Introduction
Renal cell carcinoma (RCC) comprises 2–3% of
malignant tumors in the Western world (1). It is an increasingly diagnosed
malignancy, with an increased number of cases demonstrating a shift
towards presenting with smaller renal masses, most likely due to
the increasing use of computed tomography and ultrasonography
(2). However, ~30% of patients with
RCC initially present with metastatic disease, and approximately
half of all patients with localized disease will develop metastases
over time (3).
A variety of genetic models have been proposed to
account for the development of RCC, taking into account certain
risk factors, such as cigarette smoking, hypertension and obesity
(4). However, despite the genome-wide
analyses conducted thus far, the range of molecular mechanisms
underlying the development of RCC are not yet fully understood.
Familial and sporadic forms of RCC can be associated with certain
genetic alterations, the most common of which is the von
Hippel-Lindau tumor suppressor gene mutation (5,6).
For advanced or metastatic disease, therapeutic
options remain limited. Cytotoxic chemotherapy agents have yielded
disappointing results (7). In a
previous study, the treatment of advanced and metastasized RCC was
attempted with a cytokine-based therapy with interferon-α and
high-dose interleukin-2 (IL-2), under the presumption that an
immune response could be triggered against RCC cells. However, this
therapy offered little benefit with regard to the overall survival
times of the patients (8). It is only
in the last decade that a broader understanding of RCC tumor
biology and the introduction of inhibitors of tyrosine kinase,
multikinase and mammalian target of rapamycin (mTOR) have greatly
improved the therapeutic means for metastatic RCC (9). Aside from targeting tumor
neoangiogenesis, immunomodulatory checkpoint inhibition was
recently introduced as a novel treatment approach (10). Checkpoint inhibition acts via
modulation of tumor-infiltrating T lymphocytes and shows promising
results as a second-line treatment option (10). These new therapeutic strategies
demonstrate that metastatic RCC can no longer be viewed from the
perspective of renal tubule cell biology alone, but rather within
an integrative model of tumor biology, which includes angiogenesis,
interstitial microenvironment and immunological tissue (11).
ARHGDIB belongs to the Rho protein family, whose
members are involved in a variety of cellular functions, including
proliferation, signaling, secretion, cytoskeletal organization and
proliferation; while also belonging to Ras-associated small
GTP-binding proteins (12). Since
ARHGDIB is involved in neoangiogenesis and lymphocyte function (two
key factors in the treatment of metastasized RCC), it is an area of
focus for the study of RCC, and metastatic RCC in particular
(13). However, despite serving a
role in other malignancies such bladder (14,15), the
role of ARHGDIB for RCC has not been evaluated yet. The aim of the
present study was to evaluate the role of ARHGDIB in RCC by
comparing ARHGDIB mRNA expression levels in healthy and tumor
tissue as well as by assessing the potential impact of expression
levels of ARHGDIB on survival.
Materials and methods
Tissue sampling
Renal tumor tissues and adjacent corresponding
tumor-free renal tissues of 105 patients undergoing kidney surgery
for RCC were collected between January 2001 and December 2005.
Tissue samples were obtained during surgery. All samples were
collected at Eberhard Karls University of Tübingen (Tübingen,
Germany) and samples where lymphatic tissue invasion exceeded 25%
of the specimen section were excluded. Corresponding adjacent
normal tissue was sampled 0.5–2 cm from the surgical margin of the
primary tumor and each tumor and normal tissue sample was
immediately snap frozen in liquid nitrogen and stored at −80°C.
Tumor stage and histological subtype were assessed according to the
2002 Union for International Cancer Control tumor-node-metastasis
system (16) by two independent
pathologists that were blinded to mRNA expression of ARHGDIB as
well as the clinical course of the patient. Tumor grade was defined
in accordance with the Fuhrman grading system (17), while histological subtypes were based
on the consensus classification of renal cell neoplasia (18). Localized RCC was defined as pT≤2
without organ metastasis or lymph node involvement and a grade of
≤2. Advanced RCC was defined as pT≥3 or organ metastasis or >G2
or lymph node-positive disease. Only tissue samples that included
≥75% vital tumor tissue were selected. None of the patients
received neoadjuvant treatment prior to definitive surgery; thus,
all patients in the present study were systemic therapy-naïve. Data
were gathered by physicians and passed to data managers for
evaluation and storage in a relational database. Written informed
consent was obtained from all patients prior to surgery and tissue
sampling. Approval was granted prior to the study by the Ethics
Committee of the Medical Faculty of Eberhard Karls University.
Patients
The histopathological and clinical characteristics
of the patients are summarized in Table
I. The mean age of all patients was 63.6 (±11.8) years. The
male:female ratio was 67:38 (~1.76:1; 63.8% males and 36.2%
females). Histopathological subclassification showed 77 patients
with ccRCC, 21 patients with papillary RCC (papRCC) and 5 patients
with chromophobe tumors, while 2 patients had non-classified
histology.
 | Table I.Histopathological and clinical
parameters of patients with renal cell carcinoma. |
Table I.
Histopathological and clinical
parameters of patients with renal cell carcinoma.
|
Characteristics | Value |
|---|
| Total, n | 105 |
| Mean age ± standard
deviation, years | 63.6±11.8 |
| Sex, n (%) |
|
|
Male | 67 (63.8) |
|
Female | 38 (36.2) |
| Median tumor
diameter, cm | 4.5 |
| Histology, n
(%) |
|
| Clear
cell | 77 (73.3) |
|
Papillary | 21 (20.0) |
|
Chromophobe | 5 (4.8) |
|
Other/not classified | 2 (1.9) |
| Stage, n (%) |
|
| Not
applicable | 3 (2.9) |
|
pT1 | 9 (8.6) |
|
pT1a | 32 (30.5) |
|
pT1b | 20 (19.0) |
|
pT2 | 4 (3.8) |
|
pT3 | 3 (2.9) |
|
pT3a | 10 (9.5) |
|
pT3b | 24 (22.9) |
|
pT4 | 0 (0.0) |
| Grade, n (%) |
|
| G1 | 17 (16.2) |
|
G1-2 | 14 (13.3) |
| G2 | 57 (54.3) |
|
G2-3 | 7 (6.7) |
| G3 | 10 (9.5) |
| Lymph node
metastasisa, n (%) | 11 (10.5) |
| Visceral
metastasisa, n (%) | 23 (21.9) |
| Advanced/metastatic
diseaseb, n (%) | 29 (27.6) |
Primary cells
Renal proximal tubular epithelial cells (RPTEC;
Lonza Group, Ltd., Basel, Switzerland) were cultured and prepared
according to the manufacturer's protocol as an external reference
control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Tissue was prepared from 20 cryosections of 20-µm
thickness. Using TRIzol reagent (Ambion; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), total RNA was extracted. Two sections of
each tissue sample were stained with hematoxylin-eosin and
evaluated by a pathologist. The conversion of RNA into
single-stranded complementary DNA (cDNA) was performed using the
High Capacity cDNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The TaqMan assays used were as
follows: ARHGDIB (assay ID: Hs00171288_m1), RPL13A (assay ID:
Hs0-304-3885_g1), HPRT1 (assay ID: Hs9-999-9909_m1) and GUSB (assay
ID: Hs0-093-9627_m1). For RT-qPCR, the ABI 7900 Fast Sequence
Detection System with Universal PCR Master Mix and TaqMan
Expression Assays (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) were used exactly as described in the
manufacturer's TaqMan Gene expression assay protocol PN 4333458N:
Standard 40 cycles of denaturation at 95°C (15 sec) and annealing
and extension at 60°C (60 sec) were applied for amplification of
cDNA, after standard initial denaturation and TaqPolymerase enzyme
activation at 95°C (10 min). All evaluations were duplicates
meaning that each cDNA has been analyzed twice and mean values were
used for further evaluations. For the biological control, cDNA
generated from RPTEC primary cells was used (50 ng cDNA in 1 µl per
reaction). For endogenous controls, transcripts of the human
RPL13A, HPRT1 and GUSB were used; these were combined using the
dataAssist software (version 2.0; Thermo Fisher Scientific Inc.)
and the arithmetic mean was used as a method for normalization.
Blank no-template and no RT controls were inserted for each
measurement. The method of Livak and Schmittgen (19) and reference ΔCq values originating
from the biological reference RPTEC were used for the calculation
of ΔΔCq and all relative quantity values. In total, 105
measurements for ARHGDIB expression were successfully
performed.
Statistical analysis
Data were assessed using the SDS 2.3 Manager,
dataAssist software (version 2.0; Applied Biosystems; Thermo Fisher
Scientific Inc.). Statistical programming language R 2.15–2 (R core
team; R Foundation for Statistical Computing, Vienna, Austria) was
used for all statistical calculations. The two-sided type-I error
was set to 5% for all statistical tests. All graphical plots were
generated using R statistical software (20). For statistical analysis of ARHGDIB
mRNA expression in tumor vs. paired adjacent tissue, the Wilcoxon
signed-rank test and paired t-test were used. Logistic regression
was used to assess the associations between ARHGDIB mRNA expression
and clinical parameters. Kaplan-Meier and Cox's regression analyses
were used to determine the effects of mRNA expression patterns on
RFS. Data on ARHGDIB for RCC were downloaded from the Kidney Renal
Clear Cell Carcinoma (KIRC)/The Cancer Genome Atlas (TCGA) data
portal (cancergenome.nih.gov, accessed
4/2014). The data were evaluated with a paired two-sided t-test for
paired samples and logistic regression for group comparison.
Results
Comparison of normal vs. paired tumor
tissue
Tumor kidney tissue was compared with adjacent,
histologically normal-appearing tissue in a subset of 74 paired
samples (samples in which paired tissue samples were available).
The mRNA expression levels of ARHGDIB were found to be
significantly higher in the tumor tissue compared with normal
tissue (P<0.001). Subsequently, a paired analysis was performed
with the 74 samples. The characteristics of the patients included
in the paired analysis are shown in Table II. Significantly increased mRNA
expression levels for ARHGDIB were observed for all RCC tissues
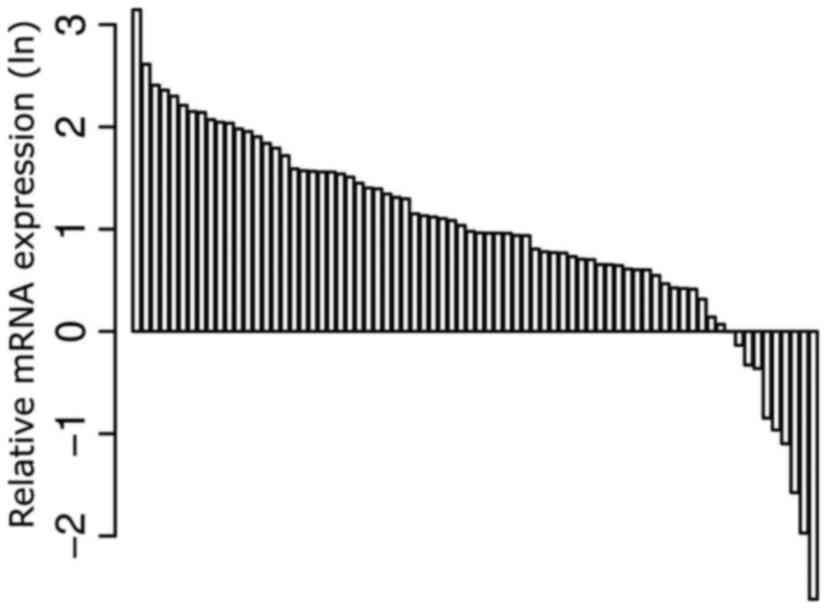
combined (P<0.001; Fig. 1), and
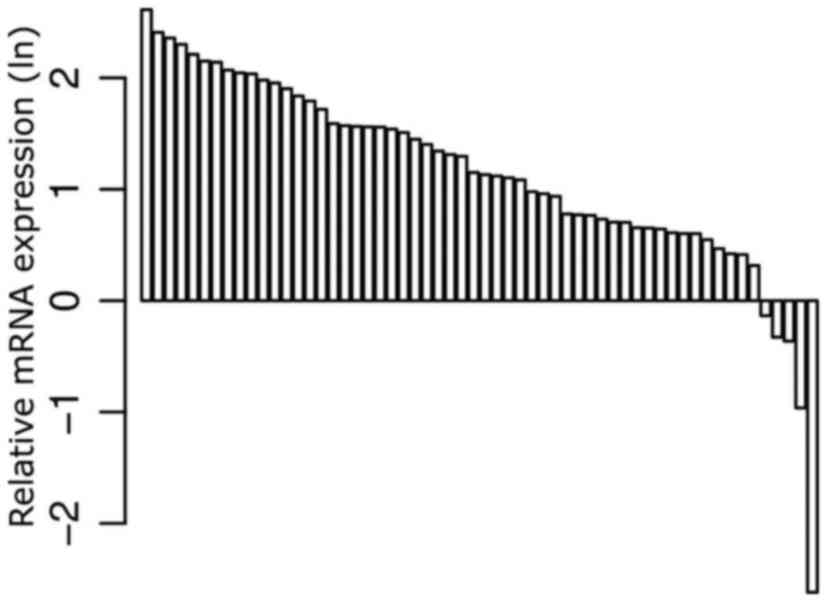
also for the subgroup of ccRCC tissues (P<0.001; Fig. 2). The mRNA expression levels of
ARHGDIB were also more pronounced in ccRCC tissues when compared
with tissues from papRCC [P<0.001; odds ratio (OR), 0.228 (95%
confidence interval, 0.118–0.444)]. The histopathological and
clinical parameters and relative expression levels (ΔΔCq) of
ARHGDIB mRNA are summarized in Table
III.
 | Table II.Histopathological and clinical
parameters of the patients included in the paired analysis of Rho
GDP dissociation inhibitor-β mRNA expression in tumors vs. adjacent
normal kidney tissues. |
Table II.
Histopathological and clinical
parameters of the patients included in the paired analysis of Rho
GDP dissociation inhibitor-β mRNA expression in tumors vs. adjacent
normal kidney tissues.
|
Characteristics | Value |
|---|
| Total, n | 74 |
| Mean age ± standard
deviation, years | 66.5±11.8 |
| Sex, n (%) |
|
|
Male | 45 (60.8) |
|
Female | 29 (39.2) |
| Histology |
|
| Clear
cell | 58 (78.4) |
|
Papillary | 11 (14.9) |
|
Chromophobe | 4 (5.4) |
|
Other/not classified | 1 (1.4) |
| Stage, n (%) |
|
| Not
applicable | 3 (4.1) |
|
pT1 | 4 (5.4) |
|
pT1a | 24 (32.4) |
|
pT1b | 14 (18.9) |
|
pT2 | 3 (4.1) |
|
pT3 | 1 (1.4) |
|
pT3a | 6 (8.1) |
|
pT3b | 19 (25.7) |
|
pT4 | 0 (0.0) |
| Grade |
|
| G1 | 9 (12.2) |
|
G1-2 | 10 (13.5) |
| G2 | 40 (54.1) |
|
G2-3 | 6 (8.1) |
| G3 | 9 (12.2) |
| Lymph node
metastasisa, n (%) | 7 (9.5) |
| Visceral
metastasisa, n (%) | 19 (25.7) |
| Locally
advanced/metastatic diseaseb, n (%) | 23 (31.1) |
 | Table III.Histopathological parameters and
relative expression levels of ARHGDIB mRNA in all patients with
RCC. |
Table III.
Histopathological parameters and
relative expression levels of ARHGDIB mRNA in all patients with
RCC.
|
Sample/pathology | Patients, n | Mean relative
ARHGDIB expression (∆∆Cq) | Standard
deviation |
|---|
| Total | 105 |
|
|
| Sex |
|
|
|
|
Male | 67 | −0.43 | 1.04 |
|
Female | 38 | −0.16 | 0.76 |
| Age |
|
|
|
| Below
median (65 years) | 54 | −0.34 | 0.98 |
| Equal
to or above median (65 years) | 51 | −0.32 | 0.93 |
| Histology |
|
|
|
|
Papillary RCC | 28 | −1.29 | 0.97 |
| Clear
cell RCC | 84 | −0.05 | 0.75 |
| M stage |
|
|
|
| M0 | 82 | −0.39 | 1.04 |
| M+ | 23 | −0.12 | 0.48 |
| N stage |
|
|
|
| N0 | 94 | −0.30 | 0.96 |
| N+ | 11 | −0.56 | 0.85 |
| Disease
progressiona |
|
|
|
|
Localized RCC | 76 | −0.36 | 1.04 |
|
Advanced RCC | 29 | −0.24 | 0.68 |
| Grade |
|
|
|
| ≤2 | 88 | −0.30 | 0.91 |
|
>2 | 17 | −0.52 | 1.15 |
Recurrence-free survival
When analyzing clinicopathological parameters in
univariate Cox regression analysis, RFS was significantly
associated with the occurrence of metastases, locally advanced
disease and tumor grade (P=0.018, P=0.002 and P<0.001,
respectively). For RCC, including all pathological subtypes,
bivariate Cox regression did not show any association of ARHGDIB
mRNA expression with RFS (Table
IV).
 | Table IV.Cox's regression analyses of clinical
parameters for recurrence-free survival. |
Table IV.
Cox's regression analyses of clinical
parameters for recurrence-free survival.
| Variable | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Metastasis
(RCC) | 5.71 | 1.91–17.1 | 0.018 |
| Locally advanced
disease (RCC) | 5.47 | 1.83–16.4 | 0.002 |
| Tumor grade
(RCC) | 12.7 | 3.89–41.3 | <0.001 |
| ARHGDIB mRNA
expression (ccRCC) | 0.11 | 0.03–0.46 | 0.002 |
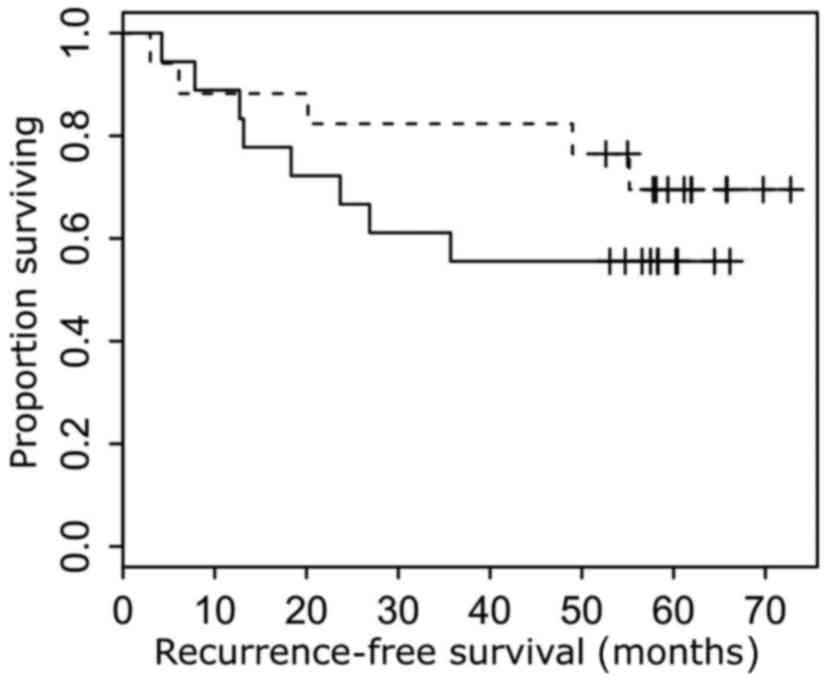
For the subgroup of ccRCC, higher expression levels
(threshold-0.45 ∆∆Cq) of ARHGDIB were associated with longer RFS
time on univariate Cox regression analysis [hazard ratio, 0.11; 95%
confidence interval (CI), 0.03–0.46; P=0.002]. A bivariate Cox
regression model, adjusted for metastatic status (P=0.001), tumor
diameter (P=0.043), advanced disease (P=0.030) and lymph node
metastasis (P=0.006) was also able to identify increased ARHGDIB
mRNA expression as a positive prognostic factor for RFS in patients
with ccRCC histology (Fig. 3).
Comparison with TCGA data
Upon interrogation of the TCGA/KIRC database
(http://cancergenome.nih.gov/) for 479
tested tissue samples, similar results were observed. ARHGDIB mRNA
expression was associated with RCC (P<0.001), but not with the
presence of metastasis (P=0.586; OR, 1.11), positive lymph nodes
(P=0.141; OR, 1.22), tumor stage (P=0.053; OR, 1.32) or grade
(P=0.463, OR, 1.11).
Discussion
The majority of members of the Ras family of GTPases
act as molecular regulators, switching between an active GTP-bound
state with protein localization at the cellular membrane, and the
cytosolic inactive protein bound to GDP (21). Rho GTPases can be regulated by GDP
dissociation inhibitors, including ARHGDIB (22). There is evidence that ARHGDIB is
expressed differently in different tumor entities at the protein
and mRNA levels; for example, ARHGDIB mRNA expression has been
found to be elevated in adenocarcinoma of the ovaries (23) and shown to be positively correlated
with tumor progression in gastric cancer (24). Furthermore, increased mRNA expression
has been detected in breast cancer, and demonstrated to be
associated with increased cell motility and invasiveness in in
vitro experiments (25). By
contrast, low ARHGDIB expression has been shown to be associated
with the invasive capacity and clinical prognosis of bladder cancer
(14,15).
The two major therapeutic approaches to treatment of
metastasized or inoperable advanced RCC have been immunotherapy via
interferon-α and IL-2 (8), as well as
targeted antiangiogenic therapy more recently (10). ARHGDIB is involved in carcinogenesis
in a variety of malignancies, and serves roles in neoangiogenesis
and signaling in the lymphocyte immune response (12–15), which
makes it an interesting target for investigation with regard to
metastatic RCC.
Neoangiogenesis is associated with hypoxia-inducible
factor (HIF) transcription factors, which perform critical roles in
cell metabolism with regard to oxygen homeostasis (26). HIF-α accumulates under hypoxic
conditions and is degraded in the presence of oxygen (26). HIF accumulation results in the
overexpression of a variety of growth factors, including vascular
endothelial growth factor (VEGF) and platelet-derived growth factor
(PDGF), which promote neoangiogenesis (27). It is well known that the type and
density of vasculature in RCC are associated with tumor
histological grade and extent of macroscopic tumor necrosis, which
affect the clinical course of RCC (28), including the ccRCC subgroup (29).
Increased understanding of the molecular aspects of
angiogenesis in tumor tissues has led to the development of
targeted therapy for RCC (30), which
is a highly-vascularized tumor entity (29). Sunitinib, one of the first novel
therapeutic agents to be used for RCC, modulates the VEGF-C pathway
(31). It is a tyrosine kinase
inhibitor that acts against VEGF receptors 1, 2 and 3, as well as
PDGF receptors α and β (32,33). In response to therapy with sunitinib,
the plasma levels of VEGF-C decrease and patients show a markedly
improved RFS (31). Bevacizumab,
another therapeutic agent for the treatment of metastasized RCC, is
an antibody that targets VEGF, and thereby also acts against tumor
neoangiogenesis via VEGF inactivation, leading to an improved RFS
time for patients with ccRCC (34).
Recently, a functional association between ARHGDIB
and VEGF-C was detected for gastric cancer; ARHGDIB overexpression
was demonstrated to lead to elevated mRNA and protein levels of
VEGF-C (13). By contrast, a
decreased level of ARHGDIB in the gastric cancer MKN-28 cell line
was associated with markedly decreased VEGF-C expression (13). Therefore, it has been demonstrated
that VEGF-C is among the mediators that induce angiogenesis in
vivo (35), and that it also
promotes cancer cell invasion and metastasis (36,37). These
results, which functionally link VEGF-C and ARHGDIB, indicate a
possible involvement of ARHGDIB in neoangiogenesis and
carcinogenesis.
Another possible functional association between
ARHGDIB and RCC signaling is the potential connection of ARHGDIB to
the invasion of lymphocytes into tumor tissues. It has previously
been shown that ARHGDIB is strongly expressed in lymphatic tissue
(38), and that RCC tumors frequently
exhibit varying amounts of infiltrating lymphocytes (39). The interaction between RCC and the
lymphocyte-containing microenvironment is of special importance, as
almost all RCCs appear to express cluster of differentiation 70,
which drives proliferative exhaustion of lymphocytes and may be a
possible mechanism underlying the immune evasion of the tumor
(40).
A previous study observed the overexpression of
ARHGDIB in human monocytes, leading to the subsequent suppression
of Rac membrane localization, disrupting the actin cytoskeleton and
thereby negatively effecting phagocytosis (41). Groysman et al (42) were able to demonstrate that ARHGDIB, a
member of the Rho GTPases, is also pivotal for T cell signaling,
where it is involved in the regulation of intracellular signaling
pathways leading to nuclear factor of activated T cells
stimulation, which in turn leads to the biosynthesis of IL-2 and
subsequent activation and proliferation of T lymphocytes. This
association with lymphocyte metabolism poses a possible association
with the previously mentioned immune-based therapeutic approaches
for RCC with IL-2 (43), which were
used prior to the advent of tyrosine kinase inhibitors.
T cell signaling is also the mechanism by which
certain novel pharmacological agents exert their antitumor effect.
Checkpoint inhibitors have shown promising results in urological
and non-urological cancers; recently, the targeting of programmed
death receptor on T cells has shown superiority in comparison with
the mTOR inhibitor everolimus in the treatment of RCC (10,44). This
has led to the implementation of the checkpoint inhibitor nivolumab
as a second-line treatment option for metastatic RCC.
One limitation of the present study was the varying
amounts of lymphatic cell invasion, vasculature and interstitial
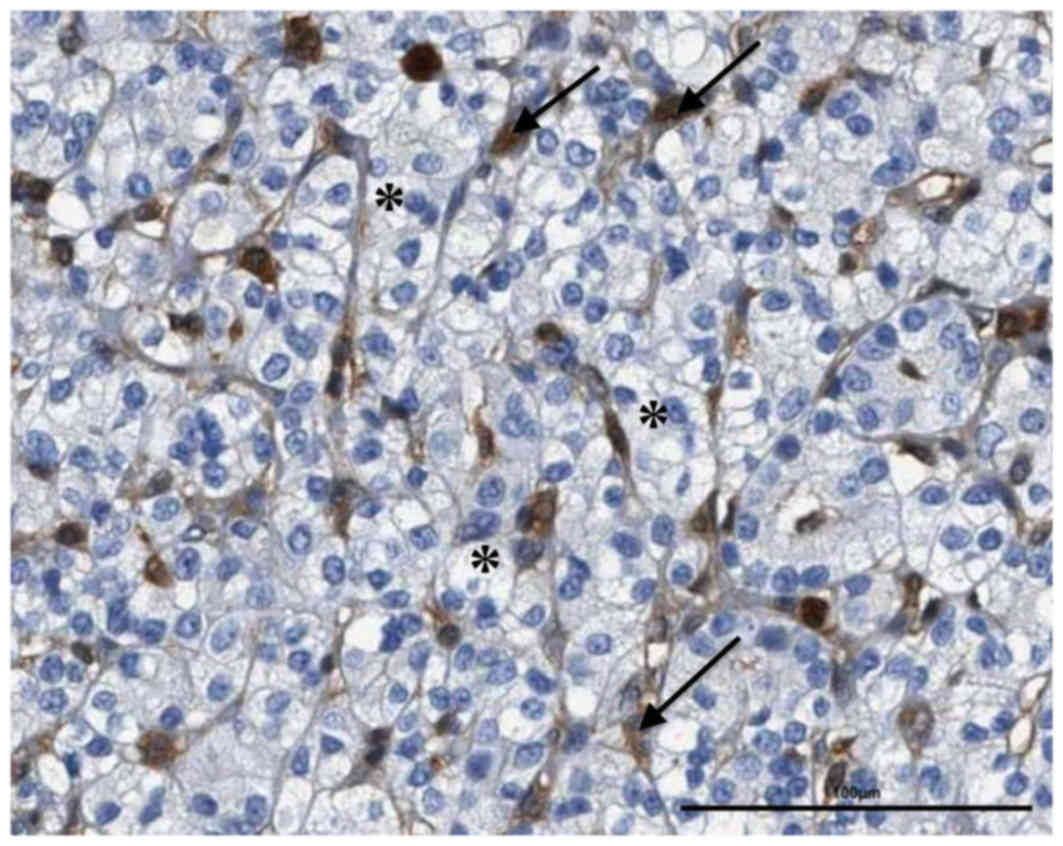
tissue among the included cases. The protein expression of ARHGDIB
in normal and tumor renal tissues has thus far only been
immunohistochemically analyzed for a small number of tissues,
showing more prominent immunopositivity in renal glomeruli, with
prominent reactivity in the surrounding lymphatic cells and
fibroblasts, but a low or no detectable reactivity in renal tubules
(Fig. 4) (45,46).
Considering that mRNA expression levels have been measured in
tissue lysates following the homogenization of the cells, the
present study cannot provide information regarding the origin of
the ARHGDIB expression and, furthermore, the results may also show
bias due to variations in lymphocyte content in the tissue samples.
Lymphocytes are often found infiltrating or surrounding RCC tissues
(47). After reviewing the
histopathological control sections in the present study, lymphatic
tissue invasion that exceeded 25% of the specimen section was
excluded. The positive correlation of ARHGDIB with RFS in the
subgroup of RCCs with clear cell histology in the present study
cannot be explained at present. However, future functional analyses
should assess the effects of ARHGDIB on the tumor-surrounding
tissues, and particularly lymphocyte function, as this may serve a
role.
In conclusion, ARHGDIB mRNA expression is highly
upregulated in RCC tissues compared with adjacent normal tissues,
and is positively associated with RFS in ccRCC. At present, the two
major modes of action for the pharmacological treatment of RCC are
angiogenesis inhibition and T-cell checkpoint inhibition.
Therefore, the effect of ARHGDIB on angiogenesis, as well as its
roles in the cell signaling of lymphatic tissue and immune therapy,
makes it an interesting target for further functional studies.
References
|
1
|
European Network of Cancer Registries.
Eurocim version 4.0, . European incidence database. V2.3, 730
entity dictionary (2001). Lyon: 2001
|
|
2
|
Katz DL, Zheng T, Holford TR and Flannery
J: Time trends in the incidence of renal carcinoma: Analysis of
Connecticut tumor registry data, 1935–1989. Int J Cancer. 58:57–63.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Mulder PH, van Herpen CM and Mulders
PA: Current treatment of renal cell carcinoma. Ann Oncol. 15 Suppl
4:iv319–iv328. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lindblad P: Epidemiology of renal cell
carcinoma. Scand J Surg. 93:88–96. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seizinger BR, Rouleau GA, Ozelius LJ, Lane
AH, Farmer GE, Lamiell JM, Haines J, Yuen JW, Collins D,
Majoor-Krakauer D, et al: Von Hippel-Lindau disease maps to the
region of chromosome 3 associated with renal cell carcinoma.
Nature. 332:268–269. 1988. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nickerson ML, Jaeger E, Shi Y, Durocher
JA, Mahurkar S, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko
V, et al: Improved identification of von Hippel-Lindau gene
alterations in clear cell renal tumors. Clin Cancer Res.
14:4726–4734. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yagoda A, Petrylak D and Thompson S:
Cytotoxic chemotherapy for advanced renal cell carcinoma. Urol Clin
North Am. 20:303–321. 1993.PubMed/NCBI
|
|
8
|
Gore ME, Griffin CL, Hancock B, Patel PM,
Pyle L, Aitchison M, James N, Oliver RT, Mardiak J, Hussain T, et
al: Interferon alfa-2a versus combination therapy with interferon
alfa-2a, interleukin-2 and fluorouracil in patients with untreated
metastatic renal cell carcinoma (MRC RE04/EORTC GU 30012): An
open-label randomised trial. Lancet. 375:641–648. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Patard JJ, Rioux-Leclercq N and Fergelot
P: Understanding the importance of smart drugs in renal cell
carcinoma. Eur Urol. 49:633–643. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus everolimus in advanced
renal-cell carcinoma. N Engl J Med. 375:1803–1813. 2015. View Article : Google Scholar
|
|
11
|
Rahat MA and Shakya J: Parallel aspects of
the microenvironment in cancer and autoimmune disease. Mediators
Inflamm. 2016:43751202016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moon SY and Zheng Y: Rho GTPase-activation
proteins in cell regulation. Trends Cell Biol. 13:13–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho HJ, Kim IK, Park SM, Baek KE, Nam IK,
Park SH, Ryu KJ, Choi J, Ryu J, Hong SC, et al: VEGF-C mediates
RHoGDI2-induced gastric cancer cell metastasis and cisplatin
resistance. Int J Cancer. 135:1553–1563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Theodorescu D, Sapinoso LM, Conaway MR,
Oxford G, Hampton GM and Frierson HF Jr: Reduced expression of
metastasis suppressor RHoGDI2 is associated with decreased survival
for patients with bladder cancer. Clin Cancer Res. 10:3800–3806.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gildea JJ, Seraj MJ, Oxford G, Harding MA,
Hampton GM, Moskaluk CA, Frierson HF, Conaway MR and Theodorescu D:
RHoGDI2 is an invasion and metastasis suppressor gene in human
cancer. Cancer Res. 62:6418–6423. 2002.PubMed/NCBI
|
|
16
|
Sobin LH and Compton CC: TNM seventh
edition: What's new, what's changed: Communication from the
international union against cancer and the American joint committee
on cancer. Cancer. 116:5336–5339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stenzl A and deKernion JB: Pathology,
biology, and clinical staging of renal cell carcinoma. Semin Oncol.
16 1 Suppl 1:S3–S11. 1989.
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta c(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
R core team (2013. r: A language and
environment for statistical computing, . r foundation for
statistical computing. vienna, austria: http://www.R-project.org/
|
|
21
|
Schmidt A and Hall A: Guanine nucleotide
exchange factors for RHo GTPases: Turning on the switch. Genes Dev.
16:1587–1609. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H,
Araki S, Ueda T, Kikuchi A and Takai Y: Molecular cloning and
characterization of a novel type of regulatory protein (GDI) for
the RHo proteins, ras p21-like small GTP-binding proteins.
Oncogene. 5:1321–1328. 1990.PubMed/NCBI
|
|
23
|
Tapper J, Kettunen E, El-Rifai W, Seppälä
M, Andersson LC and Knuutila S: Changes in gene expression during
progression of ovarian carcinoma. Cancer Genet Cytogenet. 128:1–6.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cho HJ, Baek KE, Park SM, Kim IK, Choi YL,
Cho HJ, Nam IK, Hwang EM, Park JY, Han JY, et al: RhoGDI2
expression is associated with tumor growth and malignant
progression of gastric cancer. Clin Cancer Res. 15:2612–2619. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y and Zhang B: D4-GDI, a RHo GTPase
regulator, promotes breast cancer cell invasiveness. Cancer Res.
66:5592–5598. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jaakkola P, Mole DR, Tian YM, Wilson MI,
Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji
M, Schofield CJ, et al: Targeting of HIF-alpha to the von
Hippel-Lindau ubiquitylation complex by 02-regulated prolyl
hydroxylation. Science. 292:468–472. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Patel PH, Chadalavada RS, Chaganti RS and
Motzer RJ: Targeting von Hippel-Lindau pathway in renal cell
carcinoma. Clin Cancer Res. 12:7215–7220. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sabo E, Boltenko A, Sova Y, Stein A,
Kleinhaus S and Resnick MB: Microscopic analysis and significance
of vascular architectural complexity in renal cell carcinoma. Clin
Cancer Res. 7:533–537. 2001.PubMed/NCBI
|
|
29
|
Qian CN, Huang D, Wondergem B and Teh BT:
Complexity of tumor vasculature in clear cell renal cell carcinoma.
Cancer. 115 10 Suppl:S2282–S2289. 2009. View Article : Google Scholar
|
|
30
|
Rini BI and Small EJ: Biology and clinical
development of vascular endothelial growth factor-targeted therapy
in renal cell carcinoma. J Clin Oncol. 23:1028–1043. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rini BI, Michaelson MD, Rosenberg JE,
Bukowski RM, Sosman JA, Stadler WM, Hutson TE, Margolin K, Harmon
CS, DePrimo SE, et al: Antitumor activity and biomarker analysis of
sunitinib in patients with bevacizumab-refractory metastatic renal
cell carcinoma. J Clin Oncol. 26:3743–3748. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chow LQ and Eckhardt SG: Sunitinib: From
rational design to clinical efficacy. J Clin Oncol. 25:884–896.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Motzer RJ, Hutson TE, Tomczak P,
Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik
C, Kim ST, et al: Sunitinib versus interferon alfa in metastatic
renal-cell carcinoma. N Engl J Med. 356:115–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Escudier B, Bellmunt J, Négrier S, Bajetta
E, Melichar B, Bracarda S, Ravaud A, Golding S, Jethwa S and
Sneller V: Phase III trial of bevacizumab plus interferon alfa-2a
in patients with metastatic renal cell carcinoma (AVOREN): Final
analysis of overall survival. J Clin Oncol. 28:2144–2150. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao Y, Linden P, Farnebo J, Cao R,
Eriksson A, Kumar V, Qi JH, Claesson-Welsh L and Alitalo K:
Vascular endothelial growth factor c induces angiogenesis in vivo.
Proc Natl Acad Sci USA. 95:pp. 14389–14394. 1998, View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su JL, Yang PC, Shih JY, Yang CY, Wei LH,
Hsieh CY, Chou CH, Jeng YM, Wang MY, Chang KJ, et al: The
VEGF-C/Flt-4 axis promotes invasion and metastasis of cancer cells.
Cancer Cell. 9:209–223. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dias S, Choy M, Alitalo K and Rafii S:
Vascular endothelial growth factor (VEGF)-c signaling through Flt-4
(VEGFR-3) mediates leukemic cell proliferation, survival, and
resistance to chemotherapy. Blood. 99:2179–2184. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Scherle P, Behrens T and Staudt LM:
Ly-GDI, a GDP-dissociation inhibitor of the RHoA GTP-binding
protein, is expressed preferentially in lymphocytes. Proc Natl Acad
Sci USA. 90:pp. 7568–7572. 1993, View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bromwich EJ, McArdle PA, Canna K, McMillan
DC, McNicol AM, Brown M and Aitchison M: The relationship between
t-lymphocyte infiltration, stage, tumour grade and survival in
patients undergoing curative surgery for renal cell cancer. Br J
Cancer. 89:1906–1908. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang QJ, Hanada KI, Robbins PF, Li YF and
Yang JC: Distinctive features of the differentiated phenotype and
infiltration of tumor-reactive lymphocytes in clear cell renal cell
carcinoma. Cancer Res. 72:6119–6129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mehta P, Wavreille AS, Justiniano SE,
Marsh RL, Yu J, Burry RW, Jarjoura D, Eubank T, Caligiuri MA,
Butchar JP and Tridandapani S: LyGDI, a novel ship-interacting
protein, is a negative regulator of FcγR-mediated phagocytosis.
PLoS One. 6:e211752011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Groysman M, Hornstein I, Alcover A and
Katzav S: Vav1 and Ly-GDI two regulators of RHo GTPases, function
cooperatively as signal transducers in T cell antigen
receptor-induced pathways. J Biol Chem. 277:50121–50130. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Garcia-Cozar FJ, Okamura H, Aramburu JF,
Shaw KT, Pelletier L, Showalter R, Villafranca E and Rao A:
Two-site interaction of nuclear factor of activated t cells with
activated calcineurin. J Biol Chem. 273:23877–23883. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
VON Klot CA, Merseburger AS and Kuczyk MA:
Novel therapeutic options for second-line therapy in metastatic
renal cell carcinoma. Mol Clin Oncol. 4:903–908. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based human protein atlas. Nat
Biotechnol. 28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Image (43)/ARHGDIB/data available from
v16.1, . proteinatlas.org. https://www.proteinatlas.org/ENSG00000111348-ARHGDIB/cancer/tissue/renal+cancer#img
|
|
47
|
Balch CM, Riley LB, Bae YJ, Salmeron MA,
Platsoucas CD, von Eschenbach A and Itoh K: Patterns of human
tumor-infiltrating lymphocytes in 120 human cancers. Arch. Surg.
125:200–205. 1990.
|