Introduction
Renal cell carcinoma (RCC), the most common type of
kidney cancer, accounts for 2–3% of all malignancies in adults
(1). However, RCC is highly resistant
to conventional anticancer treatments, including radio- and
cytotoxic chemotherapy. Previous studies have demonstrated that RCC
cell lines are resistant to the apoptosis induced by chemical or
immunological preparations and radiation (2). Apoptosis, the most well-known form of
programmed cell death, is a controlled, energy-dependent process.
Strategies to combat the resistance of tumors to apoptosis through
extrinsic and intrinsic pathways are being investigated. Once the
glutathione protease (caspase) family is activated, apoptosis is
triggered (3). Inhibitors of
apoptosis proteins (IAP) directly bind to and inhibit the caspases,
serving a vital role in the regulation of cell apoptosis (4). The X-linked IAP, the most potent and
ubiquitous caspase inhibitor in the IAP family, is the downstream
inhibitor of apoptosis (5). XIAP is
able to restrain apoptosis through suppression of the apoptosis
initiation factor, caspase-9, and the implementation factors,
caspase-3 and −7 (6). XIAP is highly
expressed in various malignancies and is weakly expressed or absent
in healthy cells (7). A previous
study demonstrated that XIAP expression is markedly upregulated in
RCC compared with that in healthy kidney cells (8). Additionally, in our previous study, an
upregulation and a stage- and grade-dependent increase in
anti-apoptotic XIAP expression in RCC were observed through
analysis of XIAP expression in the primary tumor tissue of 66 RCC
patients (9). Given its role in
apoptosis and its frequently elevated expression in malignant
cells, XIAP may be a promising therapeutic target in cancer
(10). Numerous studies have
demonstrated that if XIAP expression were to be inhibited by
antisense oligonucleotides or small interfering RNA (siRNA), the
proliferation of tumor cells may be suppressed, cell apoptosis
induced and malignant cells sensitized to chemotherapeutic agents
(11,12). However, a previous study on the
biological functions of XIAP focused on the direct competitive
inhibition of caspases by its baculoviral IAP repeat (BIR) 1, BIR2
and BIR3 domains (13). Further
studies have demonstrated that the RING domain of XIAP, which has
E3 ubiquitin ligase activity, is able to promote the
self-ubiquitination degradation of XIAP and its negative regulatory
protein, second mitochondria-derived activator of caspase/direct
inhibitor of apoptosis-binding protein with low pl. As a result of
the E3 ubiquitin ligase activity of its specific RING domain, XIAP
is able to attach to target proteins to determine their specificity
to ubiquitination by recognizing target substrates and promoting
protein degradation (14). A previous
study demonstrated that XIAP mediated rapid mouse double minute 2
homolog (Mdm2) degradation through intrinsic E3 ubiquitin ligase
activity and regulation of the Mdm2-tumor protein p53 (p53)
signaling pathway (15). A small
number of reports have demonstrated that the XIAP RING domain
influences certain signaling cascades associated with cell death,
inflammation and cell migration (16,17).
Therefore, it remains unclear whether or not XIAP also participates
in the regulation of other pathways by means of ubiquitin-mediated
degradation of specific substrates by its RING domain. It may be
beneficial to elucidate the currently unknown biological functions
of XIAP in RCC.
In the present study, the Caki-1 cell line with XIAP
overexpression was selected and a stably transfected Caki-1 cell
line with XIAP-knockdown was established using RNA interference
technology. The general dynamic difference in proteins between the
2 cell lines was determined through proteomic analysis, using
isobaric tag for relative and absolute quantitation (iTRAQ)
technology, during apoptosis induced by etoposide. Through
determination of the significantly differentially expressed
proteins, certain features in the potential downstream regulatory
proteins of XIAP were observed. These results suggest that XIAP may
serve important roles in a diverse set of non-apoptotic signaling
pathways in RCC and may have potential value in tumor gene
therapy.
Materials and methods
Establishing Caki-1 cell lines with a
deficiency in XIAP expression
Caki-1 cells were purchased from China
Infrastructure of Cell Line Resources (Beijing, China). The cells
were grown in minimum essential medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Zhejiang Kangyuan Biological Technology Co.,
Ltd., Zhuji, China) and maintained at 37°C in a humidified
incubator with 5% CO2. The cells were split twice weekly
and cells that were in the logarithmic growth phase were used for
experiments.
The XIAP interference target of the present study
was based upon that in the study by Bilim et al (18), which had verified that there were no
homologous sequences between this fragment and other genes, and
that the short hairpin RNA (shRNA) containing this fragment
effectively downregulated the expression of XIAP. The target
sequence was 5′-AGGTGAAGGTGATAAAGTA-3′. The BLOCK-iT™ U6 RNAi Entry
Vector kit (Invitrogen; Thermo Fisher Scientific, Inc.) was applied
for construction. Based upon the selected target sequence, the
oligonucleotide sequences of shRNA used were: Top strand,
5′-CACCGAGGTGAAGGTGATAAAGTACGAATACTTTATCACCTTCACC-3′, and bottom
strand, 5′-AAAAGGTGAAGGTGATAAAGTATTCGTACTTTATCACCTTCACCTC-3′.
These shRNA-encoding complementary single stranded
oligonucleotides were hybridized to give XhoI- and
HindIII-compatible overhangs prior to being ligated into
pENTR™/U6. The constructed plasmids were confirmed by sequencing
and were termed XIAP-shRNA-pENTR/U6.
Transfection was performed using
Lipofectamine® (250 ng/µl) 2000 transfection reagent
(Gibco; Thermo Fisher Scientific, Inc.) and a BLOCK-iT U6 RNAi
Entry Vector kit (Invitrogen; Thermo Fisher Scientific, Inc.). In
order to generate stably transfected clones, the transfected cells
were selected with Geneticin (KangWei Technology, Beijing, China)
for 3–4 weeks (the successfully transfected Caki-1 cell exhibited
Geneticin resistance). The stably transfected Caki-1 cell clones
were cloned and collected, and were termed Caki-1/XIAP-shRNA-pENTR,
with Caki-1/pENTR as the control cells. Due to the fact that the
PENTR/U6 plasmid possesses the GFP gene, it is able to produce
green fluorescence under a certain wavelength laser and thus, a
fluorescence microscope (Thermo Fisher Scientific, Inc.) was used
to identify transfected cells. Following formation of the clone
cells, the monoclonal cell colony with green fluorescence was
selected and transferred onto a 24-well plate for
amplification.
Whole cell proteins were isolated using a
radioimmunoprecipitation assay reagent (Beyotime Institute of
Biotechnology, Haimen, China) and an equal mass of total protein
(40 µg) from each lysate was loaded onto 10% SDS-PAGE. Following
electrophoresis, separated proteins were transferred onto
polyvinylidene difluoride membranes and were blocked with TBST
containing 10% powdered skimmed milk for 2 h at room temperature
prior to being incubated with antibodies against XIAP (cat no.
PRS3331; 1:2,000) and GAPDH (cat no. G9545; 1:20,000; both
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) overnight at 4°C.
Next, the substrate was catalyzed to emit light, the exposure
stripe was scanned, and the ratio of the grey values of XIAP and
GAPDH was taken as the relative content of XIAP protein.
Cell viability was determined by MTT assay. An MTT
kit (Gibco; Thermo Fisher Scientific, Inc.) was used to detect the
inhibition rate of cells at varying drug concentrations (1, 5, 10,
50, 100 µg/ml). Etoposide was used as an apoptosis inducer. DMSO
was used to dissolve the purple formazan, and viability was
subsequently analyzed at a wavelength of 570 nm. In an additional
experiment, in order to improve the experimental efficiency, flow
cytometry with BD FACSDiva Software 6.0 (BD Biosciences, Franklin
Lakes, NJ, USA) was used to detect early apoptosis according to the
manufacturer's protocol. The percentage of apoptotic cells was
detected by flow cytometry using an Annexin V/FITC and PI Apoptosis
Detection kit (BD Biosciences; cat. no. 556547; Annexin V-FITC
Apoptosis Detection kit). BD Cellquest Pro software (version 5.1;
BD Biosciences) was used to analyze the experimental data.
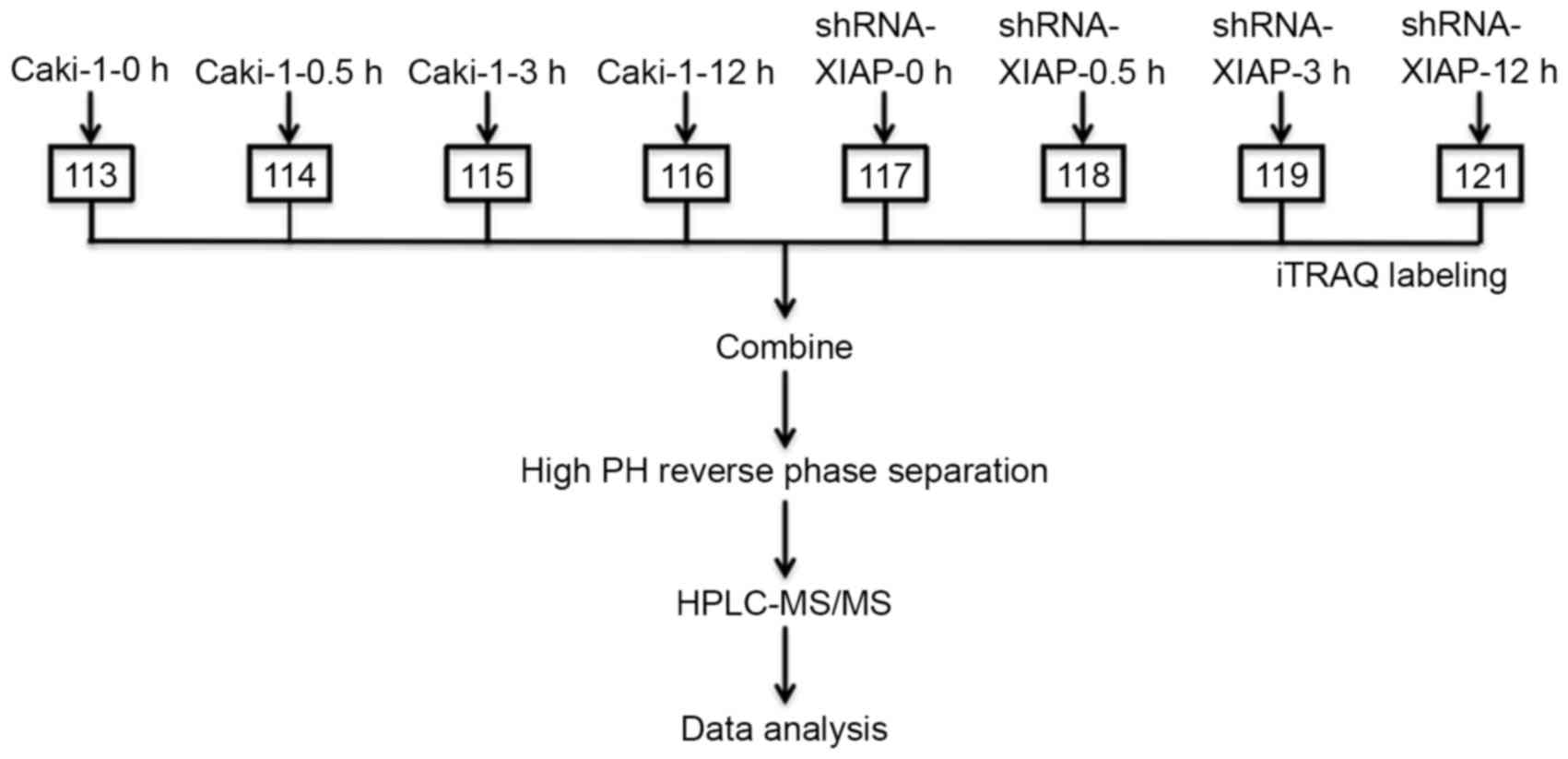
iTRAQ and mass spectrometry
The Caki-1 (overexpression of XIAP) and
XIAP-shRNA-pENTR/U6 (XIAP-knockdown) renal cell lines were
harvested. The two cell lines were cultured with 10% FBS McCoy's 5A
medium (cat no. SH30200.01; Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA), and the cells were digested with 0.25% trypsin
(cat no. REK3011; OXOID; Thermo Fisher Scientific, Inc.). Each of
the cell lines was divided into 8 T-75 cell culture bottles, each
with 15 ml cell suspension. Caki-1 cells and XIAP-shRNA cells were
treated with 60 µg/ml etoposide for 0.5, 3 and 12 h at 37°C. The
control group was set at 0 h. Samples were ground in liquid
nitrogen.
Cells were washed with phosphate-buffered saline and
were collected by incubation (5% CO2 and 37°C for 1 h)
with 2 ml cellular lysis buffer (8 M urea, 30 mM HEPES, 1 mM PMSF,
2 mM EDTA and 10 mM DTT). Ultrasonic processing (pulse on for 2
sec, pulse off for 3 sec; power, 180 W) of this solution was then
performed using an ultrasonic processor and centrifugation at
20,000 × g for 30 min at 4°C. The supernatant was transferred to a
fresh tube and stored at −80°C until use. For each sample, proteins
were precipitated with ice-cold acetone prior to being redissolved
in dissolution buffer (50% TEAB and 0.1% SDS). The proteins were
quantified by the Bradford protein assay (Promega Corporation,
Madison, WI, USA). Subsequently, 100 mg protein was tryptically
digested and the resultant peptide mixture was labeled using the
iTRAQ reagent (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The protein solution was further digested with sequence-grade
modified trypsin. For labeling, each iTRAQ reagent was dissolved in
70 µl isopropanol and added to the respective peptide mixture.
Protein samples were labeled with iTRAQ isobaric tags: 113, 114,
115, 116, 117, 118, 119 and 121.
To reduce sample complexity during liquid
chromatography tandem mass spectrometry (LC-MS/MS) analysis (Strong
cation exchange chromatography; SCX), the iTRAQ labeled peptide
samples were reconstituted in Buffer A (25% ACN, 10 mM
KH2PO4; pH 3.0) and fractionated using
Ultremex SCX column (Phenomenex, Torrance, CA, USA; Luna SCX 100A)
via the LC-20AB HPLC Pump system (Shimadzu Corporation, Kyoto,
Japan) at a flow rate of 1.0 ml/min with a gradient of Buffer B
(25% ACN, 2 M KCL, 10 mM KH2PO4; pH 3.0).
Buffer B reached 100% in 10 min. The column flow rate was
maintained at 400 nl/min, and the column temperature was maintained
at room temperature and at a pressure of 1,000 psi. The collected
fractions were desalted using Strata X C18 column (Phenomenex), and
were vacuum centrifuged (4°C and 20,000 × g for 30 min) and
reconstituted in 0.1% formic acid for subsequent LC-MS/MS analysis.
The mass spectroscopy (MS) analysis was performed using a Q
Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo
Fisher Scientific, Inc.). Nanoflow electrospray ionization MS/MS
analysis of peptide samples was performed using an Eksigent
NanoLC-2D system (AB SCIEX, Framingham, MA, USA).
MS and MS/MS data searches were performed using the
Proteome Discoverer 1.3 (PD; Thermo Fisher Scientific, Inc.) based
upon the workflow with a spectrum selector and reporterion
quantifier. The proteins were identified and quantified using the
Mascot search engine (version 2.3.0; Matrix Science Inc., Boston,
MA, USA). To minimize false positive results, a strict cutoff for
protein identification was applied with an Unused Prot Score ≥1.3,
which corresponds to a confidence limit of 95%, and two peptides
with 95% confidence were considered for protein quantification.
The functional annotations were obtained from the
Gene Ontology database (http://www.geneontology.org/). The metabolic pathways
associated with the differentially expressed proteins were
classified using the Kyoto Encyclopedia of Genes and Genomes (KEGG)
(19).
Statistical analysis
Using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA)
statistical software for analysis, the difference between two
groups was compared using Student's t-test, and the difference
among more than two groups was compared using a one-tailed analysis
of variance followed by Duncan's multiple-range test. P<0.05 was
considered to indicate a statistically significant difference.
Results
RNA interference effectively decreases
XIAP expression
In order to decrease the expression of XIAP in
Caki-1 cells, sequence-specific siRNA of XIAP was synthesized in
the aforementioned manner. In order to generate stably transfected
clones, the transfected cells were selected for 3–4 weeks. Three of
the stably transfected cell clones were screened out and one was
selected for further experiments. The interference efficiency of
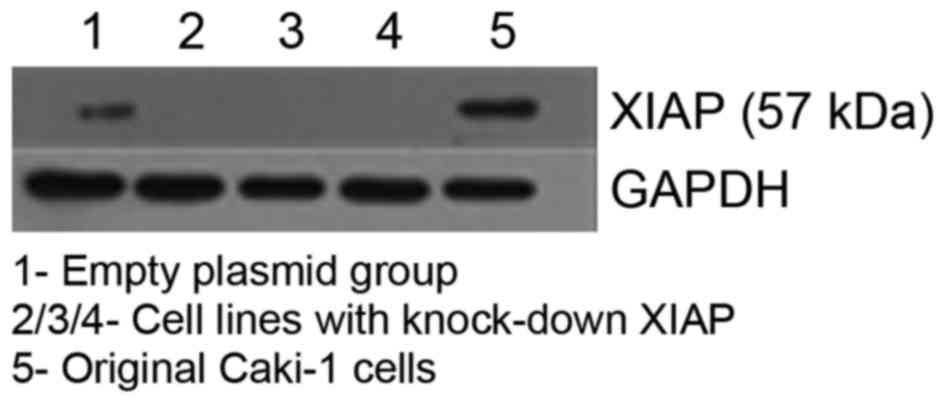
XIAP-knockdown cells was confirmed by western blot analysis. As
demonstrated in Fig. 1, XIAP shRNA
transfection markedly reduced the expression of XIAP, while no
significant difference was observed between XIAP expression in the
Caki-1 control group and that in the pENTR/U6 group.
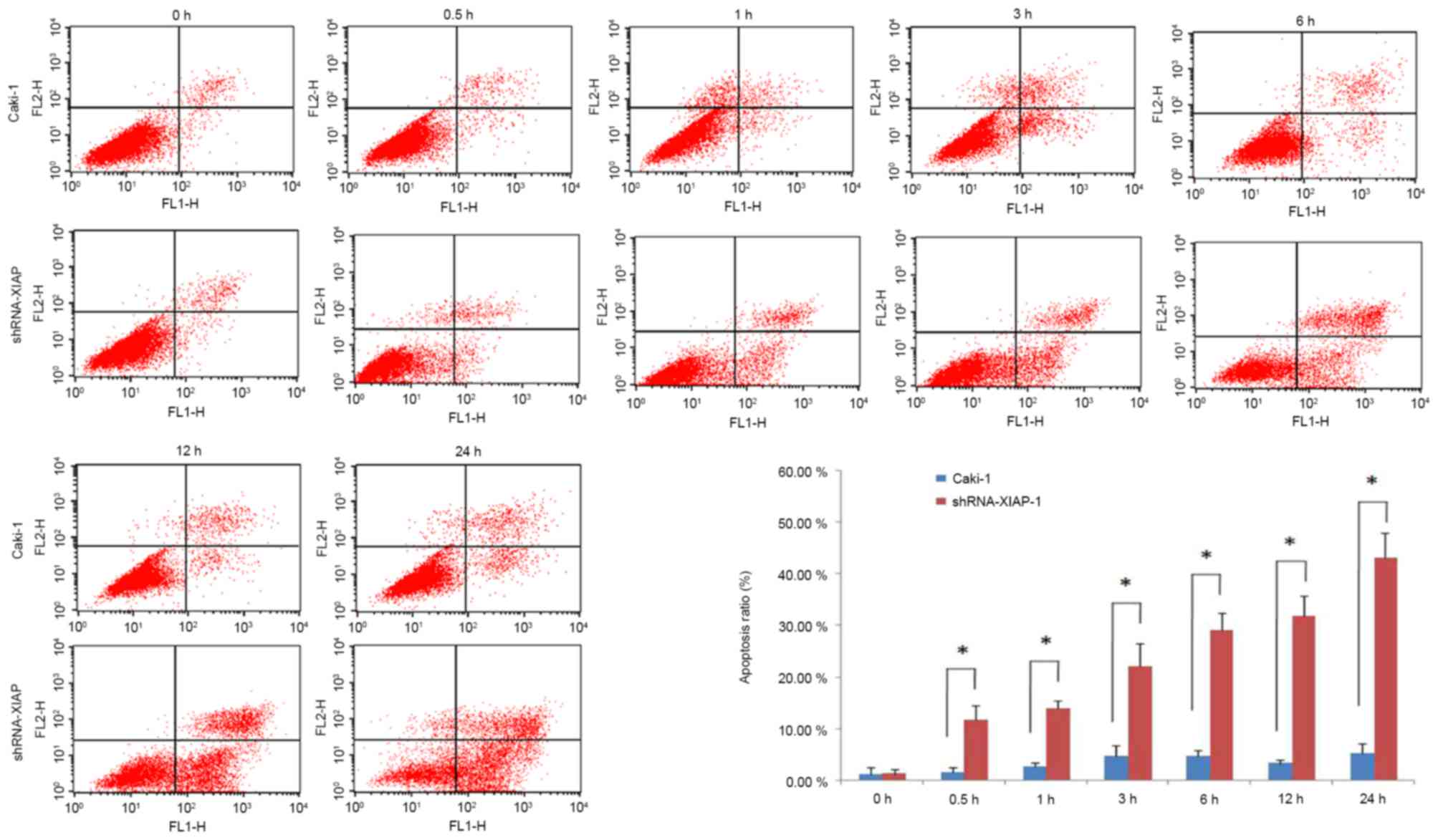
Stably transfected XIAP-knockdown
Caki-1 cell line is more sensitive to apoptosis induction than the
original Caki-1 cell line
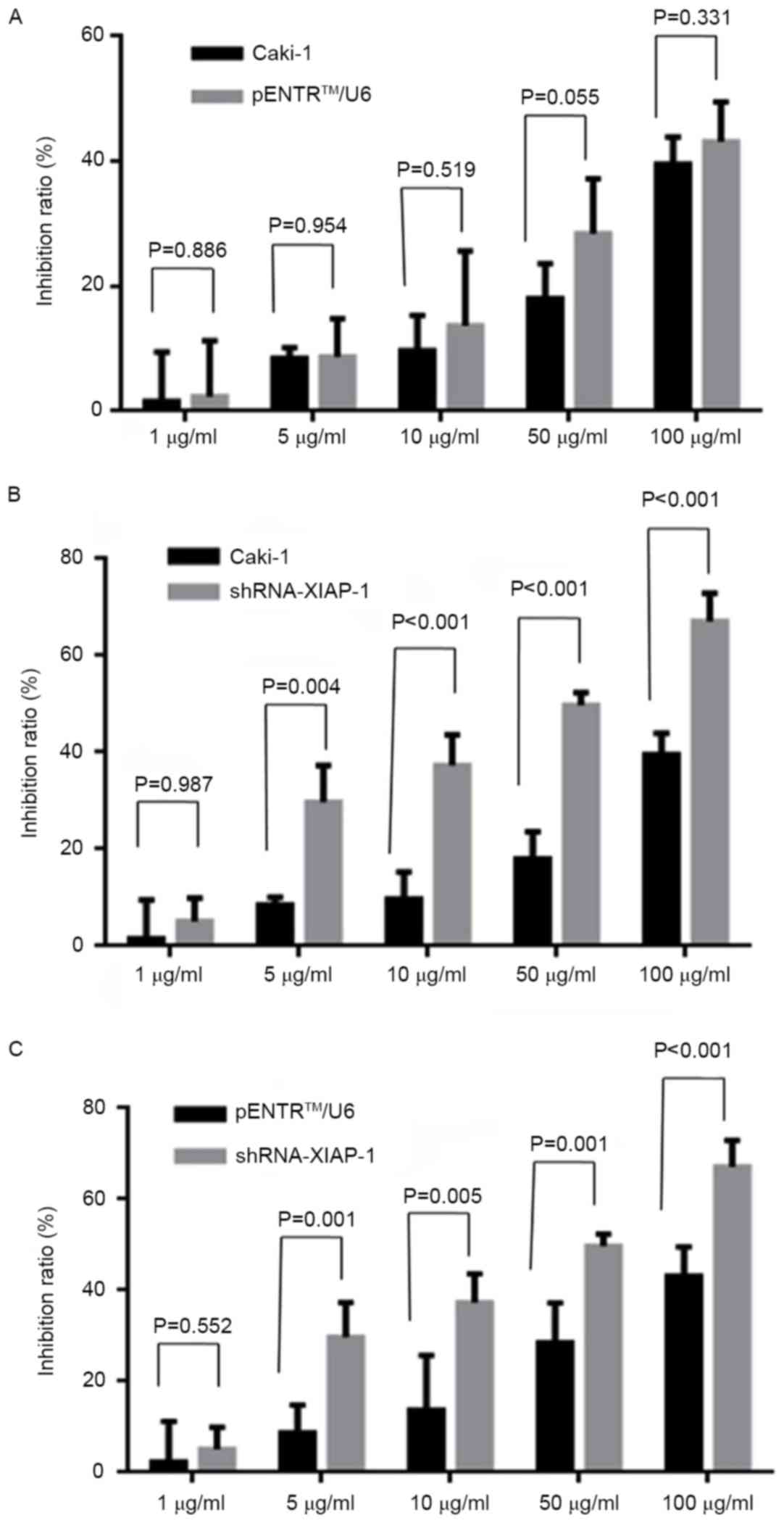
To further evaluate whether XIAP-knockdown would
influence the anti-apoptotic ability of Caki-1 cells, the MTT
method was used to detect the cell death rate under different drug
concentrations (1, 5, 10, 50 and 100 µg/ml) and flow cytometry was
used to detect the apoptosis rate at 0, 0.5, 1, 3, 6, 12 and 24 h.
As demonstrated in Fig. 2, following
XIAP-knockdown, the cell death rate was significantly increased
under all drug concentrations ≥5 µg/ml, while there was no
statistically significant difference in the cell death rate between
the original Caki-1 group and the pENTR/U6 group. Additionally, the
inhibition ratio of the 50 µg/ml treatment group was significantly
greater compared with that of the 0 µg/ml group between shRNA and
Caki-1 cell lines. The results of the flow cytometry further
demonstrated that the occurrence of apoptosis in the cell line with
XIAP-knockdown was significantly higher than that in the original
Caki-1 cell line (Fig. 3).
Number of differentially expressed
proteins determined by iTRAQ profiling
To investigate the effect of XIAP on apoptosis,
iTRAQ-based quantitative proteomics were performed on Caki-1 cells
and XIAP-knockdown cells treated with etoposide for 0, 0.5, 3 and
12 h (Fig. 4).
A total of 1,783 proteins were identified and
quantified by the iTRAQ technique. Statistical calculations for
iTRAQ-based detection and relative quantification were then
performed using PD software. The protein identification threshold,
an Unused Prot Score of >1.3, was used to obtain a confidence
level of 95%. The differentially expressed proteins were defined as
the relative ratio of >1.5 or <0.5 (calculated as a ratio of
Caki-1 cells/XIAP-knockdown cells; P<0.05). Based upon these
criteria, 87 proteins were significantly altered at 0 h. Following
apoptosis induction, there were 178 significantly altered proteins
at 0.5 h and 169 proteins at 3 h. However, no difference was
observed in the level of protein expression between the two cell
lines at 12 h, indicating that the change in cell protein levels
mainly occurs in the early stages of apoptosis.
Differentially expressed proteins are
implicated in a number of pathways and biological processes
In order to further understand the role of XIAP in
cell death and to screen for currently unknown biological behaviors
of XIAP, the differentially expressed proteins were dynamically
analyzed at 0, 0.5, 3 and 12 h. At the 12-h stage of apoptosis, no
differential proteins were observed (under the conditions of a
ratio >1.5 or <0.5; P<0.05). Therefore, the focus was on
the early stage of apoptosis between 0 and 3 h. As demonstrated in
Table I, the ratio
(Caki-1/shRNA-XIAP-1) of 22 proteins was significantly altered at
0, 0.5 and 3 h in response to etoposide treatment. Subsequently, 22
differentially expressed proteins associated with XIAP were
classified using the Protein Analysis through Evolutionary
Relationships classification system (www.pantherdb.org) to obtain a better understanding of
their molecular and functional characteristics. The results
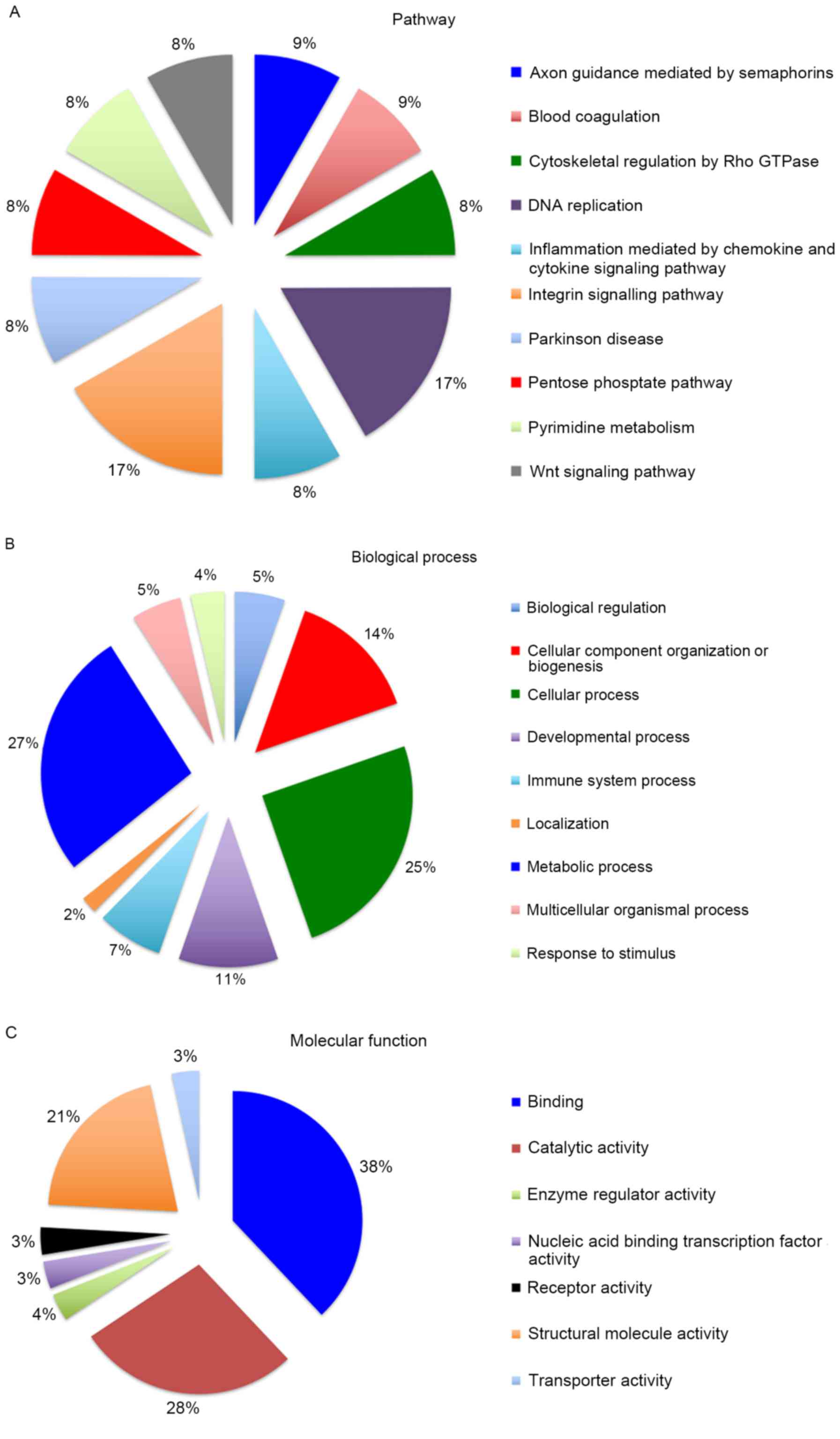
revealed the distribution of differentially expressed proteins in
pathways, biological processes and molecular functions (Fig. 5). These results indicate that the
differentially expressed proteins are involved in a number of
pathways and biological processes, suggesting that XIAP has a
number of potential physiological functions. The relevant
literature and the existing data were then combined to screen out
the biological behaviors associated with the onset and progression
of cancer. In general, differentially expressed proteins were
implicated in the p53 pathway, cytoskeletal regulation, histone
regulation, the Wnt signaling pathway, glucose metabolism and
endoplasmic reticulum (ER) stress. Representative MS/MS spectra of
select peptides with their reporter ions are presented as the 6
proteins, reticulocalbin 3 (RCN3), lactate dehydrogenase A,
UDP-glucose 6-dehydrogenase (UGDH), histone H3.3, S100
calcium-binding protein A4 (S100A4) and collagen, type 1, α-1
(COL1A1) (Fig. 6).
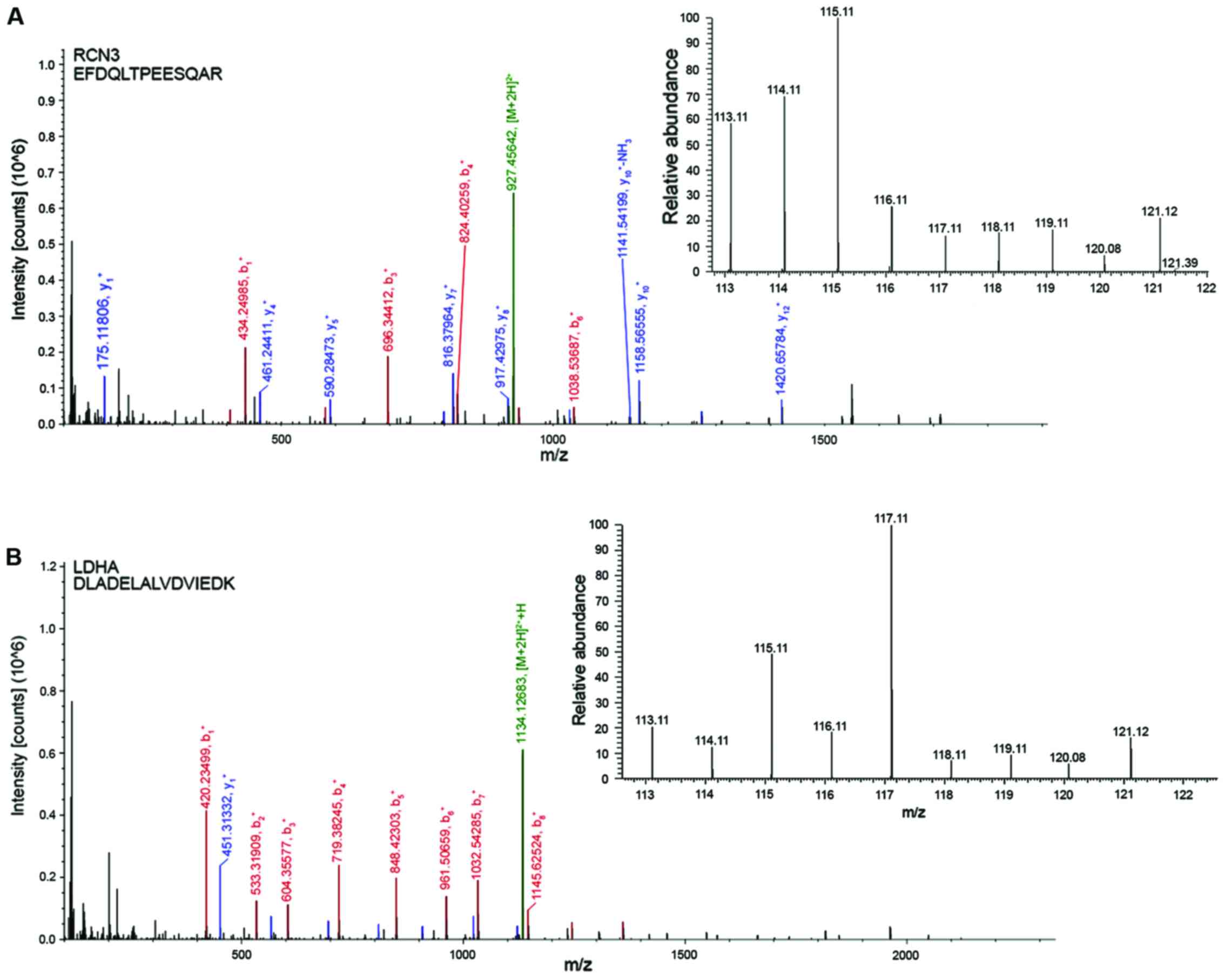 | Figure 6.Tandem mass spectrometry of select
peptides with their reporter ions for the 6 proteins. (A) RCN3, (B)
LDHA, (C) UGDH and (D) histone H3.3. 117. (E) S100A4 and (F)
COL1A1. Representative peptide sequencing and quantification using
iTRAQ with indicated amino acid sequences, annotated b-ion and
y-ion series, and an expanded view of the reporter ion region
demonstrating representative relative abundances of signature iTRAQ
ions at m/z 113, 114, 115, 116, 117, 118, 119 and 121. RCN3,
reticulocalbin 3; LDHA, lactate dehydrogenase; UGDH,
UDP-glucose-6-dehydrogenase; S100A4, S100 calcium-binding protein
A4; COL1A1, collagen, type 1, α1; iTRAQ, isobaric tag for relative
and absolute quantitation. |
 | Table I.Differentially expressed
proteins. |
Table I.
Differentially expressed
proteins.
|
|
| Ratio at different
time points (Caki-1/XIAP knockdown) |
|---|
|
|
|
|
|---|
| Accession no. | Description | 0 h | 0.5 h | 3 h | 12 h |
|---|
| P26447 | Protein S100-A4
OS=Homo sapiens GN=S100A4 [S10A4_HUMAN] | 0.219 | 0.588 | 0.548 | 1.076 |
| P08729 | Keratin, type II
cytoskeletal 7 GN=KRT7 [K2C7_HUMAN] | 0.336 | 0.469 | 0.452 | 1.021 |
| P25815 | Protein S100-P
OS=Homo sapiens GN=S100P [S100P_HUMAN] | 0.215 | 0.424 | 0.501 | 1.022 |
| P00338 | L-lactate
dehydrogenase A chain GN=LDHA [LDHA_HUMAN] | 0.429 | 1.538 | 1.895 | 1.027 |
| P29401 | Transketolase
GN=TKT [TKT_HUMAN] | 0.503 | 1.545 | 1.502 | 1.016 |
| P08195 | 4F2 cell-surface
antigen heavy chain GN=SLC3A2 [4F2_HUMAN] | 0.560 | 0.463 | 0.548 | 1.020 |
| P09493 | Tropomyosin a-1
chain GN=TPM1[TPM1_HUMAN] | 1.488 | 2.556 | 2.575 | 1.061 |
| Q99584 | Protein S100-A13
GN=S100A13 [S10AD_HUMAN] | 1.509 | 1.666 | 2.154 | 1.103 |
| P84243 | Histone H3.3
OS=Homo sapiens GN=H3F3A [H33_HUMAN] | 1.527 | 0.268 | 0.390 | 0.845 |
| Q9NR12 | PDZ and LIM domain
protein 7 GN=PDLIM7 [PDLI7_HUMAN] | 1.557 | 1.873 | 1.568 | 1.022 |
| Q9ULV4 | Coronin-1C
GN=CORO1C [COR1C_HUMAN] | 1.592 | 1.668 | 1.564 | 1.003 |
| P12814 | α-actinin-1
GN=ACTN1 [ACTN1_HUMAN] | 1.758 | 2.184 | 2.353 | 1.036 |
| O60701 | UDP-glucose
6-dehydrogenase GN=UGDH [UGDH_HUMAN] | 1.865 | 1.703 | 1.682 | 1.043 |
| P50914 | Spermine synthase
GN=SMS [SPSY_HUMAN] | 1.901 | 2.450 | 2.603 | 0.926 |
| P68431 | Histone H3.1
OS=Homo sapiens GN=HIST1H3A [H31_HUMAN] | 1.914 | 0.295 | 0.327 | 0.699 |
| P02452 | Collagen α-1(I)
chain GN=COL1A1 [CO1A1_HUMAN] | 1.974 | 1.930 | 1.819 | 1.070 |
| Q14195 |
Dihydropyrimidinase-related protein 3
GN=DPYSL3 [DPYL3_HUMAN] | 2.017 | 2.152 | 1.970 | 1.154 |
| P23528 | Cofilin-1 GN=CFL1
[COF1_HUMAN] | 2.040 | 2.056 | 3.164 | 1.082 |
| P01023 | α-2-macroglobulin
GN=A2M [A2MG_HUMAN] | 2.054 | 2.212 | 1.722 | 1.023 |
| P09936 | Ubiquitin
carboxyl-terminal hydrolase isozyme L1 GN=UCHL1 [UCHL1_HUMAN] | 2.661 | 2.392 | 2.683 | 1.112 |
| P02795 | Metallothionein-2
OS=Homo sapiens GN=MT2A [MT2_HUMAN] | 3.839 | 3.102 | 3.176 | 1.158 |
| Q96D15 | Reticulocalbin-3
GN=RCN3 [RCN3_HUMAN] | 4.607 | 3.306 | 3.580 | 1.193 |
p53 pathway
As demonstrated in Table
I, S100A4 and S100 calcium-binding protein P (S100P) were
downregulated in the Caki-1 cells. The S100 protein family belongs
to the calcium-binding protein family, members of which have a
similar structure, but a different function. S100A4 may be combined
with the regulatory region of the p53 protein C terminal, and
through protein kinase C, may inhibit the phosphorylation reaction
of the full-length p53 protein (20).
S100A4 may also promote tumor invasion and metastasis (21). The results of the present study also
demonstrated that there was a significant change (P=0.002) in the
members of the S100 family following apoptotic stimulation
(Fig. 7A), which indicates that XIAP
may affect the p53 signaling pathway by regulating associated S100
family proteins.
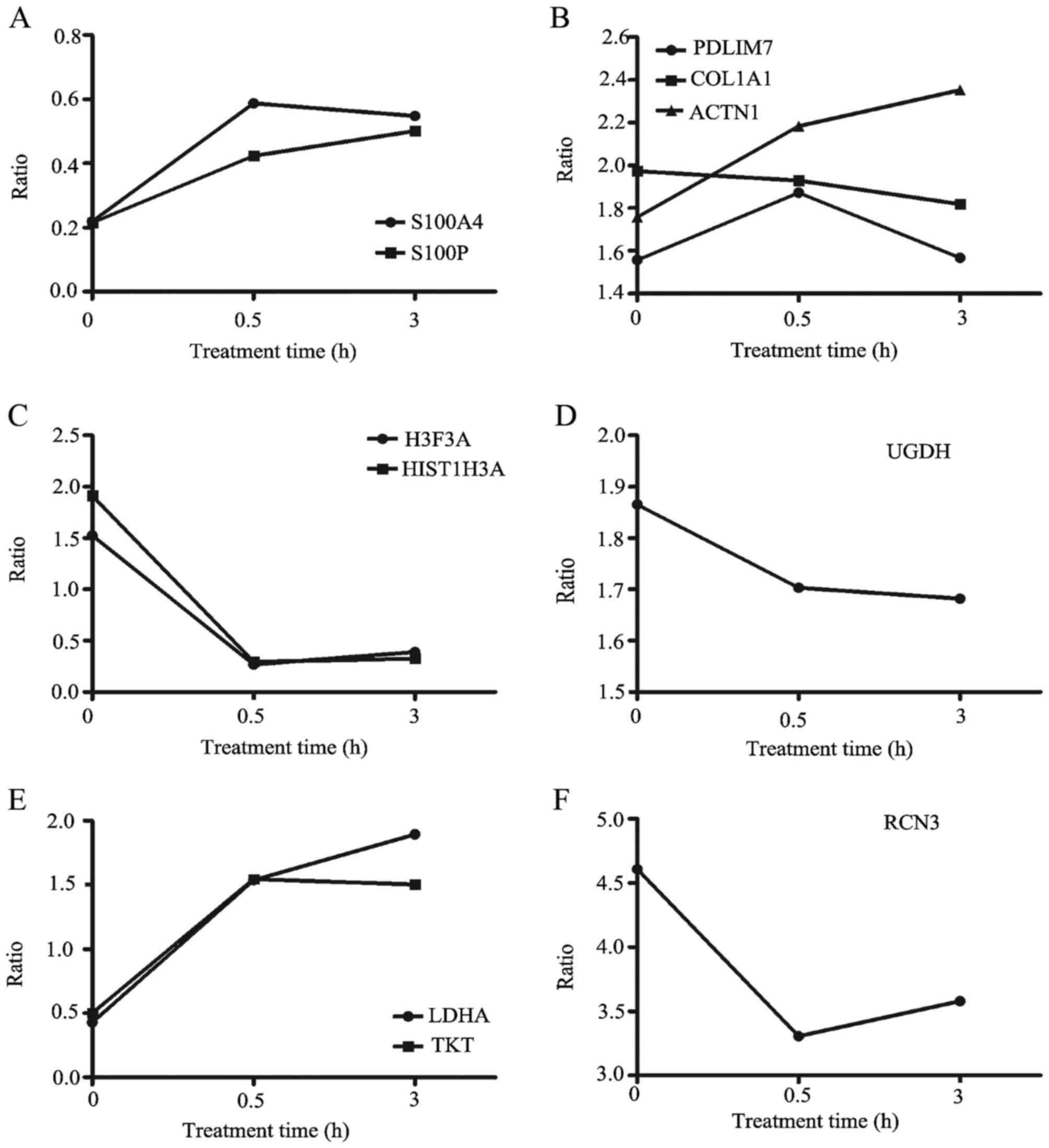 | Figure 7.Changes in differentially expressed
proteins. The x-axis represents the time of apoptosis induction (0,
0.5 and 3 h) and the y-axis represents the relative ratio of
proteins (Caki-1/shRNA-XIAP-1). The relative ratio fold-change of
(A) S100A4 and S100P, (B) PDLIM7, COL1A1 and ACTN1, (C) H3F3A and
HIST1H3A, (D) UGDH, (E) LDHA and TKT and (F) RCN3. shRNA, short
hairpin RNA; XIAP, X-linked inhibitor of apoptosis protein; S100A4,
S100 calcium-binding protein A4; S100P, S100 calcium-binding
protein P; COL1A1, collagen, type 1, α1; ACTN1, α-actinin-1; UGDH,
UDP-glucose-6-dehydrogenase; LDHA, lactate dehydrogenase; TKT,
transketolase; RCN3, reticulocalbin 3. |
Cytoskeletal regulation
Overall, 3 proteins involved in cytoskeletal
organization were upregulated in Caki-1 cells and the relative
ratios were all >1.5 at the three time points (Fig. 7B). Among them, α-actinin-1 (ACTN1)
cross-linked with the actin cytoskeleton, through cadherin,
integrins, vinculin, catenin or talin, and mediated the adhesion of
adjacent cells or the adhesion of cells to the extracellular matrix
(22). In addition, following the
induction of apoptosis, the level of ACTN1 differed more markedly
(Fig. 7B). Additionally, the
expression of COL1A1, which can promote cell adhesion and cell
proliferation, was also increased (23). The cytoskeletal PDZ-LIM protein family
has a PDZ domain with a scaffolding function through which the
coordinated assembly of proteins can occur. These results indicate
that XIAP participates in cytoskeletal regulation through ACTN1,
COL1A1 and PDLIM7.
Histone regulation
In the present study, two histone proteins were
altered. Not only does histone modification regulate gene
expression, but it also recruits the protein complex, affecting the
downstream proteins and thereby participating in cell division,
cell apoptosis and memory formation, as well as affecting the
immune system and inflammatory response. The phosphorylation,
acetylation, methylation and ubiquitination of histone proteins can
be used as a marker of DNA injury, and recruited repair proteins
serve an important role in the process of DNA repair (24,25). The
present results indicated that the ratios (Caki-1/shRNA-XIAP-1) of
two histone proteins were upregulated at 0 h, but were decreased at
0.5 and 3 h (Fig. 7C). These results
suggest that XIAP serves an important role in histone
regulation.
Wnt signaling pathway
The enzyme UGDH catalyzes the two-fold oxidation of
UDP-glucose into UDP-glucuronic acid, the activated nucleotide
sugar donor required for the synthesis of heparin sulfate
glycosminoglycans (26). To a large
extent, the biological functions of heparin sulfate proteoglycans
(HSPGs) depend upon the interaction of the glycosaminoglycan chains
with different protein ligands. HSPGs are present at the cell
surface and in the extracellular matrix. Through their projection
into the extracellular space, HSPGs are capable of interacting with
signaling molecules and can ultimately influence the activity route
of their recipient cell. For example, HSPGs can influence the
activities of the Wnt growth factor (27). Table I
indicates that the expression of UGDH in Caki-1 cells is
significantly higher than that in the XIAP-knockdown cells.
However, no change was observed prior to and following the
induction of apoptosis (Fig. 7D).
These results suggest that XIAP influences the Wnt pathway.
Cellular glucose metabolism
Table I demonstrates
that two proteins, LDHA and transketolase (TKT), were involved in
cell glucose metabolism. These two proteins were downregulated at 0
h (ratio <0.5) and upregulated at 0.5 and 3 h (ratio >1.5;
Fig. 7E). TKT, a thiamine
diphosphate-dependent enzyme, catalyzes several key reactions of
the non-oxidative branch of the pentose phosphate pathway. TKT
catalyzes the conversion of D-pentose (xylulose and ribose)
5-phosphate into D-glyceraldehyde 3-phosphate and D-sedoheptulose
7-phosphate (28). LDHA is a
glycolytic enzyme that is catalyzed into a pyruvate lactic acid
enzyme, existing in almost all tissues; it serves a key role in the
regulation of the conversion of glucose fermentation and aerobic
oxidation. In a previous study, the tumor cells exhibited the
characteristics of anaerobic fermentation and inhibition of
mitochondrial aerobic oxidation (29).
ER stress
RCN3 is a novel ER-resident calcium-binding protein
with multiple EF-hand motifs and a carboxyl-terminal HDEL sequence
(30). There is crosstalk between the
unfolded protein reaction and calcium signals in the ER. The
destruction of the calcium pool in the ER may lead to ER stress
(31). Table I demonstrates that the ratio of RCN3
is markedly upregulated prior to and following the induction of
apoptosis (Fig. 7F). Therefore, XIAP
may be associated with ER stress.
Discussion
XIAP has been revealed to be an important regulator
of cell apoptosis. The overexpression of XIAP may reduce the
sensitivity of RCC cells to apoptosis induced by extrinsic or
intrinsic factors, and may provide favorable conditions for tumor
cell survival and development. Inhibition of XIAP by a chemical
inhibitor or siRNA has been reported to reduce the growth of tumor
cells (32). Additionally, in recent
years, with the continuous development of gene knockout technology
in animals and cells, our knowledge regarding the functions of XIAP
has expanded (33). In addition to
participating in the regulation of cell apoptosis, XIAP is also
involved in other cancer cell biological behaviors. In a previous
study, Cao et al (34)
discovered that XIAP and its E3 ligase serve an important function
with regards to regulating cyclin D1 expression in tumor cells.
Through the E3 ligase activity, XIAP may be a generalist in cancer
development. This suggests that when XIAP is used as a target
molecule for the treatment of RCC, the possible synergistic effects
and side effects of the non-apoptotic biological functions of XIAP
in tumor therapy require consideration.
Our previous study revealed that XIAP is
overexpressed in the Caki-1 cell line (9). It is easier to display the
differentially expressed proteins if only one cell line is used for
the in vitro experiments. Therefore, the Caki-1 cell line
with XIAP overexpression was selected for the present study.
Through the construction of a plasmid, along with cell
transfection, the stably transfected XIAP-knockdown Caki-1 cell
line was obtained. Additionally, iTRAQ proteomic bio mass
spectrometry was used to identify the proteins that were
differentially expressed between the Caki-1 cells with XIAP
overexpression and those with XIAP-knockdown. Combining the GO
database and the KEGG pathway, the proteins that were significantly
altered prior to and following apoptosis stimulation at 0, 0.5 and
3 h were classified. According to the biological processes and
pathways involved, the biological behaviors of RCC cells, in which
XIAP is possibly involved, were summarized.
XIAP may affect cell transformation and
tumorigenesis through the p53 and Wnt pathways in RCC. p53 protein
serves an important role in coping with different types of stress,
including DNA damage, oncogene activation and hypoxia. However, the
mutant p53 gene has emerged as a proto-oncogene that can promote
the occurrence and development of a tumor (35). The Wnt signaling pathway also serves
an important role in embryonic cell development, proliferation,
transformation, cell adhesion, cell survival and apoptosis. A
recent study demonstrated that inappropriate activation of the Wnt
signaling pathway is involved in the occurrence and metastasis of
tumor invasion (36). The present
study revealed that the expression of the protein, S100A4, which
can regulate the p53 pathway, was significantly different in the
two cell lines. Furthermore, the fold-change of UDGH demonstrated
that XIAP has other biological functions through the Wnt signaling
pathway.
XIAP may be involved in cytoskeletal regulation,
which is associated with the adhesion and invasion of tumor cells.
The cytoskeleton is associated with a variety of cellular
functions, including cell migration, ion channel activity, cell
secretion, apoptosis and cell survival (37). Actin contraction promotes migration of
tumor cells. One study indicated that the major actin regulatory
factors change during the development of cancer and are upregulated
in mobile cancer cells (38).
Phosphorylation of myosin II light chain (MLC) is one of the main
mechanisms of the regulation of myosin contraction. In addition to
phosphorylating MLC, calcium ions may promote tumor cell metastasis
through binding to S100A4 (39).
ACTN1, COL1A1, PDL1M7 and S100A4, which were all associated with
cytoskeletal regulation, were significantly altered in the two
groups in the present study.
Histone H3.1 and H3.3 exhibited a common trend of
change in the two groups at different time points, indicating that
XIAP may participate in DNA repair and replication through
influencing histone modification in RCC cells. Certain studies have
proposed the concept of the ‘code histone’, in which there are
various covalent modifications that may affect each other, change
the chromatin structure, and affect gene transcription and
replication (40). Additionally,
histone modification serves an important role in the formation and
maintenance of DNA methylation and has been revealed to be
associated with DNA damage repair (41).
The results of the present study demonstrated the
likely association between LDHA or TKT and XIAP. Therefore, XIAP
also influenced glucose metabolism in RCC cells. A series of
complex factors, including the microenvironment and gene mutations,
may cause metabolic changes in tumor cells. The expression of the
enzyme inducing the glycolytic pathway is increased and the
inhibition of mitochondrial aerobic oxidation, including the
reduction of pyruvate utilization or damage to the electron
transfer chain, further increases the activity of the glycolytic
pathway (42).
ER stress is not conducive to cell survival. Hypoxia
activates the unfolded protein response, which adapts tumor cells
to stress conditions, thereby increasing the malignancy of the
cancer (43). In addition, ER stress
serves an important role in cell apoptosis, autophagy and
necroptosis-3 main cell programmed death pathways. Therefore, we
hypothesized that XIAP serves a critical role in the regulation of
programmed cell death in RCC cells.
In summary, in the present study, Caki-1 cells with
XIAP-knockdown were successfully constructed and the differential
proteins between the XIAP overexpression and the XIAP-knockdown
cell lines were analyzed. According to these results, it was
concluded that XIAP may have a number of other biological functions
in addition to participating in the mechanism of apoptosis. For
example, XIAP may be involved in the p53 pathway, the Wnt signal
pathway, glucose metabolism, ER stress, and DNA repair and
replication. These biological behaviors are closely associated with
the progression and aggression of malignancies. Additionally, XIAP
affected the cytoskeletal regulation, migration and metastasis of
RCC cells. These findings may provide novel concepts and directions
for research regarding XIAP. However, these conclusions are solely
an inference based upon the differential expression of proteins in
two cell lines and require verification through further
experiments. With regards to the E3 ubiquitin ligase activity of
the XIAP RING domain, studies on the other potential biological
behaviors of XIAP are not comprehensive and further studies are
required to fully understand the role served by XIAP.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81441073)
and the Beijing Municipal Commission of Education Science and
Technology Plan Projects (grant no. KM201310025017).
References
|
1
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yagoda A, Abi-Rached B and Petrylak D:
Chemotherapy for advanced renal-cell carcinoma: 1983–1993. Semin
Oncol. 22:42–60. 1995.PubMed/NCBI
|
|
3
|
Chang HY and Yang X: Proteases for cell
suicide: Functions and regulation of caspases. Microbiol Mol Biol
Rev. 64:821–846. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burz C, Berindan-Neagoe I, Balacescu O and
Irimie A: Apoptosis in cancer: Key molecular signaling pathways and
therapy targets. Acta Oncol. 48:811–821. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Holcik M, Lefebvre C, Yeh C, Chow T and
Korneluk RG: A new internal-ribosome-entry-site motif potentiates
XIAP-mediated cytoprotection. Nat Cell Biol. 1:190–192. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Holcik M and Korneluk RG: XIAP, the
guardian angel. Nat Rev Mol Cell Biol. 2:550–556. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Devi GR: XIAP as target for therapeutic
apoptosis in prostate cancer. Drug News Perspect. 17:127–134. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mizutani Y, Nakanishi H, Li YN, Matsubara
H, Yamamoto K, Sato N, Shiraishi T, Nakamura T, Mikami K, Okihara
K, et al: Overexpression of XIAP expression in renal cell carcinoma
predicts a worse prognosis. Int J Oncol. 30:919–925.
2007.PubMed/NCBI
|
|
9
|
Yan Y, Mahotka C, Heikaus S, Shibata T,
Wethkamp N, Liebmann J, Suschek CV, Guo Y, Gabbert HE, Gerharz CD
and Ramp U: Disturbed balance of expression between XIAP and
Smac/DIABLO during tumour progression in renal cell carcinomas. Br
J Cancer. 91:1349–1357. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang C, Yi XP, Shen H and Li YX:
Targeting X-linked inhibitor of apoptosis protein inhibits
pancreatic cancer cell growth through p-Akt depletion. World J
Gastroenterol. 18:2956–2965. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kwatra SG: Targeting x-linked inhibitor of
apoptosis protein for melanoma therapy: The need for more
homogeneous samples and the importance of cell lines. J Invest
Dermatol. 131:7972011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schimmer AD, Dalili S, Batey RA and Riedl
SJ: Targeting XIAP for the treatment of malignancy. Cell Death
Differ. 13:179–188. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun H, Nikolovska-Coleska Z, Lu J, Meagher
JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, et al:
Design, synthesis, and characterization of a potent, nonpeptide,
cell-permeable, bivalent Smac mimetic that concurrently targets
both the BIR2 and BIR3 domains in XIAP. J Am Chem Soc.
129:15279–15294. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu J, Zhang D, Luo W, Yu J, Li J, Yu Y,
Zhang X, Chen J, Wu XR and Huang C: E3 ligase activity of XIAP RING
domain is required for XIAP-mediated cancer cell migration, but not
for its RhoGDI binding activity. PLoS One. 7:e356822012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang X, Wu Z, Mei Y and Wu M: XIAP
inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J.
32:2204–2216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suzuki Y, Nakabayashi Y and Takahashi R:
Ubiquitin-protein ligase activity of X-linked inhibitor of
apoptosis protein promotes proteasomal degradation of caspase-3 and
enhances its anti-apoptotic effect in Fas-induced cell death. Proc
Natl Acad Sci USA. 98:pp. 8662–8667. 2001; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gyrd-Hansen M and Meier P: IAPs: From
caspase inhibitors to modulators of NF-kappaB, inflammation and
cancer. Nat Rev Cancer. 10:561–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bilim V, Yuuki K, Itoi T, Muto A, Kato T,
Nagaoka A, Motoyama T and Tomita Y: Double inhibition of XIAP and
Bcl-2 axis is beneficial for retrieving sensitivity of renal cell
cancer to apoptosis. Br J Cancer. 98:941–949. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tong W, Harris S, Cao X, Fang H, Shi L,
Sun H, Fuscoe J, Harris A, Hong H, Xie Q, et al: Development of
public toxicogenomics software for microarray data management and
analysis. Mutat Res. 549:241–253. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li CL, Chang L, Guo L, Zhao D, Liu HB,
Wang QS, Zhang P, Du WZ, Liu X, Zhang HT, et al: β-elemene induces
caspase-dependent apoptosis in human glioma cells in vitro through
the upregulation of Bax and Fas/FasL and downregulation of Bcl-2.
Asian Pac J Cancer Prev. 15:10407–10412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kriajevska M, Fischer-Larsen M, Moertz E,
Vorm O, Tulchinsky E, Grigorian M, Ambartsumian N and Lukanidin E:
Liprin beta 1, a member of the family of LAR transmembrane tyrosine
phosphatase-interacting proteins, is a new target for the
metastasis-associated protein S100A4 (Mts1). J Biol Chem.
277:5229–5235. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Simpson PT, Shoker BS, Barraclough R,
Halliwell N, Rudland PS, Sibson DR and Davies MP: Examination of
tumour histopathology and gene expression in a neu/S100A4
transgenic model of metastatic breast cancer. Int J Exp Pathol.
84:173–184. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Knudsen KA, Soler AP, Johnson KR and
Wheelock MJ: Interaction of alpha-actinin with the cadherin/catenin
cell-cell adhesion complex via alpha-catenin. J Cell Biol.
130:67–77. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Prockop DJ and Kivirikko KI: Collagens:
Molecular biology, diseases, and potentials for therapy. Annu Rev
Biochem. 64:403–434. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Misri S, Pandita S, Kumar R and Pandita
TK: Telomeres, histone code, and DNA damage response. Cytogenet
Genome Res. 122:297–307. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xie A, Odate S, Chandramouly G and Scully
R: H2AX post-translational modifications in the ionizing radiation
response and homologous recombination. Cell Cycle. 9:3602–3610.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Campbell RE, Sala RF, van de Rijn I and
Tanner ME: Properties and kinetic analysis of UDP-glucose
dehydrogenase from group A streptococci. Irreversible inhibition by
UDP-chloroacetol. J Biol Chem. 272:3416–3422. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
García-García MJ and Anderson KV:
Essential role of glycosaminoglycans in Fgf signaling during mouse
gastrulation. Cell. 114:727–737. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schenk G, Duggleby RG and Nixon PF:
Properties and functions of the thiamin diphosphate dependent
enzyme transketolase. Int J Biochem Cell Biol. 30:1297–1318. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ozawa M and Muramatsu T: Reticulocalbin, a
novel endoplasmic reticulum resident Ca(2+)-binding protein with
multiple EF-hand motifs and a carboxyl-terminal HDEL sequence. J
Biol Chem. 268:699–705. 1993.PubMed/NCBI
|
|
32
|
Wang R, Li B, Wang X, Lin F, Gao P, Cheng
SY and Zhang HZ: Inhibiting XIAP expression by RNAi to inhibit
proliferation and enhance radiosensitivity in laryngeal cancer cell
line. Auris Nasus Larynx. 36:332–339. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang S, Li SS, Yang XM, Yin DH and Wang L:
Embelin prevents LMP1-induced TRAIL resistance via inhibition of
XIAP in nasopharyngeal carcinoma cells. Oncol Lett. 11:4167–4176.
2016.PubMed/NCBI
|
|
34
|
Cao Z, Zhang R, Li J, Huang H, Zhang D,
Zhang J, Gao J, Chen J and Huang C: X-linked inhibitor of apoptosis
protein (XIAP) regulation of cyclin D1 protein expression and
cancer cell anchorage-independent growth via its E3 ligase-mediated
protein phosphatase 2A/c-Jun axis. J Biol Chem. 288:20238–20247.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Horn HF and Vousden KH: Coping with
stress: Multiple ways to activate p53. Oncogene. 26:1306–1316.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gaston-Massuet C, Andoniadou CL, Signore
M, Jayakody SA, Charolidi N, Kyeyune R, Vernay B, Jacques TS,
Taketo MM, Le Tissier P, et al: Increased Wingless (Wnt) signaling
in pituitary progenitor/stem cells gives rise to pituitary tumors
in mice and humans. Proc Natl Acad Sci USA. 108:pp. 11482–11487.
2011; View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Papakonstanti EA and Stournaras C: Actin
cytoskeleton architecture and signaling in osmosensing. Methods
Enzymol. 428:227–240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li A, Zhou T, Guo L and Si J: Collagen
type I regulates beta-catenin tyrosine phosphorylation and nuclear
translocation to promote migration and proliferation of gastric
carcinoma cells. Oncol Rep. 23:1247–1255. 2010.PubMed/NCBI
|
|
39
|
Garrett SC, Varney KM, Weber DJ and
Bresnick AR: S100A4, a mediator of metastasis. J Biol Chem.
281:677–680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Turner BM: Cellular memory and the histone
code. Cell. 111:285–291. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wen KX, Miliç J, El-Khodor B, Dhana K,
Nano J, Pulido T, Kraja B, Zaciragic A, Bramer WM, Troup J, et al:
The role of DNA methylation and histone modifications in
neurodegenerative diseases: A systematic review. PLoS One.
11:e01672012016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:703–707. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fels DR and Koumenis C: The
PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and
tumor growth. Cancer Biol Ther. 5:723–728. 2006. View Article : Google Scholar : PubMed/NCBI
|