Introduction
Colorectal cancer (CRC), also known as colon cancer,
rectal cancer or bowel cancer, is the third most common type of
cancer in males and the second most common in females worldwide in
2015 (1). In total 1,200,000
individuals develop CRC worldwide and 600,000 patients succumbed to
the disease annually in 2014 (2). In
total >50% of the western population develops a colorectal tumor
by the age of 70 years and for ~10% of these individuals the tumors
become malignant (3). Treatment
approaches for patients with CRC have undergone marked changes in
the last decade yet, despite the decline in mortality rates, CRC
remains the third leading cause of cancer-associated mortality
worldwide (4,5). Previous research has identified the
potential exploitation of microRNAs (miRNAs) as CRC biomarkers and
demonstrated that aberrantly expressed miRNAs serve a role in CRC
(4). Furthermore, it has been
demonstrated previously that miRNAs may be involved in
carcinogenesis (6).
miRNAs are a class of non-coding small
single-stranded RNAs of between 18 and 25 nucleotides in length,
which regulate gene expression by binding to specific sites at the
3′ untranslated region (UTR) of target mRNAs. miRNAs are involved
in numerous cellular and pathological processes including
regulating cell proliferation, differentiation and apoptosis.
MiRNAs also participate in tumor initiation and the progression of
several human malignancies including CRC (7). It is becoming clear that miRNA
expression is altered in numerous types of cancer, associates with
tumor cell proliferation and may serve an oncogenic role in the
cellular processes of cervical cancer (8–10).
miRNA-532-5p is located at human chromosome Xp11.23
and mature miRNA-532-5p consists of 22 nucleotides, with its
sequence conserved between numerous species, indicating its
importance (11). miR-532-5p is a
regulatory factor of runt-related transcription factor 3 (RUNX3)
expression and RUNX3 is a known tumor suppressor gene of
several types of carcinoma (12).
Furthermore, miR-532-5p and RUNX3 are defined as cancer markers and
therapeutic targets for cancer diagnosis, prognosis and treatment
(13). To the best of our knowledge,
the functional and regulatory role of miR-532-5p in human CRC
remains unknown.
The present study was undertaken to reveal the
regulatory role of miR-532-5p in CRC proliferation and to identify
its association with the tumor suppressor gene RUNX3. The
results of the present study suggest that miR-532-5p promotes tumor
cell growth via direct targeting of the 5′UTR of RUNX3 and
functions in an oncogenic role in human CRC.
Materials and methods
CRC specimens, cell culture and
reagents
The present study included 63 patients with CRC, who
were registered at the Department of Colorectal and Anal Surgery of
The First Affiliated Hospital of Jilin University (Changchun,
China). The use of CRC samples was approved by the institutional
review board of The First Affiliated Hospital of Jilin University
according to the guidelines of the Dutch Federation of Medical
Research Associations (14). Written
informed consent was obtained from all 63 patients for the use of
their CRC samples for research purposes. A total of 63 CRC tissues
and 63 peri-CRC tissues (1 cm away from the edge of tumors) were
collected during eradication surgery from the patients with CRC who
were registered in The First Affiliated Hospital of Jilin
University between August 12th 2012 and October 24th 2014. No
patients were subject to any radical resection or chemotherapy
prior to surgery. Clinicopathological characteristics including
age, sex, location, stage, differentiation and vessel invasion are
presented in Table I.
 | Table I.Association of miRNA-532-5p level with
clinicopathological characteristics of colorectal cancer. |
Table I.
Association of miRNA-532-5p level with
clinicopathological characteristics of colorectal cancer.
|
| miR-532-5p |
|---|
|
|
|
|---|
| Characteristic | n | Mean ± standard error
of the mean | P-value |
|---|
| Sex |
|
| 0.1711 |
| Male | 35 | 1.823±0.1273 |
|
|
Female | 28 | 1.586±0.1140 |
|
| Age, years |
|
| 0.6342 |
| ≤55 | 30 | 1.735±0.1284 |
|
|
>55 | 33 | 1.652±0.1158 |
|
| Location |
|
| 0.1964 |
|
Colon | 36 | 1.788±0.1239 |
|
|
Rectum | 27 | 1.563±0.1098 |
|
| T stage |
|
| 0.0125a |
|
T1+T2 | 19 | 1.371±0.1022 |
|
|
T3+T4 | 44 | 1.830±0.1083 |
|
| N stage |
|
| 0.0145a |
| ≤N1 | 21 | 1.399±0.1183 |
|
|
>N1 | 42 | 1.838±0.1078 |
|
| Differentiation |
|
| 0.0818 |
| High | 24 | 1.502±0.1463 |
|
| Low | 39 | 1.808±0.1019 |
|
| Vessel invasion |
|
| 0.1803 |
|
Positive | 66 | 1.366±0.1667 |
|
|
Negative | 7 | 1.732±0.0929 |
|
The human HT-29CRC cell line was purchased from the
American Type Culture Collection (Manassas, VA, USA). Cells were
cultured in McCoy's 5a medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), which was supplemented with
10% fetal bovine serum (FBS; GE Healthcare Life Sciences, Logan,
UT, USA) and incubated at 37°C with 5% CO2. For the
miR-532-5p manipulation, 20 or 40 nM miR-532-5p mimic or control
miRNA (Ctrl miRNA; Merck KGaA, Darmstadt, Germany) were transfected
with Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific,
Inc.) into HT-29 cells at 85% confluence.
Preparation and quantitative analysis
of mRNA and miRNA samples
mRNA and miRNA samples from CRC tissues or from
HT-29 cells were prepared respectively with the Recover All Total
Nucleic Acid Isolation kit (Ambion; Thermo Fisher Scientific, Inc.)
or with the mirPremier™ microRNA Isolation kit (Merck KGaA,
Darmstadt, Germany), according to the manufacturer's protocol. For
the quantitative analysis of miR-532-5p, the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
performed with the mirVana qRT-PCR miRNA Detection kit (Invitrogen;
Thermo Fisher Scientific, Inc.) for each miRNA sample, with 5S rRNA
as internal control. For the quantitative analysis of mRNA levels
of RUNX3, RT-qPCR was performed with the QuantiTect SYBR
Green PCR kit (Qiagen GmbH, Hilden, Germany), with GAPDH as an
internal control. The ∆∆Cq method was used for relative
quantification (15).
Western blot analysis for RUNX3
Cellular protein in HT-29 cells was prepared using a
NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Pierce;
Thermo Fisher Scientific, Inc.) and was treated with a Protease
Inhibitor Cocktail (catalog no. ab65621; Abcam, Cambridge, MA,
USA). The protein level of RUNX3 was evaluated using western
blot analysis. In brief, protein samples were separated by SDS-PAGE
(11% gel) and transferred onto a hydrophobic polyvinylidene
difluoride membrane. The RUNX3 band on the membrane was
detected using the enhanced chemiluminescence detection system (GE
Healthcare, Chicago, IL, USA), following the first incubation with
rabbit anti-human RUNX3 antibody (1:300; cat. no. ab49117;
Abcam) or GAPDH (1:1,000; cat. no. ab9485; Abcam) at 4°C for 8 h
and the second incubation with the horseradish
peroxidase-conjugated anti-rabbit immunoglobulin G antibody
(1:2,000; cat. no. A16104SAMPLE; Thermo Fisher Scientific, Inc.) at
room temperature for 1 h. Prior to each incubation, membranes were
washed four times in PBS.
Luciferase activity assay
miR-523-5p-targeted and control sequences were
inserted into the 5′UTRs of the cytomegalovirus promoter of the
luciferase reporter plasmid. For the luciferase reporter assay, 85%
confluent 293T cells, which were isolated from human embryonic
kidneys and transformed with large T antigen (American Type Culture
Collection, Rockville, MD, USA), were transfected for 6 h at 37°C
with 20 or 40 nM miRNA-523-5p or Ctrl miRNA. Medium was replaced
with fresh Dulbecco's modified Eagle's medium containing 2% FBS for
a 24 h incubation at 37°C. Cells were then harvested with a cell
scraper (Corning Incorporated, Corning, NY, USA) for luciferase
analysis. Luciferase activity was detected using a Rapid Detection
of Firefly Luciferase Activity kit (cat. no. E1500; Promega
Corporation, Madison, WI, USA).
Cell proliferation and colony
formation assay
To investigate the regulatory action of miRNA-523-5p
on the proliferation of CRC cells, a Cell Counting Kit-8 (CCK-8)
assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) and
colony forming assay were performed for the HT-29 cells with
miRNA-523-5p upregulation. HT-29 cells at 85% confluence were
transfected with 0 or 40 nM miRNA-523-5p mimic or Ctrl miRNA and
incubated at 37°C for another 0, 12, 24 and 48 h. Cells were then
incubated in CCK-8 reagent (Dojindo Molecular Technologies, Inc.).
The 450 nm absorbance of each cell well was detected after the
occurrence of visual color. For the cell colony formation assay,
100 HT-29 cells were incubated in 12-well plates and then
transfected with 0 or 40 nM miR-523-5p mimic or Ctrl miRNA for 72 h
of incubation. Colonies were stained with 0.002% crystal violet
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and were counted
using the naked eye.
Statistical analysis
SPSS software (version 17.0; SPSS Inc., Chicago, IL,
USA) was used for statistical analysis. Data are presented as the
mean ± standard error of the mean. Student's t-test was performed
to evaluate the difference between two groups. Multiple comparisons
between the groups were performed using the Student-Newman-Keuls
method. Spearman's rank correlation coefficient was used to assess
correlation between variables. P<0.05 was considered to indicate
a statistically significant difference.
Results
miR-532-5p and RUNX3 mRNA levels are
increased and decreased, respectively, in human CRC tissues
compared with peri-CRC tissues
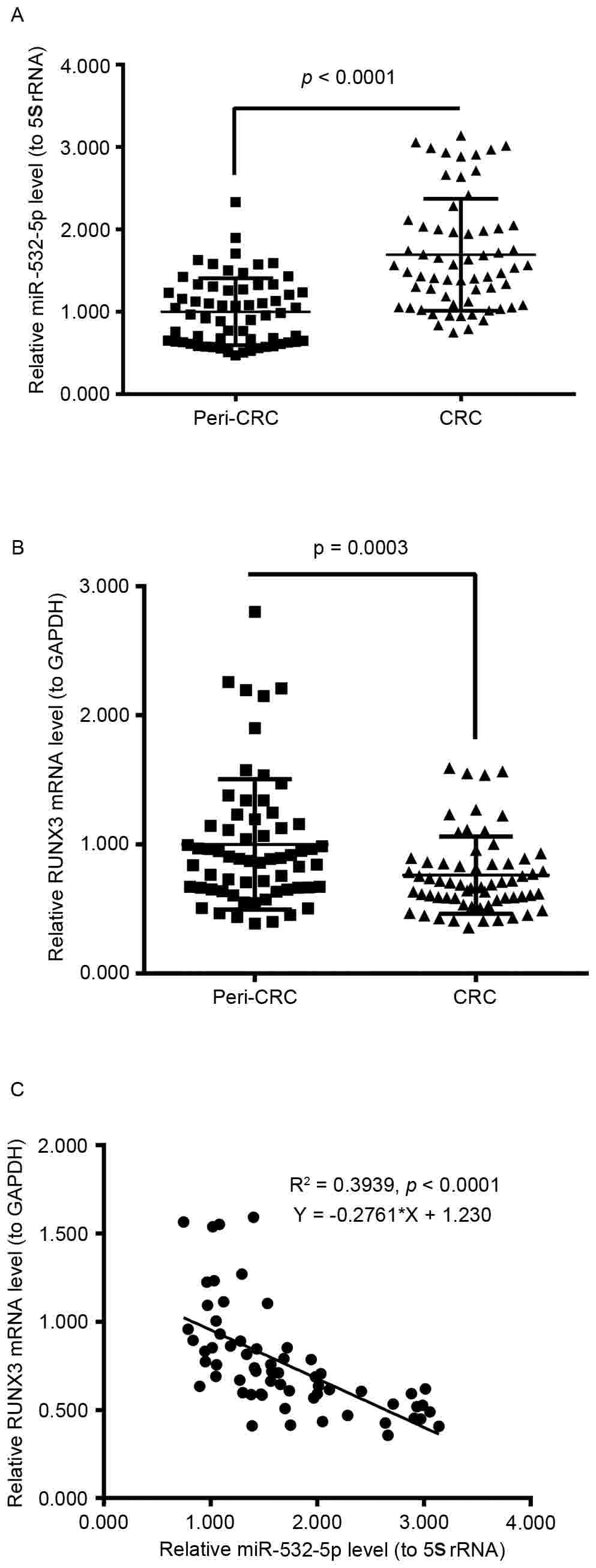
To investigate miR-532-5p and RUNX3 mRNA
levels in human CRC tissues, RT-qPCR was performed. The relative
miR-532-5p levels to 5S rRNA in 63 human CRC tissues were
significantly increased compared with 63 peri-CRC tissues
(P<0.0001; Fig. 1A); and the
relative RUNX3 mRNA levels in 63 human CRC tissues compared
with 63 peri-CRC tissues were significantly decreased (P=0.0003;
Fig. 1B). Furthermore, the scatter
plot of the relative level association between miR-532-5p and
RUNX3 mRNA from the 63 CRC tissue samples revealed a
negative association via Spearman's correlation
(R2=0.3939; P<0.0001; Fig.
1C). These results demonstrated that the expression level of
miR-532-5p was increased and the expression level of RUNX3
mRNA was decreased in human CRC tissues.
RUNX3 mRNA and protein levels are
associated inversely with miR-532-5p levels in HT-29 CRC cell
lines
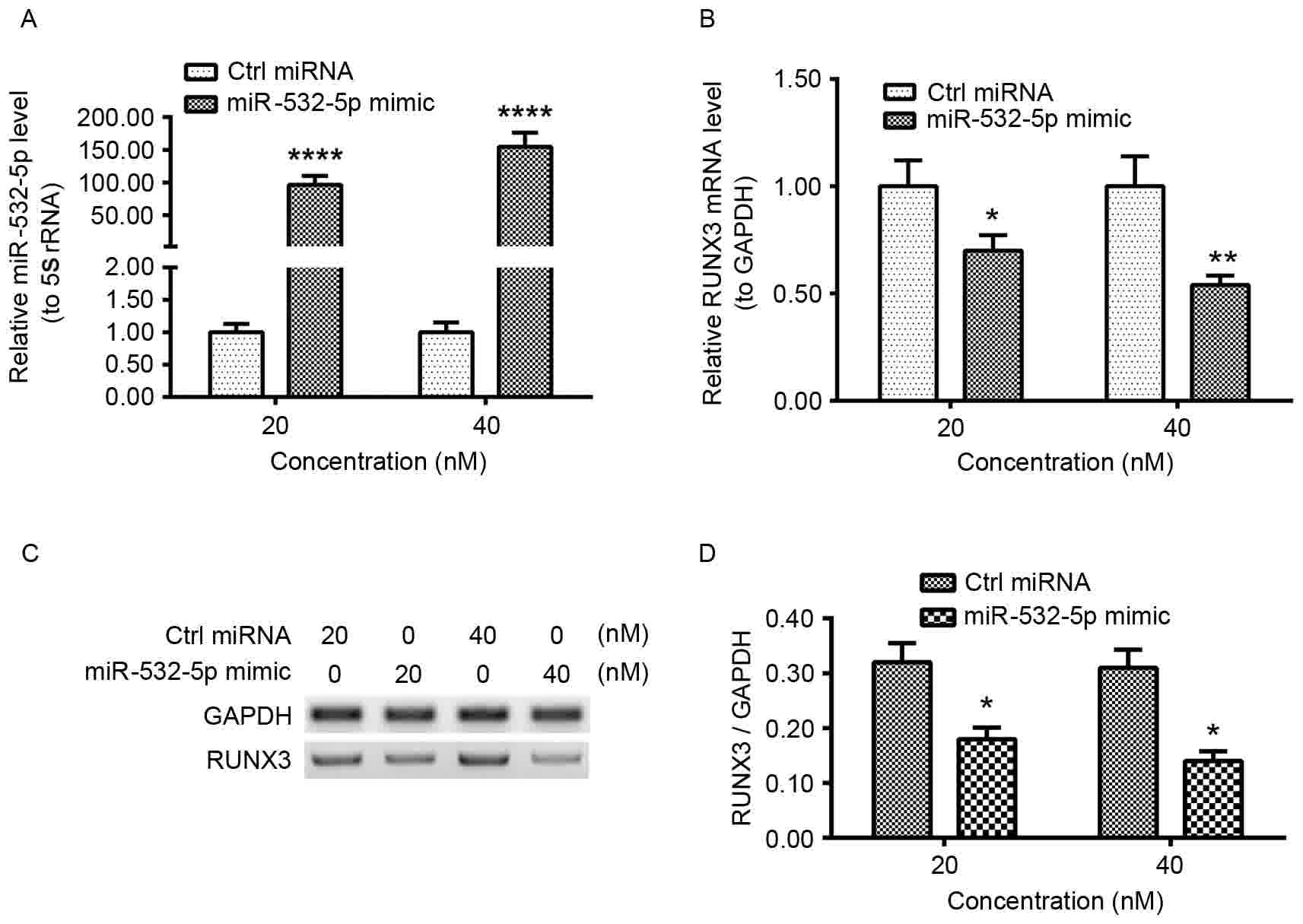
To investigate whether transfected miR-532-5p mimic
affects RUNX3 levels, HT-29 cells were transfected with 20
or 40 nM miR-532-5p mimic or Ctrl miRNA for 12 h. The relative
miR-532-5p and RUNX3 mRNA levels were examined using
RT-qPCR. Significantly increased levels of miR-532-5p were observed
in 20 or 40 nM miR-532-5p mimic-transfected HT-29 cells (Fig. 2A). The level of RUNX3 mRNA was
significantly decreased in the transfected cell lines (Fig. 2B). To determine whether miR-532-5p had
an effect on RUNX3 protein levels, western blot analysis was used.
There was a decrease in RUNX3 protein quantities in 20 or 40 nM
miR-532-5p mimic-transfected cells (Fig.
2C). Quantification and statistical analysis of expression
levels determined the decrease to be significant (Fig. 2D). These results demonstrated that
miR-532-5p downregulated the mRNA and protein levels of RUNX3 in
HT-29 CRC cell lines.
Targeting inhibition on RUNX3
expression by miR-532-5p
We hypothesize that RUNX3 may be a target of
miR-532-5p. Homo sapiens miR-532-5p aligns with the target
sequences with in the 5′UTR of H. sapiens RUNX3 (Fig. 3A). The luciferase reporter assay
revealed a significant decrease in the relative activity value of
the 5′UTR of RUNX3 in HT-29 cells following transfection
with miR-532-5p mimic when compared with those transfected with
Ctrl miRNA. The mutant 5′UTR of RUNX3 group exhibited no
significant difference between the relative activity values of
cells transfected with miR-532-5p and cells transfected with Ctrl
miRNA (Fig. 3C). These data suggest
that the 5′ UTR of RUNX3 is a functional target site for
miR-532-5p in HT-29 CRC cells.
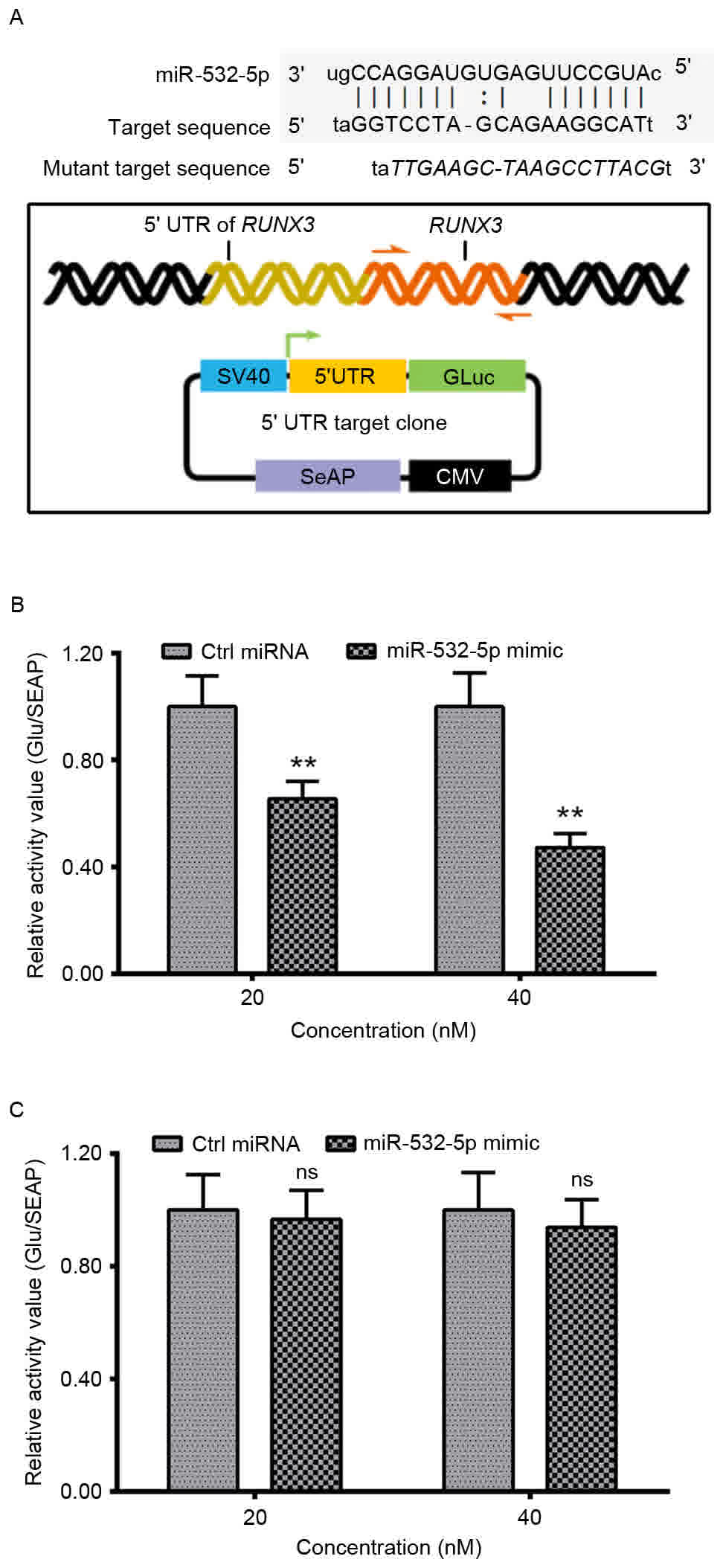 | Figure 3.Targeting inhibition by miR-532-5p on
the expression of RUNX3, with the luciferase reporting assay
in HT-29 cells. (A) Alignment of Homo sapiens miR-532-5p
with the target sequences within the 5′UTR of H. sapiens
RUNX3 and a schematic diagram of the luciferase reporter with
the 5′UTR of RUNX3 (wild-type or mutant 5′UTR of
RUNX3). Relative luciferase activity of the reporter with
the (B) 5′UTR of RUNX3 or (C) mutant 5′UTR of RUNX3
in the HT-29 cells, following transfection with miR-532-5p mimic or
with Ctrl miRNA. Experiments were performed in triplicate.
**P<0.01, ns, not significant. SV40, simian virus 40; GLuc,
Gaussia luciferase; SeAP, secreted embryonic alkaline phosphatase;
CMV, cytomegalovirus; miR, microRNA; RUNX3, runt-related
transcription factor 3; Ctrl, control; UTR, untranslated
region. |
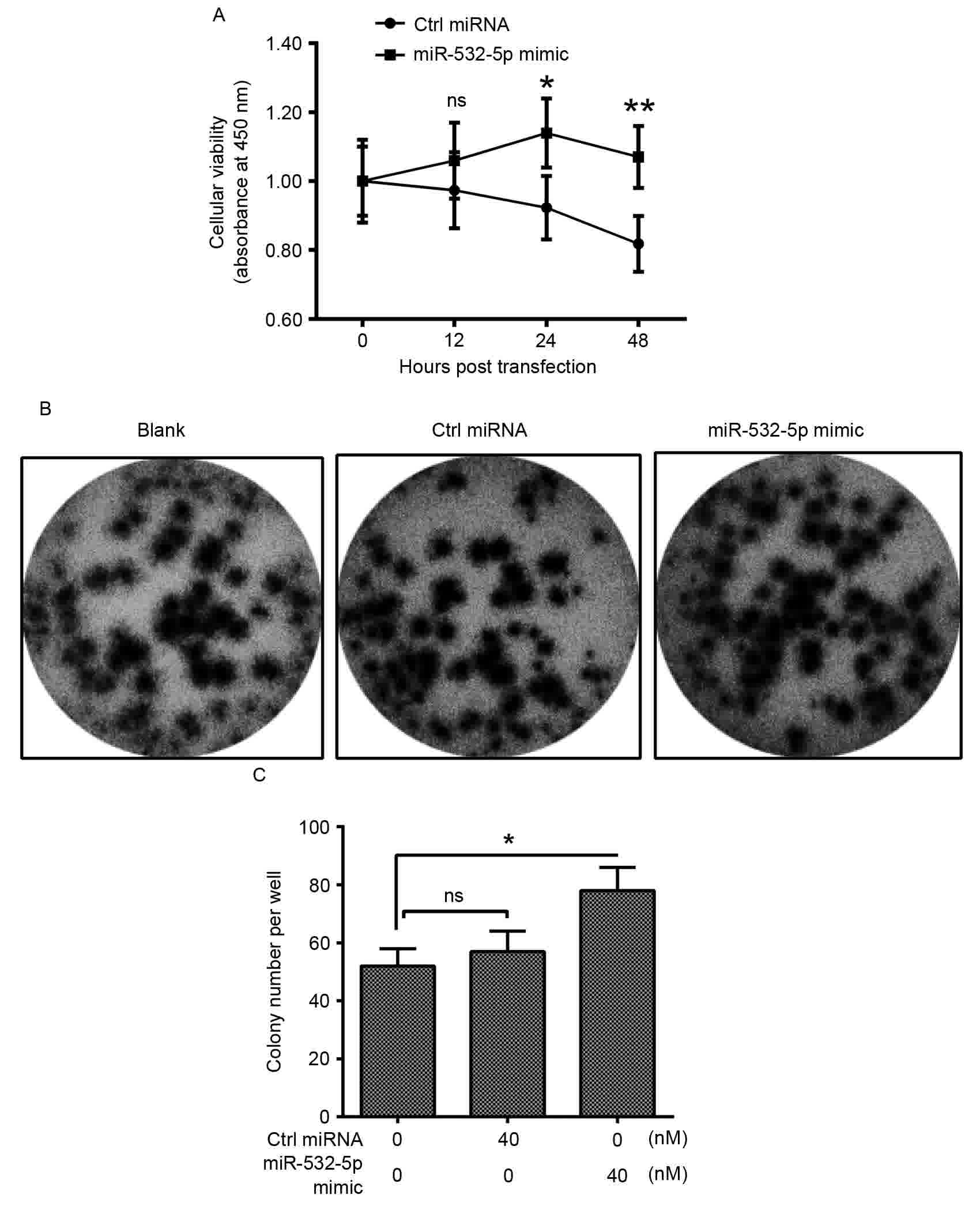
miR-532-5p promotes the growth of
HT-29 CRC cells
miR-532-5p is upregulated in CRC, therefore it may
serve a role in the proliferation of CRC cells. An MTT assay and
colony formation assay were performed. The relative cellular
viability was significantly increased in cells transfected with
miR-532-5p mimic compared with Ctrl miRNA from 24 h after
transfection onwards (Fig. 4A).
Additionally, the colony formation assay analysis revealed that
following transfection with miR-532-5p and Ctrl miRNA for 48 h, the
visible cultures of cells transfected with miR-532-5p covered more
of the plate compared with those transfected with Ctrl miRNA
(Fig. 4B). Quantification of the
colony counts of the HT-29 cells also identified significantly
upregulated quantities inmiR-532-5p-transfected cells (Fig. 4C). These results suggest that
miR-532-5p may serve an oncogenic role in the proliferation and
colony formation of CRC cells.
Discussion
miRNAs are small non-coding RNA molecules, involved
in numerous cellular processes. There have been >200 mammalian
miRNAs identified to date, with evidence suggesting that aberrantly
expressed miRNAs are associated with a variety of diseases and
serve roles in cell growth and apoptosis, including CRC (4,16–18). RUNX3 has been demonstrated to be a
tumor suppressor and exhibits anti-cancer activity (19). Previous studies have demonstrated that
aberrantly expressed miR-532-5p promoted cell growth in
vitro (11) and regulated RUNX3
expression (12). However, little is
known about the underlying molecular mechanism of miR-532-5p in
CRC.
In the present study, aberrant expression of
miR-532-5p and RUNX3, the association between the two
biomarkers, the target of miR-532-5p in CRC tissues and influence
of miR-532-5p on the HT-29 cell viability and colony formation were
investigated. Using RT-qPCR, it was identified that miR-532-5p was
upregulated in CRC tissue; however, the RUNX3 miRNA level
was downregulated in CRC tissue.miR-532-5p (Table I) and RUNX3 (Table II) were associated with several
clinicopathological characteristics of CRC, including tumor stage,
lymph node involvement, differentiation, vessel invasion and tumor
recurrence.
 | Table II.Association of RUNX3 mRNA level with
clinicopathological characteristics of colorectal cancer. |
Table II.
Association of RUNX3 mRNA level with
clinicopathological characteristics of colorectal cancer.
|
| Relative RUNX3 |
|---|
|
|
|
|---|
| Characteristic | n | Mean ± standard error
of the mean | P-value |
|---|
| Sex |
|
| 0.0623 |
| Male | 35 | 0.8251±0.0553 |
|
|
Female | 28 | 0.6842±0.0458 |
|
| Age, years |
|
| 0.3925 |
| ≤55 | 30 | 0.7284±0.0492 |
|
|
>55 | 33 | 0.7935±0.0564 |
|
| Location |
|
| 0.4270 |
|
Colon | 36 | 0.7887±0.0550 |
|
|
Rectum | 27 | 0.7277±0.0488 |
|
| T stage |
|
| 0.0355a |
|
T1+T2 | 19 | 0.8822±0.0838 |
|
|
T3+T4 | 44 | 0.7108±0.0381 |
|
| N stage |
|
| 0.0592 |
|
≤N1 | 21 | 0.8718±0.0591 |
|
|
>N1 | 42 | 0.7079±0.0477 |
|
|
Differentiation |
|
| 0.0079a |
|
High | 24 | 0.8878±0.0681 |
|
|
Low | 39 | 0.6854±0.0399 |
|
| Vessel
invasion |
|
| 0.0359a |
|
Positive | 66 | 0.7348±0.0358 |
|
|
Negative | 7 | 0.9846±0.1695 |
|
| Tumor
recurrence |
|
| 0.0613 |
|
Positive | 43 | 0.7125±0.0461 |
|
|
Negative | 20 | 0.8700±0.0665 |
|
Furthermore, it was demonstrated that both the mRNA
and protein level of RUNX3 were significantly downregulated in
miR-532-5p mimic-transfected HT-29 cells. Therefore, as previously
reported, miR-532-5p is a regulatory factor of RUNX3
expression (20). The specific target
site of miR-532-5p was also investigated in the present study. The
5′UTR of RUNX3 was demonstrated to be a functional target
site for miR-532-5p in HT-29 cells using an in vitro
luciferase reporting assay. Additionally, tumor cell viability and
colony formation ability were promoted in overexpressed miR-532-5p
HT-29 cells. Taken together, the results of the present study
demonstrate that the tumor suppressor RUNX3 is negatively regulated
by miR-532-5p at the 5′UTR of RUNX3. The present study is, to the
best of our knowledge, the first to investigate the underlying
molecular mechanism of miR-532-5p in CRC cells.
To conclude, it was revealed that miR-532-5p was
positively regulated in CRC cells; by contrast, RUNX3 mRNA levels
were negatively regulated in CRC cells. Furthermore, it was
demonstrated that miR-532-5p significantly downregulated mRNA and
protein levels of RUNX3. It was also identified that miR-532-5p may
serve an oncogenic role in CRC proliferation and colony formation
via targeting of the specific site at the 5′UTR of RUNX3.
The results of the present study may provide insight into novel
therapeutic approaches towards CRC.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer: Lancet. 383:1490–1502. 2014.
|
|
3
|
Kinzler KW and Vogelstein B: Lessons from
hereditary colorectal cancer. Cell. 87:159–170. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Okugawa Y, Toiyama Y and Goel A: An update
on microRNAs as colorectal cancer biomarkers: Where are we and
what's next? Expert Rev Mol Diagn. 14:999–1021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moriarity A, O'Sullivan J, Kennedy J,
Mehigan B and McCormick P: Current targeted therapies in the
treatment of advanced colorectal cancer: A review. Ther Adv Med
Oncol. 8:276–293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, Prosper F and Garcia-Foncillas J:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bovell L, Putcha BDK, Devadasan D, Bae S,
Grizzle WE and Manne U: Abstract 1944: Evaluation of the prognostic
value of miRNA-181b and its target identification and validation in
colorectal cancers. Cancer Res. 73 8 Suppl:19442013. View Article : Google Scholar
|
|
8
|
Yao Q, Xu H, Zhang QQ, Zhou H and Qu LH:
MicroRNA-21 promotes cell proliferation and down-regulates the
expression of programmed cell death 4 (PDCD4) in HeLa cervical
carcinoma cells. Biochem Biophys Res Commun. 388:539–542. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Motoyama K, Inoue H, Takatsuno Y, Tanaka
F, Mimori K, Uetake H, Sugihara K and Mori M: Over- and
under-expressed microRNAs in human colorectal cancer. Int J Oncol.
34:1069–1075. 2009.PubMed/NCBI
|
|
11
|
Xu X, Zhang Y, Liu Z, Zhang X and Jia J:
miRNA-532-5p functions as an oncogenic microRNA in human gastric
cancer by directly targeting RUNX3. J Cell Mol Med. 20:95–103.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kitago M, Martinez SR, Nakamura T, Sim MS
and Hoon DS: Regulation of RUNX3 tumor suppressor gene expression
in cutaneous melanoma. Clin Cancer Res. 15:2988–2994. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoon DSB and Kitago M: Use of runx3 and
mir-532-5p as cancer markers and therapeutic targets. US: 2009
|
|
14
|
Kloth JN, Kenter GG, Spijker HS, Uljee S,
Corver WE, Jordanova ES, Fleuren GJ and Gorter A: Expression of
Smad2 and Smad4 in cervical cancer: Absent nuclear Smad4 expression
correlates with poor survival. Mod Pathol. 21:866–875. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen X, Guo X, Zhang H, Xiang Y, Chen J,
Yin Y, Cai X, Wang K, Wang G, Ba Y, et al: Role of miR-143
targeting KRAS in colorectal tumorigenesis. Oncogene. 28:1385–1392.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li QL, Ito K, Sakakura C, Fukamachi H,
Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al: Causal
relationship between the loss of RUNX3 expression and gastric
cancer. Cell. 109:113–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|