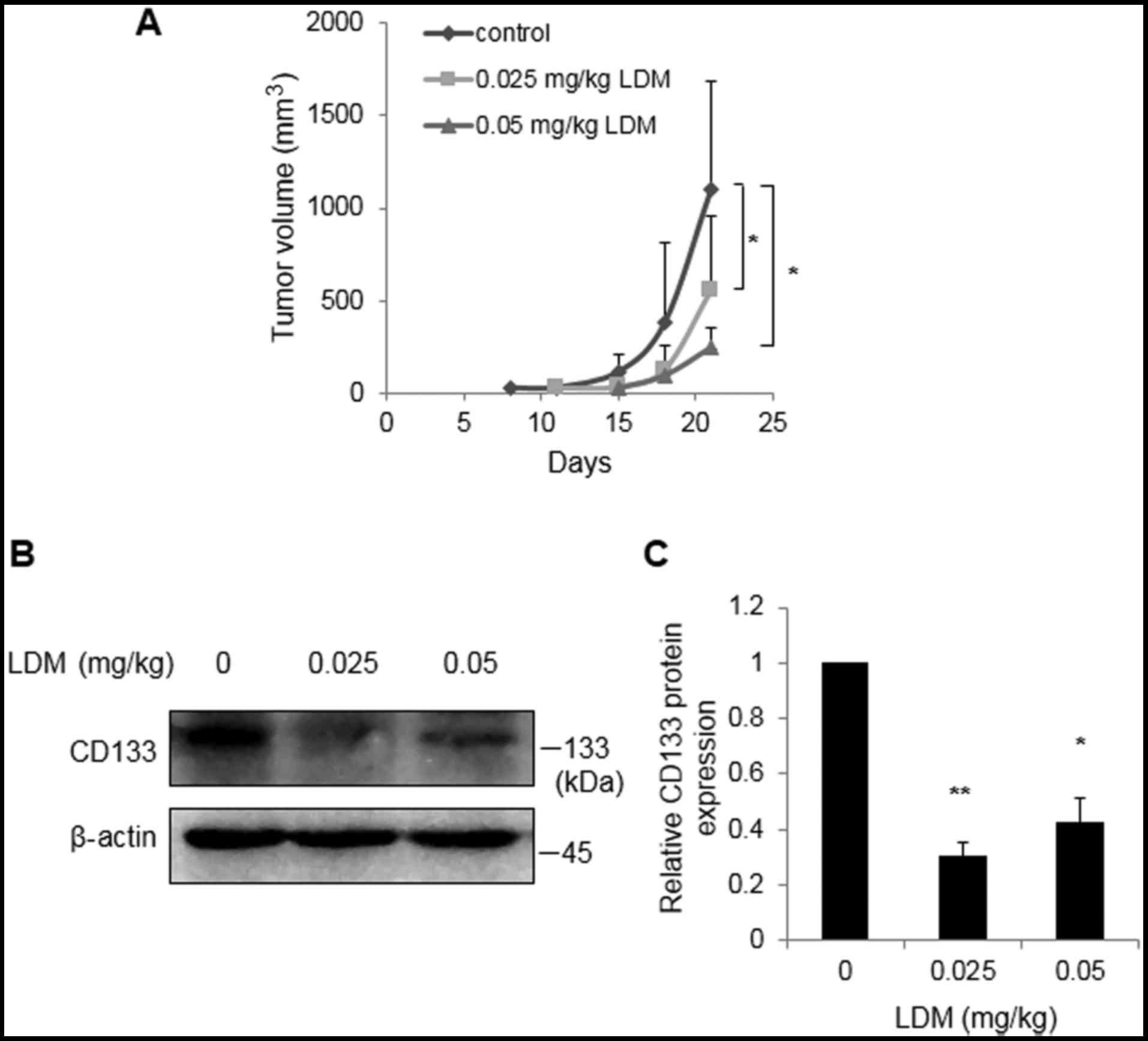
LDM suppresses CD133 in vivo
To investigate the effect of LDM on CD133 in
vivo, enriched CD133+ cells were injected into the
subcutaneous tissues of nude mice. Tumor volumes were restricted at
21 days following 0.025 and 0.05 mg/kg LDM treatment, compared with
control group (P<0.05; Fig. 3A).
In vivo western blot analysis demonstrated that 0.025 and
0.05 mg/kg LDM effectively inhibited CD133 protein expression
compared with the saline control, consistent with the
aforementioned in vitro data (P<0.05; Fig. 3B). Notably, there was no marked
difference between the two concentrations of LDM in
vivo.
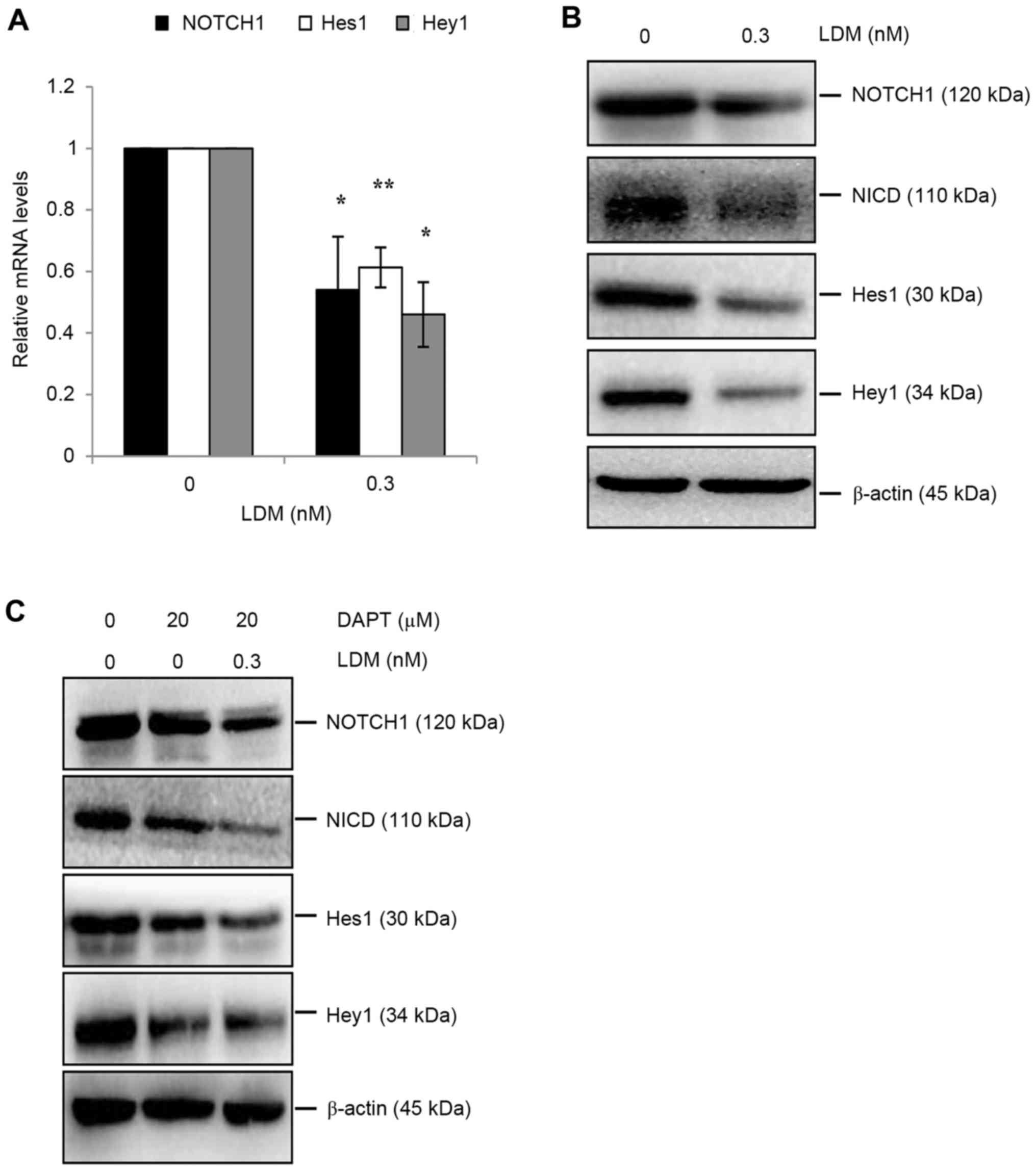
Notch signaling pathway downregulation
by LDM
The Notch signaling pathway has been revealed to be
upregulated in CSCs and has subsequently been demonstrated as an
effective target to eliminate CSCs (4,19–21). The Notch signaling pathway has also
been suggested to be involved in CD133 signaling (22). To investigate the mechanism of LDM on
CD133 in HCC, activation of the Notch signaling pathway was
evaluated. LDM decreased the mRNA level of NOTCH1, Hes1 and Hey1
genes (P<0.05; Fig. 4A).
Additionally, NOTCH1 protein levels decreased following LDM
treatment, decreasing the expression of downstream proteins,
including NICD, Hes1 and Hey1 (Fig.
4B). DAPT is a γ-secretase inhibitor, and is commonly used to
inhibit Notch signaling. Therefore, DAPT was used with or without
LDM separately. NOTCH1 was downregulated only by LDM and not by
DAPT, as compared with the control. However, DAPT-induced abatement
of NICD was enhanced by LDM via downregulating NICD protein
expression. Thus, Hes1 and Hey1 proteins were restricted only by
DAPT, or by DAPT combined with LDM (Fig.
4C). These data suggest that the inhibitory effect of LDM on
CD133 may result from inhibited activity of the Notch signaling
pathway.
Discussion
LDM has been demonstrated to exhibit potential
antitumor effects in HCC; a previous study revealed that LDM
decreased the levels of embryonic stem cell-like genes (23). In addition, LDM inhibits
EpCAM+ tumor initiating cells through the
GSK3β/β-catenin signaling pathway in HCC (15). The effects of LDM on CD133 in HCC were
investigated. In the present study, LDM markedly inhibited CD133
expression of HCC in vitro and in vivo.
Although the function of CD133 is not entirely
known, CD133 is considered an important marker of CSCs with
numerous potential applications; direct downregulation of CD133 may
be an effective strategy for the elimination of CSCs (12,24). A
previous study demonstrated that using antisense
oligodeoxynucleotides to knock-down CD133 expression leads to a
decrease in colony formation and to cell cycle arrest (24). Another group generated an
anti-CD13/anti-CD133 bispecific antibody (BsAb) with
cytokine-induced killer (CIK) cells to target CD133 CSCs, where by
BsAb-CIK significantly inhibited tumor growth in HCC (12). In addition to direct targeting of
CD133, the CD133-associated signaling pathway may be regulated, and
it has been demonstrated that CD133 was decreased through the
blockade of the Notch signaling pathway (25). The results of the present study
suggested that LDM-mediated suppression of CD133 may proceed via
the downregulation of Notch signaling, with data suggesting that,
direct or indirect, the inhibition of CD133 may be an effective
target for CSCs.
The Notch signaling pathway serves an important
function in cell proliferation, self-renewal and differentiation.
In canonical signaling, Notch ligands, including Jagged1, Jagged2
or Delta-like 4, bind to Notch receptors (Notch1-4). Subsequently,
intracellular cleavage is promoted in Notch receptors by
γ-secretase, and NICD, which is the active form of Notch, is
released and translocates to the nucleus where it binds to
transcription factors. Thus, downstream genes are expressed,
including Hes1 and Hey1 (26). DAPT
is a Notch signaling pathway inhibitor that decreases NICD levels
via the inactivation of γ-secretase (27). Consistent with a previous study, the
results of the present study demonstrated that LDM decreases the
mRNA and protein levels of NOTCH1, compared with the control.
Expression of NICD protein and the mRNA and protein levels of Hes1
and Hey1 were similarly decreased compared with the control.
Additionally, LDM-induced decrease of NICD expression was enhanced
in the presence of DAPT through the downregulation of NICD protein
expression. Consequently, Hes1 and Hey1 protein were markedly
decreased by DAPT combined with LDM. The results of the present
study suggested that the inhibitory effect of LDM may suppress the
activity of the canonical Notch signaling pathway.
In a previous study, LDM was revealed to
downregulate embryonic stem cell-like genes: Octamer-binding
transcription factor 4, SRY-box 2 and MYC proto-oncogene (23). Additionally, LDM has been demonstrated
to decreases the expression of EpCAM, aldehyde
dehydrogenase-positive cells, sphere formation in vitro, and
inhibit tumor volume and incidence in vivo; this suggests
that LDM may suppress Huh7 tumor initiating cells (15). The results of the present study
demonstrated that LDM decreases the expression of CD133 in
vitro and in vivo, supporting the function of LDM as an
inhibitor of tumor growth and CSCs, and indicated the potential
mechanism of LDM on CSCs as well as suggesting the benefits and
clinical applications for HCC. CD133 and EpCAM are the main CSC
markers; however, there are many other markers, including CD90,
CD13, ABCG2 and CD44, in which the effects exhibited by
LDM require further investigation.
In conclusion, the results of the present study
demonstrated that LDM decreases the CD133+ cell
population, inhibits CD133 protein expression and suppresses the
sphere formation of sorted CD133+ cells. In vivo
experiments revealed that Huh7 tumor growth, formed from enriched
CD133+ cells, was suppressed following LDM treatment.
LDM also inhibits CD133 expression in tumor tissue. Furthermore,
downregulation of the Notch signaling pathway may be a potential
underlying molecular mechanism between LDM and CD133. These results
support the requirement for further clinical evaluation of LDM.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81402441 and
81773984), the China Postdoctoral Science Foundation (grant no.
2014M562272) and Fundamental Research Funds for the Central
Universities (grant nos. XDJK2016C070 and XDJK2017D153).
Glossary
Abbreviations
Abbreviations:
|
LDM
|
lidamycin
|
|
HCC
|
hepatocellular carcinoma
|
|
CSCs
|
cancer stem cells
|
|
NICD
|
Notch intracellular domain
|
References
|
1
|
Dudeck O and Ricke J: Advances in regional
chemotherapy of the liver. Expert Opin Drug Deliv. 8:1057–1069.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen Y, Yu D, Zhang H, He H, Zhang C, Zhao
W and Shao RG: CD133(+)EpCAM(+) phenotype possesses more
characteristics of tumor initiating cells in hepatocellular
carcinoma Huh7 cells. Int J Biol Sci. 8:992–1004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma S, Lee TK, Zheng BJ, Chan KW and Guan
XY: CD133+ HCC cancer stem cells confer chemoresistance by
preferential expression of the Akt/PKB survival pathway. Oncogene.
27:1749–1758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamashita T, Ji J, Budhu A, Forgues M,
Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chiba T, Iwama A and Yokosuka O: Cancer
stem cells in hepatocellular carcinoma: Therapeutic implications
based on stem cell biology. Hepatol Res. 46:50–57. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wright MH, Calcagno AM, Salcido CD,
Carlson MD, Ambudkar SV and Varticovski L: Brca1 breast tumors
contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell
characteristics. Breast Cancer Res. 10:R102008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Soner BC, Aktug H, Acikgoz E, Duzagac F,
Guven U, Ayla S, Cal C and Oktem G: Induced growth inhibition, cell
cycle arrest and apoptosis in CD133+/CD44+ prostate cancer stem
cells by flavopiridol. Int J Mol Med. 34:1249–1256. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farhana L, Antaki F, Anees MR,
Nangia-Makker P, Judd S, Hadden T, Levi E, Murshed F, Yu Y, Van
Buren E, et al: Role of cancer stem cells in racial disparity in
colorectal cancer. Cancer Med. 5:1268–1278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weng CC, Kuo KK, Su HT, Hsiao PJ, Chen YW,
Wu DC, Hung WC and Cheng KH: Pancreatic tumor progression
associated with CD133 overexpression: involvement of increased TERT
expression and epidermal growth factor receptor-dependent Akt
activation. Pancreas. 45:443–457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koren A, Rijavec M, Kern I, Sodja E,
Korosec P and Cufer T: BMI1, ALDH1A1, and CD133 transcripts connect
epithelial-mesenchymal transition to cancer stem cells in lung
carcinoma. Stem Cells Int. 2016:97143152016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma S, Chan KW, Lee TK, Tang KH, Wo JY,
Zheng BJ and Guan XY: Aldehyde dehydrogenase discriminates the
CD133 liver cancer stem cell populations. Mol Cancer Res.
6:1146–1153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang J, Li C, Wang Y, Lv H, Guo Y, Dai H,
Wicha MS, Chang AE and Li Q: Cytokine-induced killer (CIK) cells
bound with anti-CD3/anti-CD133 bispecific antibodies target
CD133(high) cancer stem cells in vitro and in vivo. ClinImmunol.
149:156–168. 2013.
|
|
13
|
Shao RG and Zhen YS: Enediyne anticancer
antibiotic lidamycin: Chemistry, biology and pharmacology.
Anticancer Agents Med Chem. 8:123–131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi YK, Wu SY, Huang YH and Zhen YS:
Chemosensitivity of mdr1 gene overexpressed multidrug resistant
cancer cells to lidamycin. Yao Xue Xue Bao. 41:1146–1151. 2006.(In
Chinese). PubMed/NCBI
|
|
15
|
Chen Y, Yu D, Zhang C, Shang B, He H, Chen
J, Zhang H, Zhao W, Wang Z, Xu X, et al: Lidamycin inhibits tumor
initiating cells of hepatocellular carcinoma Huh7 through
GSK3β/β-catenin pathway. Mol Carcinog. 54:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Griffioen AW, Mans LA, de Graaf AMA,
Nowak-Sliwinska P1, de Hoog CLMM, de Jong TAM, Vyth-Dreese FA, van
Beijnum JR, Bex A and Jonasch E: Rapid angiogenesis onset after
discontinuation of sunitinib treatment of renal cell carcinoma
patients. Clin Cancer Res. 18:3961–3971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Zhang S, He H, Zhao W, Ren K,
Chen J and Shao RG: RasGAP-derived peptide 38GAP potentiates the
cytotoxicity of cisplatin through inhibitions of Akt, ERK and NF-κB
in colon carcinoma HCT116 cells. Cancer Lett. 308:62–70. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang
R, Li J, Zhang Y, Chen L, Qian H, et al: Sphere-forming cell
subpopulations with cancer stem cell properties in human hepatoma
cell lines. BMC Gastroenterol. 11:712011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Sousa EM, Vermeulen L, Richel D and
Medema JP: Targeting Wnt signaling in colon cancer stem cells. Clin
Cancer Res. 17:647–653. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Zhang T, Korkaya H, Liu S, Lee HF,
Newman B, Yu Y, Clouthier SG, Schwartz SJ, Wicha MS and Sun D:
Sulforaphane, a dietary component of broccoli/broccoli sprouts,
inhibits breast cancer stem cells. Clin Cancer Res. 16:2580–2590.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu W, Lin H, Zhang Y, Chen X, Hua B, Hou
W, Qi X, Pei Y, Zhu X, Zhao Z and Yang L: Compound kushen injection
suppresses human breast cancer stem-like cells by down-regulating
the canonical Wnt/β-catenin pathway. J Exp Clin Cancer Res.
30:1032011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu D and Pan W: GSK3: A multifaceted
kinase in Wnt signaling. Trends Biochem Sci. 35:161–168. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhen HY, He QH, Zhen YZ, Wang SL, Liu YN,
Wu WH, Zhang XY, Lu AL and Shen L: Inhibition of mouse embryonic
carcinoma cell growth by lidamycin through down-regulation of
embryonic stem cell-like genes Oct4, Sox2 and Myc. Invest New
Drugs. 29:1188–1197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yao J, Zhang T, Ren J, Yu M and Wu G:
Effect of CD133/prominin-1 antisense oligodeoxynucleotide on in
vitro growth characteristics of Huh-7 human hepatocarcinoma cells
and U251 human glioma cells. Oncol Rep. 22:781–787. 2009.PubMed/NCBI
|
|
25
|
Fan X, Khaki L, Zhu TS, Soules ME, Talsma
CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, et al: NOTCH pathway
blockade depletes CD133-positive glioblastoma cells and inhibits
growth of tumor neurospheres and xenografts. Stem Cells. 28:5–16.
2010.PubMed/NCBI
|
|
26
|
Crabtree JS, Singleton CS and Miele L:
Notch signaling in neuroendocrine tumors. Front Oncol. 6:942016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Ye Z, Zheng S, Chen L, Wan Y, Deng
Y and Yang R: Lingo-1 shRNA and Notch signaling inhibitor DAPT
promote differentiation of neural stem/progenitor cells into
neurons. Brain Res. 1634:34–44. 2016. View Article : Google Scholar : PubMed/NCBI
|