Introduction
Breast cancer is the most common malignant tumor and
the leading cause of cancer-associated mortality cases in females
in developing countries (1). Despite
major advances in biomedical research and the development of novel
therapeutic agents and treatment strategies, chemotherapy is an
irreplaceable and effective comprehensive treatment of breast
cancer. Currently, epirubicin and docetaxel are the most commonly
used drugs in breast cancer treatment (2). However, ~30% of all patients with
early-stage breast cancer develop recurrent disease due to acquired
resistance (3). In addition, due to
the presence of intrinsic resistance, numerous patients undergo
treatments that are ineffective, resulting in a delay in receiving
other more suitable therapies and in adverse side effects (4). Chemoresistance has become a major
obstacle during the treatment of breast cancer; therefore, it is
critical to understand the mechanisms underlying the development of
breast cancer chemoresistance and to develop novel treatment
strategies for this tumor.
Several mechanisms have been identified that
underlie the intrinsic and acquired chemoresistance, including
deletion of receptors, altered drug metabolism, impaired drug
uptake, various mechanisms of anti-apoptosis, increased DNA damage
repair, quantitative and qualitative alterations in drug targets,
and increased drug efflux (5).
Numerous previous studies have focused on pathways that modulate
the cancer cell sensitivity, including the transmembrane
ATP-dependent efflux pump P-glycoprotein (P-gp) (6,7), human
epidermal growth factor receptor 2/Erb-b2 receptor tyrosine kinase
2 (8), B-cell lymphoma-2 family
proteins (9) and various microRNAs
(10). The tumor microenvironment is
important for tumor cell survival at the primary lesion and distant
metastatic sites, and its role in tumor drug resistance has
received increasing attention (11).
Accumulating evidence has demonstrated that an acidic tumor
microenvironment leads to a more aggressive phenotype and increases
the drug resistance by elevating the expression of P-gp, suggesting
that management of the tumor pH value and inhibition of the
proton-sensing system blockade are important in preventing
metastasis, as well as improving drug efficacy (5,12–14). Compared with normal cells, tumor cells
have been observed to exhibit low extracellular pH (pHe)
and high intracellular pH (pHi) characteristics
(15). The particular transmembrane
pH gradient occurring between the intracellular and extracellular
spaces has a negative impact on the distribution, uptake and
bioavailability of weak base antineoplastic drugs, eventually
leading to chemoresistance (5,14,16). It has been reported that exposure to
proton pump inhibitors (PPIs) was able to resensitize
multidrug-resistant cells to chemotherapeutic drugs, suggesting
that counteracting the acidity of the tumor microenvironment or
altering the transmembrane pH gradient of tumor cells may overcome
the mechanisms of chemoresistance (5,17).
Vacuolar H+-ATPase (V-ATPase), a key
multi-subunit proton pump, serves an important role in the acidic
microenvironment of the tumor. It relies on the energy transfer
protons generated by the hydrolysis of ATP to produce the
electrochemical gradient of the transmembrane and to regulate the
transmembrane pH gradient (18).
V-ATPase has been considered to be one of the important targets for
overcoming an acidic tumor microenvironment (5,18). In
addition, several studies have demonstrated that PPIs, which
directly inhibit V-ATPase at the cellular level, reverted
chemoresistance in drug-resistant tumors and directly induced tumor
cell death, indicating that targeting V-ATPase may be an option for
reversing multidrug resistance (17).
Cluster of differentiation 147 (CD147) has been
identified as a novel tumor marker for breast cancer. It is
involved in a variety of malignant biological behaviors, including
tumor invasion, metastasis, angiogenesis, energy metabolism and
multidrug resistance (19–21). CD147 combines with a number of other
molecules and forms a polymer on the cell membrane in order to
regulate the biological function of other molecules (22,23).
Slomiany et al (24) reported
that CD147 and monocarboxylate transporters (MCTs) co-localized on
the cell membrane and participated in lactate efflux, regulating
the pH value in the tumor microenvironment and thus resulting in
chemoresistance in breast cancer (24,25). Our
earlier study (22) demonstrated that
CD147 is highly expressed in chemotherapy-resistant breast cancer.
Furthermore, CD147 was observed to form a complex with ATP-binding
cassette sub-family G member 2 (ABCG2) and regulate ABCG2
expression, in order to induce chemoresistance via affecting the
location and dimerization of ABCG2. Despite the involvement of both
CD147 and V-ATPase in chemoresistance, there are currently no
studies on the mutual interaction between CD147 and V-ATPase, and
their roles in drug sensitivity in breast cancer.
In the present study, the expression of V-ATPase in
chemotherapy-resistant breast cancer samples and its correlation
with CD147 expression were investigated. Subsequently, MCF-7 and
MDA-MB-231 breast cancer cell lines were used to investigate the
role of the interaction between CD147 and V-ATPase in breast cancer
chemoresistance. The results demonstrated that CD147 regulated the
expression and activity of V-ATPase to mediate the chemotherapy
drug resistance of breast cancer cells.
Materials and methods
Cell culture
MCF-7 and MDA-MB-231 cells were obtained from the
Shanghai Institute of Cell Biology at the Chinese Academy of
Sciences (Shanghai, China). The cells were maintained in
high-glucose Dulbecco's modified Eagle's medium (DMEM; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) 100 U/ml
penicillin-G (Jingmei Biotech Co., Ltd., Shenzhen, China) and 100
µg/ml streptomycin (Jingmei Biotech Co., Ltd.) at 37°C in a
humidified incubator with 5% CO2.
Establishment of transfected
cells
The pGC-Fu-CD147 plasmid encoding CD147 cDNA and the
pSUPER/CD147 short hairpin (sh)RNA vector targeting human CD147
mRNA were constructed and packaged with a lentivirus by Shanghai
GeneChem Co., Ltd. (Shanghai, China) as described previously
(22,26). Briefly, the cDNA containing the entire
region of human CD147 was prepared by Agel enzyme digestion
(Shanghai Genechem Co., Ltd.) and cloned into the pGCFU vector
(Shanghai GeneChem Co., Ltd.). The shRNA sequence was:
5′-GATCCCCTGACAAAGGCAAGAACGTCTTCAAGAGAGACGTTCTTGCCTTTGTCATTTTTGGAAA-3.
The sequence has no homology to other human genes, as determined by
nucleotide-nucleotide Basic Local Alignment Search Tool search in a
previous study (27). A control
scrambled sequence (5′-TTCTCCGAACGTGTACGT-3′) with no homology to
other genes was annealed and ligated into the linearized plasmid
using T4 DNA ligase (Promega Corporation, Madison, WI, USA).
Chemically competent Escherichia coli DH5α (Takara Bio., Inc.,
Otsu, Japan) were transformed, and positive transformants were
isolated using ampicillin-G (Jingmei Biotech Co., Ltd.) selection
(100 ng/ml) and amplified using the EndoFree Plasmid Maxi kit
(Qiagen China Co., Ltd., Shanghai, China) according to the
manufacturer's protocol. The successful insertion of siRNA into
pSUPER (GeneChem Co., Ltd.) was confirmed by DNA sequencing, PCR
and restriction endonuclease digestion which were performed by
Genechem Co., Ltd. Subsequently, the plasmids were successfully
packaged with the lentivirus by using the Lenti-Easy Packaging
System (LPK 001; Genechem Co., Ltd.). MCF-7 cells with
low-expression CD147 (22) were
transfected with the lentivirus vector containing the pGC-Fu-CD147
plasmid or green fluorescent protein (GFP) vector (Shanghai
GeneChem Co., Ltd.), which served as a control, and MDA-MB-231
cells overexpressing CD147 (25) were
transfected with the lentivirus vector containing pSUPER/CD147
shRNA or a GFP vector using the FuGENE 6 transfection reagent
(Roche Diagnostics GmbH, Mannheim, Germany), respectively. After 48
h transfection, the MCF-7 and MDA-MB-231 cells were then grown in
DMEM containing 1 µg/ml puromycin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 5–7 days. The cells were then maintained in
DMEM containing 0.5 µg/ml puromycin for at least 4 weeks, until the
final stable single cell clones were harvested and verified by
reverse transcription-quantitative PCR (RT-qPCR) and western blot
analyses, as described below. The 4 stable transfected cell lines
were MCF-7 control (MCF-7/CON), CD147 overexpression (MCF-7/CD),
MDA-MB-231 control (MDA-MB-231/CON) and CD147 knockdown cells
(MDA-MB-231/si).
RNA isolation and RT-qPCR
analysis
Total RNA was extracted from the cells with the
RNeasy Plus Mini kit (Qiagen China Co., Ltd.), and the total RNA
concentration was measured spectrophotometrically from the ratio of
absorbance at 260 and 280 nm using a NanoDrop ND-1000 (NanoDrop
Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
A total of 2 µg RNA samples were used to synthesize cDNA using the
Super Script VILO cDNA Synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.). Amplification was subsequently performed on an
ABI PRISM 7900HT (Applied Biosystems; Thermo Fisher Scientific,
Inc.) in a final volume of 10 µl, using 5 µl Power
SYBR®-Green PCR Master Mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.) and 0.5 µM of each primer. Thermocycling
conditions for CD147 and β-actin were as follows: Template
pre-denaturation (30 sec at 95°C), denaturation (15 sec at 95°C)
and annealing and extension (25 sec at 60°C) for 40 cycles. The
protocol for melting curve analysis was as follows: 15 sec at 95°C,
1 min at 60°C and 15 sec at 95°C. The target primers were as
follows: CD147 forward primer, 5′-GCAGCGGTTGGAGGTTGT-3′; and
reverse primer, 5′-AGCCACGATGCCCAGGAAGG-3′; β-actin forward primer,
5′-GTCATCACCATTGGCAATGAG-3′; and reverse primer,
5′-CGTCACACTTCATGATGGAGTT-3′. Target gene primers were synthesized
by Sangon Biotech Co., Ltd. (Shanghai, China), and amplification of
endogenous β-actin was used as an internal control.
2−ΔΔCq method was used to quantify as previously
described (28).
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay
buffer (Thermo Fisher Scientific, Inc.) and sonicated on ice for
three times, 5 sec at 20 KHz. The cell lysates was then centrifuged
at 14,000 × g for 20 min at 4°C, and the supernatant was collected.
Protein concentration was quantified by Bio-Rad Protein Assay (cat.
no. 5000006; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Subsequently, protein (50 µg) samples were subjected to 10%
SDS-PAGE (GenScript Biotech Corporation, Piscataway, NJ, USA) and
transferred to the polyvinylidene difluoride membranes. The samples
were then blocked with 3% bovine serum albumin (BSA) for 1 h at
room temperature, followed by incubation with monoclonal antibodies
against CD147 (1:2,000; ab212856; Abcam, Cambridge, UK) or
anti-GAPDH (1:500; sc-FL335; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA). Secondary horseradish peroxidase-conjugated
polyclonal goat anti-mouse IgG antibodies (1:5,000; sc-2005; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) were then incubated with
the membranes for 1 h at room temperature. The signal from
antibody-conjugated horseradish peroxidase was visualized by
applying SuperSignal™ West Pico Chemiluminescent Substrate (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and exposing to X-ray
film. The GAPDH expression was used as an internal control and the
grayscale ratios of CD147 to GAPDH were calculated.
Patient samples
A retrospective analysis comparing the chemotherapy
response to the protein levels of CD147 and V-ATPase was also
conducted in the present study. The study design was reviewed and
approved by the Research Ethics Board of the Research Institute at
Xiangya Hospital (Central South University, Changsha, China; no.
201403152). Informed consent was obtained from all participants
whose tissue samples were included in the study. The criteria for
inclusion into the current retrospective analysis were as follows:
i) A confirmed diagnosis of invasive ductal breast cancer by
pathologic examination (29) and
receiving treatment by neoadjuvant chemotherapy; ii) patients had
not received any previous treatment; iii) patients receiving only
four cycles of based neoadjuvant chemotherapy with the AC
(involving pirarubicin and cyclophosphamide) or EC (involving
epirubcin and cyclophosphamide) regimens prior to surgery (22); and iv) availability of complete
hospital record, including chemotherapy efficacy evaluation. The
clinical Response to AC/EC chemotherapy was evaluated by the
decrease in tumor size and classified according to the Response
Evaluation Criteria In Solid Tumors (RECIST) criteria (30). Patients demonstrating complete or
partial remission were classified as chemotherapy-sensitive cases,
while those with stable or progressive disease were classified as
chemotherapy-resistant cases. A total of 84 patients with breast
cancer met all the criteria above between February 2014 and
February 2015 in the Affiliated Xiangya Hospital of Central South
University (Changsha, China). According to the RECIST criteria, the
84 patients were divided into two groups: A chemotherapy-sensitive
group (63 cases) and a chemotherapy-resistant group (21 cases).
Immunohistochemical assay
Biopsy samples of the included patients with breast
cancer were collected and embedded in paraffin by the Department of
Pathology of the Xiangya Hospital of Central South University, and
then were stored at room temperature. The paraffin-embedded samples
were stored at temperature and subjected to immunohistochemical
assay using standard procedures to examine their CD147 and V-ATPase
content. Briefly, 5 µm tissue sections were deparaffinized and
blocked with 0.3% hydrogen peroxide for 30 min at room temperature.
Subsequent to heating for 20 min at 100°C in a microwave oven for
antigen retrieval and blocking with normal rabbit serum for 20 min
at room temperature (cat. no. ab166640; Abcam), the sections were
incubated with primary antibodies at 4°C overnight, including CD147
(1:200; cat. no. ab212856; Abcam) and V-ATPase (1:200; cat. no.
sc-69088; Santa Cruz Biotechnology, Inc. Dallas, TX, USA), or with
PBS as the negative control. A 2-step Plus Poly-HRP Anti-Mouse/Goat
IgG detection system (OriGene Technologies, Inc., Beijing, China)
was then applied according to the manufacturer's protocol, followed
by DAB visualization. Two pathologists blinded to the markers
examined the immunohistochemically stained sections independently.
Five fields-of-view at a magnification of ×400 were randomly
selected for analysis in each section. For the semi-quantitative
analysis of the immunoreactivity of CD147 and V-ATPase, H-score
(31) was used to assess the
following parameters: i) Intensity of staining, which was scored
between 0 and 3, with 0 assigned upon absence of staining, 1 for
weak staining, 2 for moderate staining, 3 for strong staining; and
ii) the percentage of positive cells. The range of possible scores
was between 0 and 300.
Immunofluorescence assay
Cells were fixed with 100% ice-cold methanol for 10
min and blocked with 10% BSA for 1 h at room temperature.
Subsequent to washing with PBS, the cells were incubated for 2 h at
room temperature with primary antibodies against CD147 (1:200; cat.
no. ab212856; Abcam) and V-ATPase (1:100; cat. no. sc-69088; Santa
Cruz Biotechnology, Inc. Dallas, TX, USA), followed by incubation
with secondary antibodies, including anti-mouse Cy3 (1:200; cat.
no. AP192C; EMD Millipore, Billerica, MA, USA) or anti-goat FITC
(1:500; cat. no. ab6881; Abcam) antibodies for 1 h at room
temperature. Cells were then rinsed with PBS for three times and
mounted in a ProLong Gold Antifade reagent with DAPI (Invitrogen;
Thermo Fisher Scientific, Inc.). Images were captured using a Leica
fluorescence microscope (Leica Microsystems GmbH, Wetzlar,
Germany).
Measurement of plasma membrane
V-ATPase activity
The transmembrane protein was initially extracted
using a transmembrane protein extraction kit (cat. no. GMS30039.2;
Genmed Pharmaceutical Technology Co., Ltd., Shanghai, China), and
the protein concentration was quantified by a Bio-Rad protein assay
kit (cat. no. 5000006; Bio-Rad Laboratories, Inc.) according to the
manufacturer's instructions. Subsequently, measurement of the
plasma membrane V-ATPase activity was performed using a V-ATPase
activity detection colorimetric kit (cat. no. GMS50247.1; Genmed
Pharmaceutical Technology Co., Ltd.), according to the
manufacturer's protocol.
Measurements of pHi and
pHe values
The pHi value was measured in the
monolayers using the pH-sensitive fluorescent probe
2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein/acetoxymethyl ester
(BCECF/AM; sc-202492; Santa Cruz Biotechnology, Inc.), as
previously described (17,32). Briefly, in order to determine the
pHi, the standard buffers were initially prepared. The
pH standard buffer A consisted of 133 mM KCl, 7 mM choline
chloride, 1 mM CaCl2, 2 mM KH2PO4,
5 mM glucose and 6 mM HEPES, and the pH values of solutions of
buffer A were adjusted to 6.2, 6.4, 6.6, 6.8, 7.0, 7.2, 7.4 and
7.6, respectively. Buffer B contained 135 mM NaCl, 5 mM KCl, 1.8 mM
CaCl2, 0.8 mM MgSO4, 5 mM glucose and 10 mM
HEPES, and the pH of this buffer was adjusted to 7.4. Subsequently,
a standard curve was established. For this, cells (MCF-7 or
MDA-MB-231 cells) were cultured for 24 h in 6-well plates at a
density of 1×105 cells per well under the aforementioned
cell culture conditions. The culture medium was removed, and the
cells were washed with buffer B twice for 5 min each time, followed
by addition of buffer B containing BCECF/AM (1 µl/1 ml; 5 µM) and
incubation for 1 h at 37°C. The supernatant was removed and the
cells were washed twice with each given pH value of buffer A,
followed by addition of buffer A containing nigericin (1 µl/1 ml; 5
µM) into each well and incubation for 15 min under normal
conditions. The cells were then trypsinized and resuspended with 1
ml of each given pH value of buffer A. Next, the BCECF fluorescence
intensity was recorded by flow cytometry (33) at excitation light and emission light
wavelengths of 490 and 530 nm, respectively. Bivariate correlation
analysis between the fluorescence intensity at 490 nm and the pH
value was performed, and then the pHi standard curve was
developed. Finally, following the measurement of the pHi
value of transfected cells as described earlier but using buffer B
without nigericin instead of buffer A, the fluorescent intensity at
490 nm was recorded and the pHi value was calculated
according to the pHi standard curve. The pHe
values of the culture medium after 24-h incubation were measured by
a Calibration Check Microprocessor pH Meter (FE28K; Mettler Toledo,
Columbus, OH, USA).
Cell sensitivity to drugs by
sulforhodamine B (SRB) assay
Cells were seeded into 96-well tissue culture plates
at a density of 5×103 cells per well. Following
overnight incubation, various concentrations of the anticancer
drugs epirubicin or docetaxel (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) was added into the medium and cultured for 72
h, and then the cell viability was measured by an SRB assay
(34,35). Briefly, the cells were fixed with 10%
trichloroacetic acid for 30 min at 4°C and stained with 0.4% (w/v)
SRB (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in 1% acetic
acid solution for 30 min. SRB was then removed and plates were
washed for 5 min with 1% acetic acid. Bound SRB was solubilized
with 10 mM Tris buffer, and the absorbance (OD) was measured at 510
nm using a microplate reader. The half maximal inhibitory
concentration (IC50) values were determined from the
growth inhibition curves.
To reverse the drug resistance, cells were seeded
into 24-well tissue culture plates at a density of 2×104
cells per well, and pantoprazole (PPZ) (D-78467; Altana Pharma AG,
Konstanz, Germany) was added into the cells after 24-h incubation
at a final concentration of 10 µg/ml, as described in a previous
study (32). After a further 24 h,
docetaxel was added to MCF-7/CON and MCF-7/CD cells at the final
docetaxel concentration of 70 nM, while MDA-MB-231/CON and
MDA-MB-231/si cells were treated with final concentration of
docetaxel of 15 nM. All cells were incubated for 48 h, and cell
viability was then measured by the SRB assay. Independent
experiments were conducted at least in triplicates.
Statistical analysis
A Mann-Whitney U test was used to compare CD147 and
V-ATPase H-scores in the chemotherapy-sensitive and
chemotherapy-resistance groups of invasive ductal breast cancer.
The correlation between CD147 and V-ATPase was evaluated using
Spearman's rank correlation coefficient test. All other values were
expressed as the mean ± standard error of the mean, and the Student
t-test was used to determine statistical differences between the
groups. Values of P<0.05 were considered to indicate differences
that were statistically significant. These analyses were conducted
using the SPSS version 19.0 statistical software (IBM Corp.,
Armonk, NY, USA).
Results
V-ATPase is highly expressed in
chemotherapy-resistant breast cancer and is correlated with CD147
expression
According to the RECIST criteria, 84 invasive ductal
breast cancer patients who had accepted neoadjuvant chemotherapy
with four cycles of the AC/EC regimen were divided into two groups,
including the chemotherapy-sensitive (61 cases) and
chemotherapy-resistant (23 cases) groups. The V-ATPase and CD147
expression levels in these samples were detected by
immunohistochemical analysis. It was observed that both V-ATPase
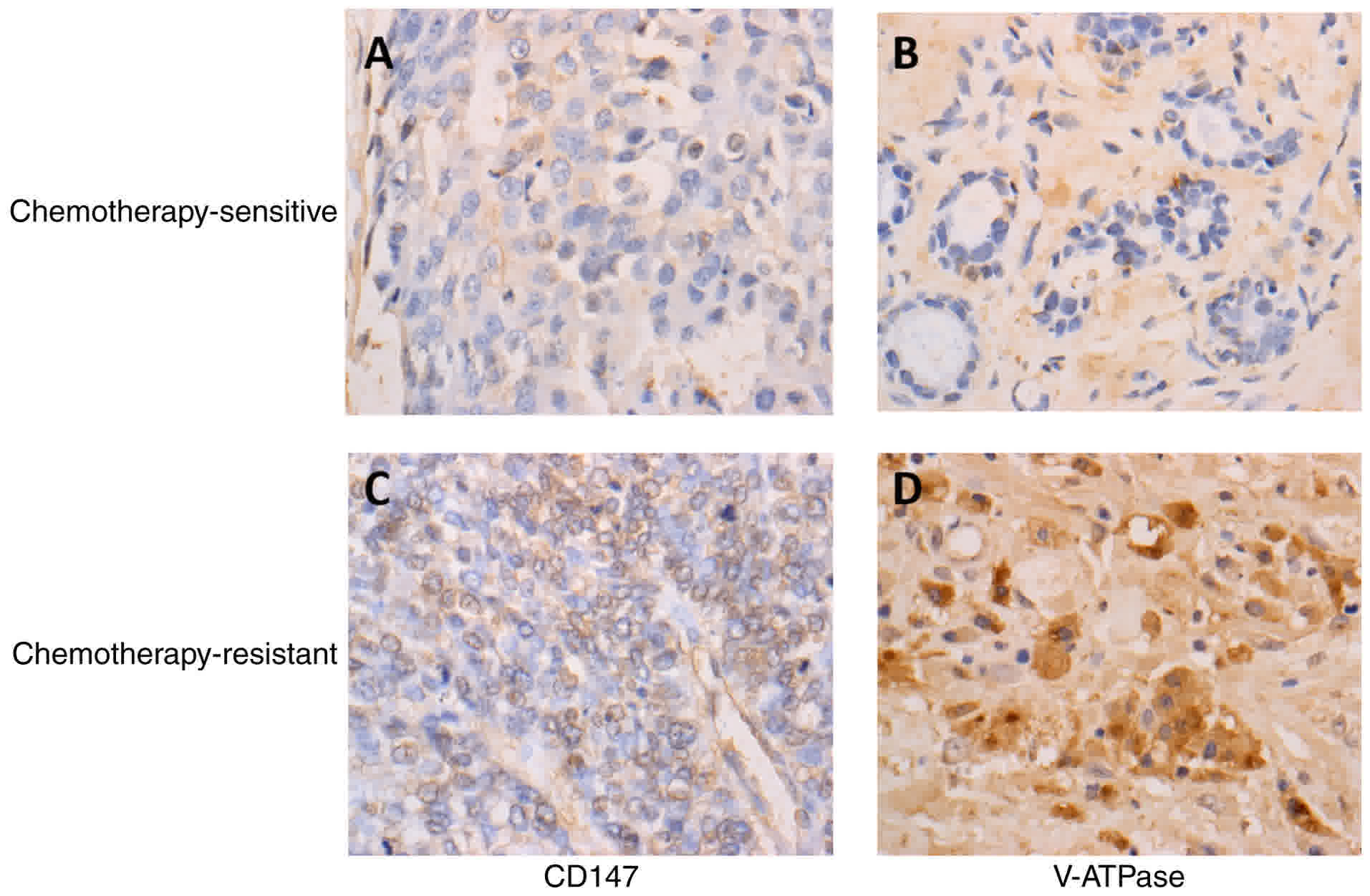
and CD147 were expressed in invasive breast cancer. CD147 was
mainly expressed on the cell membrane, while V-ATPase was located
in the cell membrane and cytoplasm (Fig.
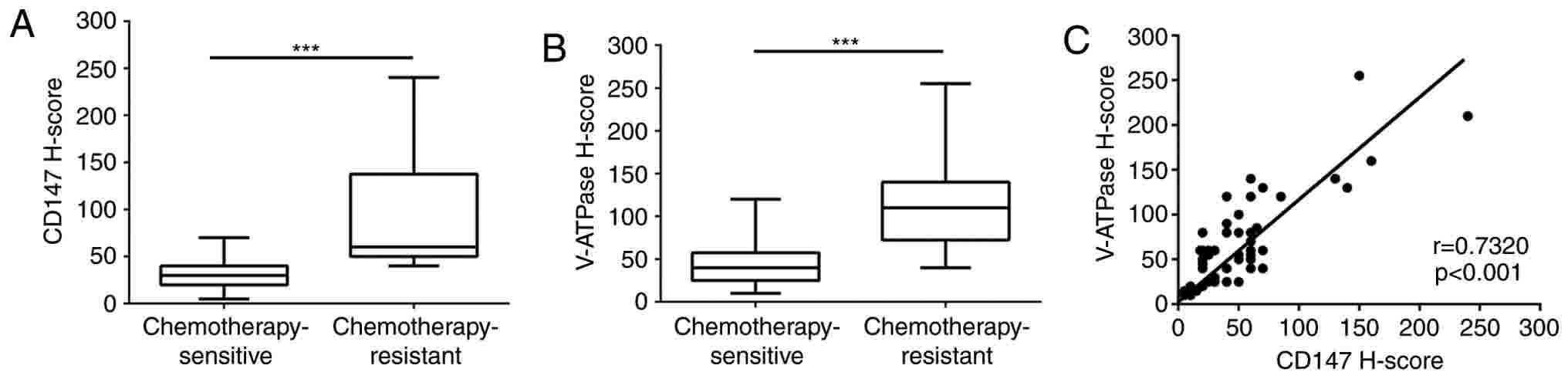
1). Furthermore, V-ATPase and CD147 expression levels in the
chemotherapy-resistant group were significant higher in comparison
with those in the chemotherapy-sensitive group (P<0.001;
Fig. 2A and B). In addition, there
was a significant correlation between CD147 and V-ATPase expression
levels in invasive ductal breast cancer (r=0.732, P<0.001;
Fig. 2C). These results suggested
that CD147 and V-ATPase may have synergistic effects in breast
cancer drug resistance.
Establishment of MCF-7/CD and
MDA-MB-231/si cell lines
Four cell lines were established in the current
study, including the MCF-7/CON, MCF-7/CD, MDA-MB-231/CON and
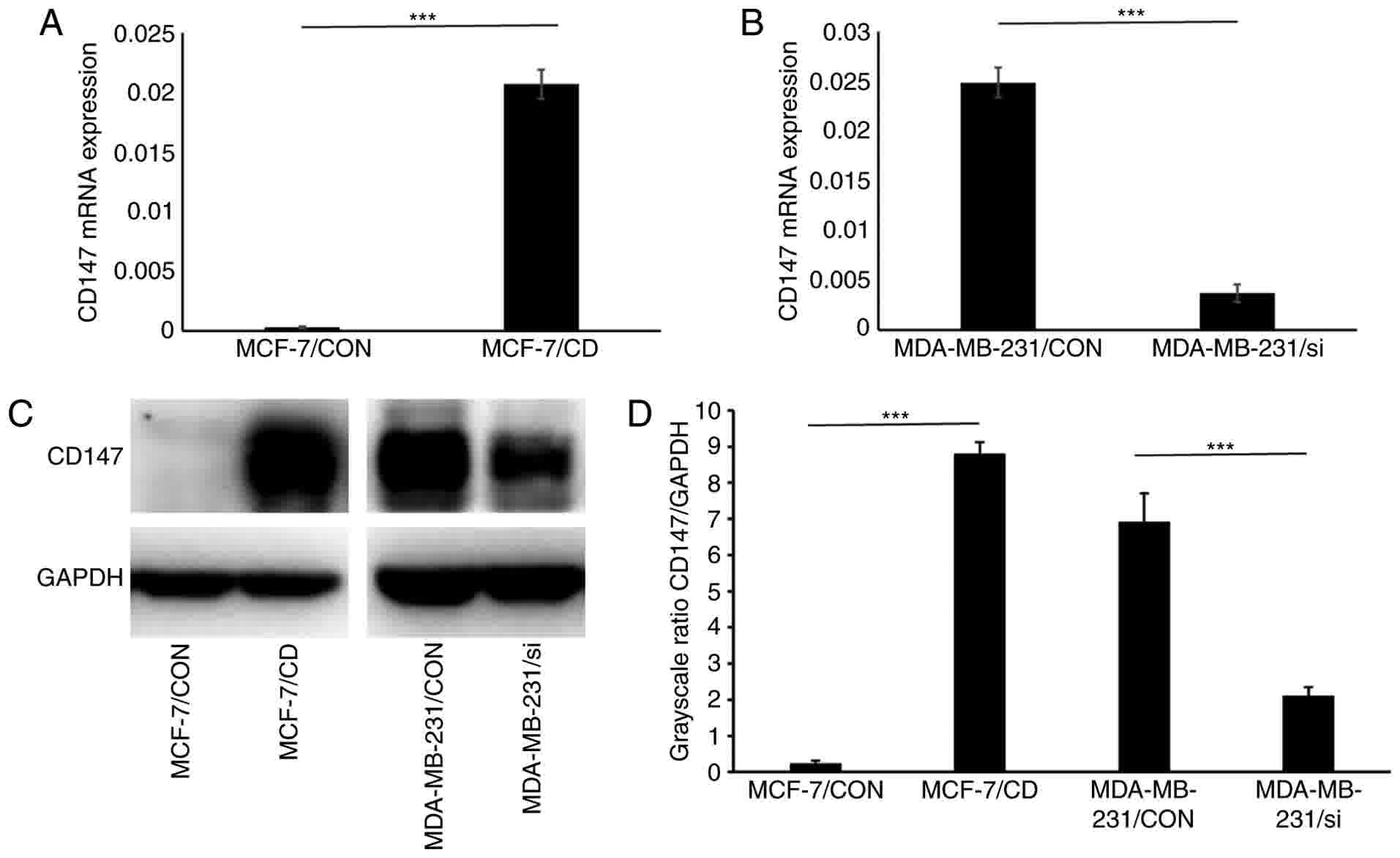
MDA-MB-231/si. Total RNA and protein levels from the transfected
cells were extracted and analyzed by RT-qPCR and western blot
analysis, respectively. The mRNA and protein expression levels of
CD147 were significantly upregulated in MCF-7/CD cells as compared
with the MCF-7/CON. By contrast, the mRNA and protein expression
levels of CD147 were significantly downregulated in MDA-MB-231/si
cells as compared with those in MDA-MB-231/CON cells (P<0.001;
Fig. 3A-D). These results
demonstrated the successful establishment of the transfected cell
lines with CD147 overexpression and downregulation that were used
in subsequent experiments.
CD147 affects V-ATPase expression and
activity
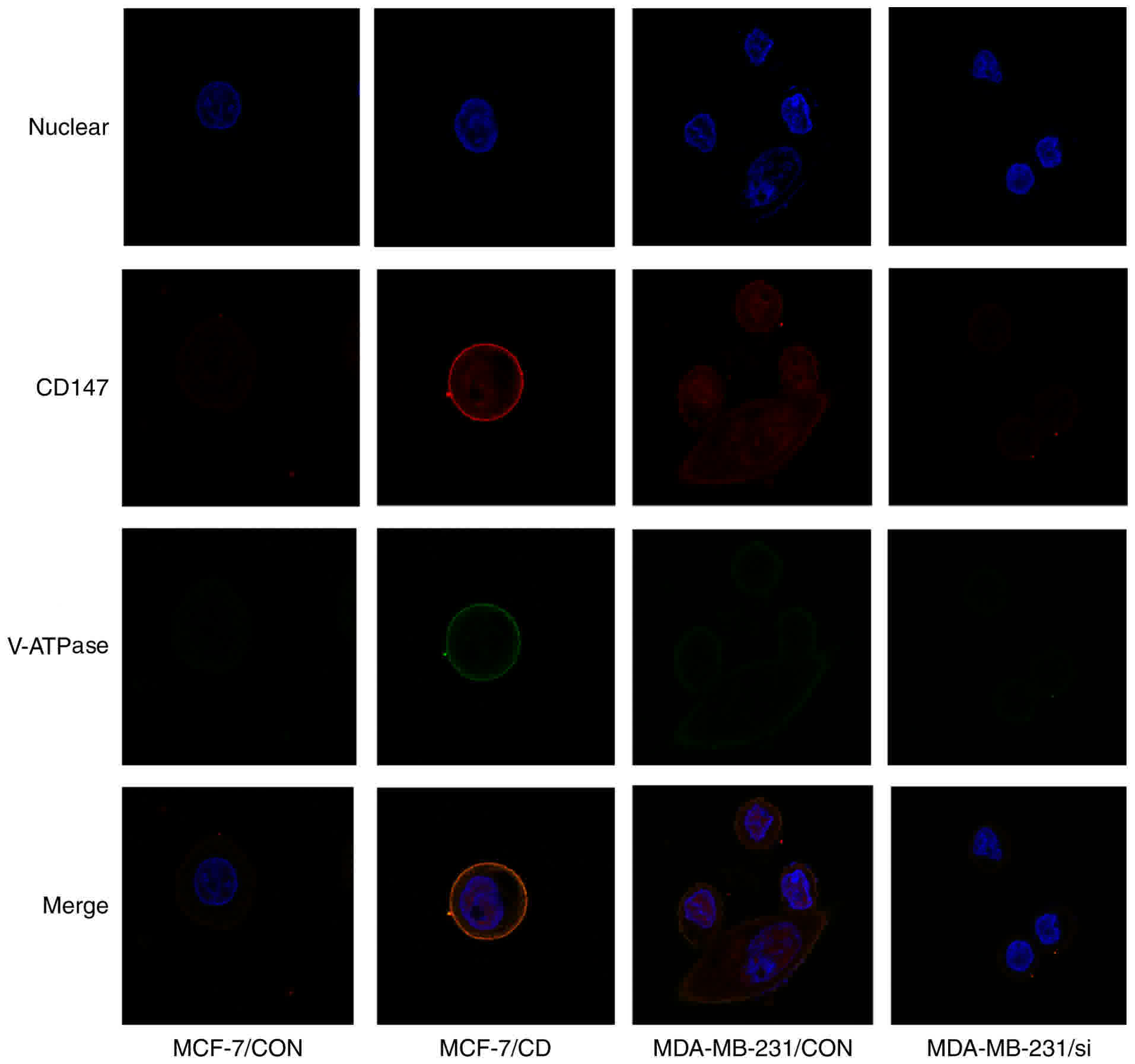
To determine the interaction between CD147 and
V-ATPase, the V-ATPase expression was detected in the four cell
lines by fluorescence staining. Low CD147 and V-ATPase expression
levels were observed in MCF-7/CON cells, while V-ATPase expression
was significantly elevated on the cell membrane of MCF-7/CD cells,
suggesting that overexpression of CD147 in MCF-7 cells enhanced
V-ATPase expression. By contrast, V-ATPase expression was
significantly decreased on the cell membrane of MDA-MB-231/si cells
compared with MDA-MB-231/CON cells, indicating that CD147 knockdown
also decreased V-ATPase expression. These data demonstrated that
CD147 affected the V-ATPase expression on the cell membrane
(Fig. 4). The V-ATPase activity was
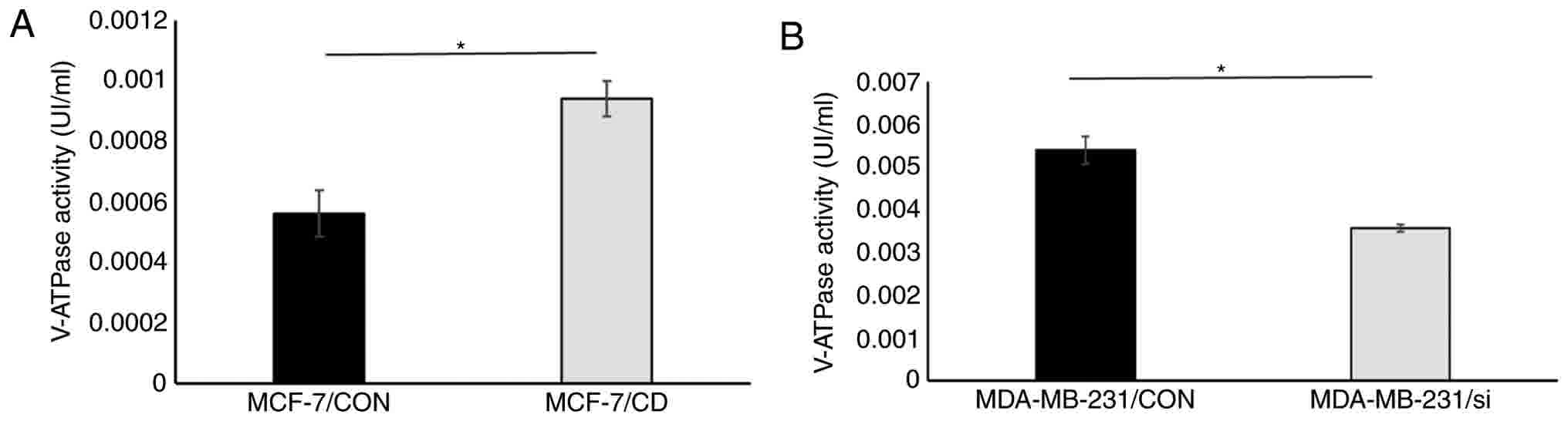
also detected, and it was observed that CD147 overexpression was
able to enhance V-ATPase activity in breast cancer cells, while
CD147 knockdown inhibited the activity of V-ATPase (Fig. 5).
CD147 regulates the transmembrane pH
gradient in breast cancer cells
Since V-ATPase serves a critical role in regulating
the H+ efflux of cancer cells and CD147 affects membrane
V-ATPase expression and activity, the pHi and
pHe of the transfected cells were determined. The
standard curves of the pHi value for MCF-7 and
MDA-MB-231 cells are demonstrated in Fig.
6A and B, respectively. The pHi values of the four
cell lines after 24 h of incubation were calculated according to
the standard curve, and are shown in Fig.
6C and D. Similarly, the pHe values of these cells
are shown in Fig. 6E and F. The
pHi value of MCF-7/CD cells was significantly higher in
comparison with that in MCF-7/CON cells (7.78±0.06 vs. 7.41±0.24,
respectively; P=0.001), while the pHe value of MCF-7/CD
cells was lower compared with that in the MCF-7/CON group
(7.12±0.04 vs. 7.27±0.03, respectively; P=0.006). These results
demonstrated that CD147 overexpression in MCF-7 cells resulted in
pHi increase and pHe decrease. By contrast,
the pHi value of MDA-MB-231/si cells was significantly
lower compared with that in MDA-MB-231/CON cells (7.58±0.01 vs.
7.74±0.02, respectively; P<0.001), and the pHe value
of MDA-MB-231/si cells was higher compared with that in the control
cells (7.34±0.05 vs. 7.20±0.04, respectively; P=0.02). These
results demonstrated that silencing CD147 in MDA-MB-231 cells
resulted in pHi decrease and pHe increase.
Taken together, it can be concluded that CD147 was able to regulate
the transmembrane pH gradient in breast cancer cells.
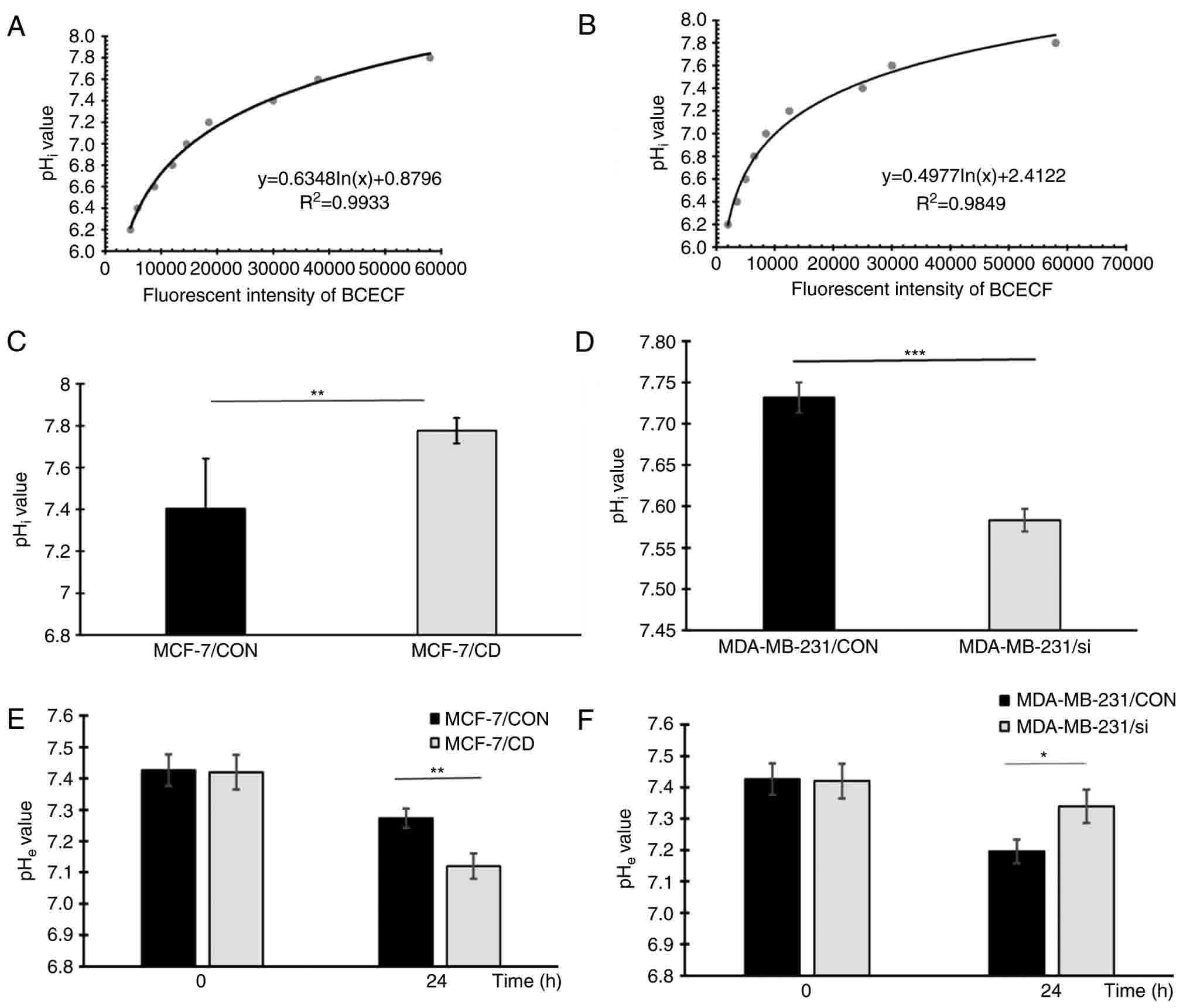 | Figure 6.CD147 affects the pHi and
pHe values in breast cancer cells. Standard curves of
the fluorescence intensity of BCECF/AM vs. the pHi value
of (A) MCF-7 cells and (B) MDA-MB-231 cells. The pHi
values of (C) MCF-7/CON and MCF-7/CD cells, and (D) MDA-MB-231/CON
and MDA-MB-231/si cells are presented. The pHe values of
(E) MCF-7/CON and MCF-7/CD cells, and (F) MDA-MB-231/CON and
MDA-MB-231/si cells are presented. Experiments were repeated at
least three times. *P<0.05, **P<0.01 and ***P<0.001.
CD147, cluster of differentiation 147; V-ATPase, vacuolar
H+-ATPase; CON, control; CD, CD147 overexpression; si,
CD147 knockdown; pHi, intracellular pH; pHe,
extracellular pH; BCECF/AM,
2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein/acetoxymethyl
ester. |
CD147 mediates drug resistance in
breast cancer cells potentially via V-ATPase
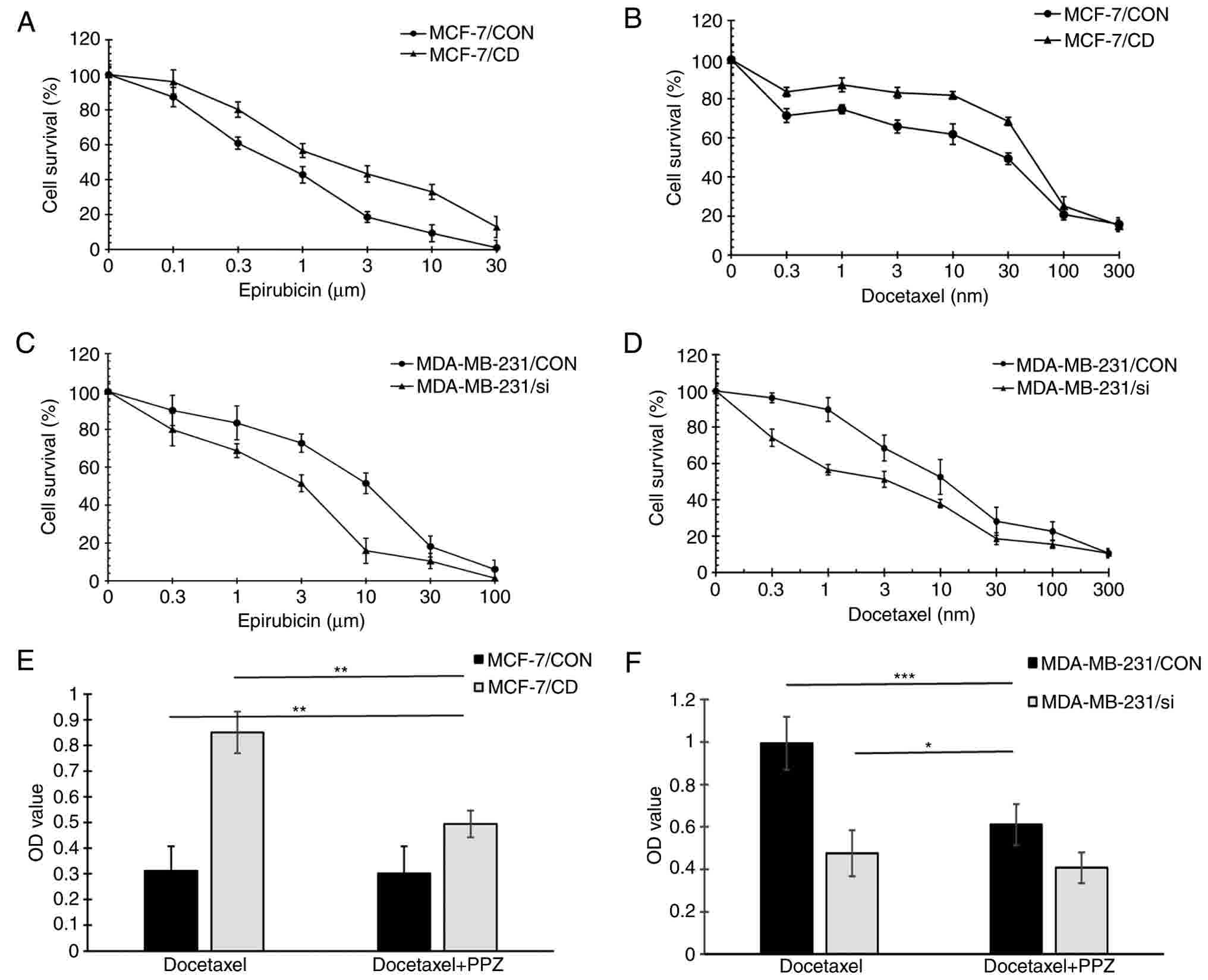
Using an SRB assay, the sensitivity to two
chemotherapeutic drugs, namely epirubicin and docetaxel, was
examined in the four cell lines (Table
I). For MCF-7/CON and MCF-7/CD cells, the IC50
values of epirubicin were 0.99±0.08 and 2.78±0.16 µM, respectively,
while the IC50 values of docetaxel were 28.54±3.51 and
72.13±4.24 nM, respectively (Fig. 7A and
B). For MDA-MB-231/CON and MDA-MB-231/si cells, the
IC50 values of epirubicin were 10.41±0.85 and 3.19±0.20
µM, whereas the IC50 values of docetaxel were 14.07±1.92
and 4.32±1.26 nM, respectively (Fig. 7C
and D). These data suggested that CD147 overexpression
increased the resistance to epirubicin and docetaxel in MCF-7
cells, while CD147 knockdown enhanced the sensitivity to epirubicin
and docetaxel in MDA-MB-231 cells.
 | Table I.Different sensitivities of four
transfected cells to epirubicin and docetaxel. |
Table I.
Different sensitivities of four
transfected cells to epirubicin and docetaxel.
| Cells | Epirubicin
IC50 (µM) | Docetaxel
IC50 (nM) |
|---|
| MCF-7/CON |
0.99±0.08 |
28.54±3.51 |
| MCF-7/CD |
2.78±0.16 |
72.13±4.24 |
| MDA-MB-231/CON |
10.41±0.85 |
14.07±1.92 |
| MDA-MB-231/si |
3.19±0.20 |
4.32±1.26 |
PPZ may reverse chemoresistance in drug-resistant
tumors by directly inhibiting V-ATPase at the cellular level
(32), and the present study
investigated whether PPZ reversed the CD147-mediated drug
resistance in breast cancer cells. The effect of PPZ treatment on
the cytotoxicity of docetaxel was also examined, according to the
method described in a previous study (32). No evident effect on cell viability was
observed at 48 h, and docetaxel was added for a further 48 h
incubation. Cell viability was assessed by the SRB assay following
treatment with docetaxel with or without PPZ. The cell viabilities
of CD147-overexpressing cells (MCF-7/CD and MDA-MB-231/CON)
(25) in the PPZ and docetaxel
combined group were evidently decreased as compared with cells in
the docetaxel alone group. Furthermore, the results revealed that
the cell viabilities of MCF-7/CON cells with docetaxel treatment
was lower compared with that of MCF-7/CD cells with PPZ and
docetaxel combined treatment. The cell viabilities of MDA-MB-231/si
cells with docetaxel treatment was also lower in comparison with
that of MDA-MB-231/CON cells with PPZ and docetaxel combined
treatment (P<0.001; Fig. 7E and
F). These observations indicated that PPZ was able to partially
reverse the CD147-mediated drug resistance in breast cancer cells.
Based on the aforementioned data, it is concluded that CD147
mediated the drug resistance in breast cancer cells via regulating
V-ATPase expression and activity.
Discussion
The acidic tumor microenvironment serves a critical
role in various biological behaviors of tumor cells, including
their proliferation, invasion and metastasis, angiogenesis and drug
resistance (36). V-ATPase is
considered to induce tumor invasion and multi-drug resistance in
several malignant tumors, due to its contribution in maintaining
the pHi under an acidic microenvironment by inducing
proton extrusion into the extracellular medium (12,15). The
plasma membrane V-ATPase is critical for the invasion and migration
of MDA-MB-231 breast cancer cells in vitro (37). V-ATPase expression was elevated in the
ellipticine-resistant UKF-NB-4ELLI cell line and mediated its
ellipticine resistance via the sequestration of ellipticine into
the subcellular compartments (38).
García-García et al (39) also
demonstrated the overexpression of the V-ATPase subunit C gene in
cisplatin-resistant tumors. In the present study, the association
of CD147 and V-ATPase in the invasive ductal breast cancer was
investigated by immunohistochemistry. All patients received four
cycles of the AC or EC chemotherapy regimen prior to surgery.
According to the RECIST criteria, 84 clinical samples were divided
into the chemotherapy-sensitive (including 61 cases) and
chemotherapy-resistant (including 23 cases) groups. It was observed
that the CD147 and V-ATPase expression levels were significant
higher in the chemotherapy-resistant group in comparison with those
in the chemotherapy-sensitive group. The current study results also
revealed that there was a significant correlation between CD147 and
V-ATPase expression in invasive ductal breast cancer, suggesting
that CD147 may interact with V-ATPase, and this interaction may
contribute to breast cancer drug resistance in clinical
practice.
Previous studies have demonstrated that CD147
combines with numerous other important molecules on the cell
membrane and serves the role of a molecular chaperone, which
mediates the biological function of other molecules (19,40). The
charged residues and leucine zipper in the transmembrane region of
CD147 are potential protein-protein interaction motifs, which
possibly mediates its signal transduction involved in the formation
of polypeptide chains or membrane transport protein ingredients
(41). CD147 is able to interact with
proteins such as integrin (42),
cyclophilins (43), MCTs (24), ABCG2 (22), hyaluronan (44) and P-gp (45). These interactions may mediate
extensive cell biological functions, including tumor drug
resistance (40).
The function of CD147 as a molecular chaperone in
tumor drug resistance prompted us to explore its association with
V-ATPase, which is another important transporter protein located on
the cell membrane along with MCT, Na+/H+
exchanger and carbonic anhydrase IX, and contributes to
dysregulated pH in the tumor microenvironment, favoring tumor
progression and metastasis (46). The
present study investigated whether CD147 interacts with V-ATPase on
the cell membrane of breast cancer cells and whether the
interaction of these two molecules mediates breast cancer drug
resistance. The results demonstrated that CD147 affected the
membrane V-ATPase expression and activity, as well as regulated the
transmembrane pH gradient in breast cancer cells, suggesting that
CD147 may regulate transmembrane pH gradient through affecting
membrane V-ATPase expression and activity. It was then observed
that the transmembrane pH gradient changed as CD147 expression was
altered, suggesting that CD147 mediated the acidic tumor
microenvironment formation, in which V-ATPase may serve an
important role. The present study results also revealed that CD147
overexpression increased the resistance to epirubicin and docetaxel
in MCF-7 cells, while CD147 knockdown enhanced the sensitivity to
epirubicin and docetaxel in MDA-MB-231 cells, suggesting that
decreased pHe value may cause the chemotherapeutic drug
resistance of epirubicin and docetaxel. This experiment was then
further verified by PPZ treatment in vitro, which is highly
effective in inhibiting V-ATPase. The results indicated that PPZ
partially reversed CD147-mediated chemoresistance in breast cancer
cells. Taken together, these data suggested that CD147 was able to
mediate drug resistance in breast cancer cells though interacting
with V-ATPase.
CD147 has also been demonstrated to specifically be
associated with cell surface expression and the appropriate
location of MCTs as a chaperone in the energy metabolism of tumors,
thus contributing to the tumor invasion, metastasis and drug
resistance (24,47–49). In
addition, CD147 have been observed to influence tumor drug
resistance through different mechanisms. These include cell
survival signaling pathways, drug transporter expression and
activity, glycolytic phenotype, and its cancer stem-like cell
characteristics (47). However, the
present study is the first to observe that CD147 regulates the
expression and activity of V-ATPase, thus regulating the
transmembrane pH and mediating tumor drug resistance. The current
study findings also suggested that the interaction CD147 and
V-ATPase is another mechanism of CD147 contribution to the tumor
acidic microenvironment. In recent years, it has been reported that
the stability of the CD147-MCT1 complex requires the co-binding of
other small molecule chaperones (49). However, it remains unclear whether
other small molecule chaperones are required in the CD147-V-ATPase
interaction, and the underlying mechanism should be further
investigated.
Breast cancer management has entered the era of
individualized multidisciplinary treatment, and it is widely
appreciated that individualized and novel strategies are required
for breast cancer treatment (50).
One strategy is the use of proton pump inhibitors which can enhance
the tumor chemosensitivity by increasing the pH of the tumor
microenvironment (5). Recent clinical
trials in animals with spontaneous tumors have indicated that
patient alkalization is capable of reversing acquired
chemoresistance in a large percentage of tumors that are refractory
to chemotherapy (51,52). The present study also used a PPI to
inhibit the function of V-ATPase, leading to alteration of the
transmembrane pH gradient, and the results revealed a certain
degree of reversal of the docetaxel resistance. Furthermore, these
results indicated that V-ATPase inhibitors have the potential to
increase the tumor sensitivity to chemotherapeutic drugs. However,
this reversal is not complete and indicates that other mechanisms
of drug resistance must exist. These underlying mechanisms are the
direction of our future studies, and multigene signatures should be
examined to comprehensively identify the drug resistance mechanism
of traditional drugs.
In conclusion, clinical and experimental approaches
were combined to demonstrate that CD147 was able to regulate
V-ATPase expression and activity to mediate drug resistance in
breast cancer. The findings of the present study suggested that the
interaction of CD147 and V-ATPase may be a good therapeutic target
for breast cancer drug resistance. To this end, drugs targeting the
CD147-V-ATPase complex, such as monoclonal antibodies, can be
specifically developed to overcome the occurrence of tumor
resistance. The cytology in vitro results of the present
study should also be confirmed in vivo in clinical
experiments.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (nos. 81101654 and
81573049).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LL and YK obtained funding to conduct the research
on the role of CD147 in breast cancer. SW, LT, JH, and GY collected
all the blood samples and clinicopathological factors of the
patients with breast cancer. LL and YK conducted all the
experiments, interpreted the results and drafted the manuscript.
All authors participated in the critical revision of the manuscript
and have read and approved the final version.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
Institutional and/or National Research Committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards. Research protocols for the use of human tissue were
approved by and conducted in accordance with the policies of the
Institutional Review Boards at Central South University (Central
South University, approval no. 201403152).
Consent for publication
All patients from Xiangya Hospital of Central South
University (Changsha, China) were informed that their resected
tumor samples may be used for medical research and their clinical
medical records may be used for publication at admission. Informed
consent was obtained from all individual participants included in
the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baguley BC: Multiple drug resistance
mechanisms in cancer. Mol Biotechnol. 46:308–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moulder S: Intrinsic resistance to
chemotherapy in breast cancer. Womens Health (Lond). 6:821–830.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taylor S, Spugnini EP, Assaraf YG,
Azzarito T, Rauch C and Fais S: Microenvironment acidity as a major
determinant of tumor chemoresistance: Proton pump inhibitors (PPIs)
as a novel therapeutic approach. Drug Resist Updat. 23:69–78. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baldini N, Scotlandi K, Barbanti-Bròdano
G, Manara MC, Maurici D, Bacci G, Bertoni F, Picci P, Sottili S,
Campanacci M, et al: Expression of P-glycoprotein in high-grade
osteosarcomas in relation to clinical outcome. N Engl J Med.
333:1380–1385. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Serra M, Scotlandi K, Manara MC, Maurici
D, Benini S, Sarti M, Campanacci M and Baldini N: Analysis of
P-glycoprotein expression in osteosarcoma. Eur J Cancer.
31A:1998–2002. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferrari S, Bertoni F, Zanella L, Setola E,
Bacchini P, Alberghini M, Versari M and Bacci G: Evaluation of
P-glycoprotein, HER-2/ErbB-2, p53, and Bcl-2 in primary tumor and
metachronous lung metastases in patients with high-grade
osteosarcoma. Cancer. 100:1936–1942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Y, Zhang CL, Zeng BF, Wu XS, Gao TT
and Oda Y: Enhanced chemosensitivity of drug-resistant osteosarcoma
cells by lentivirus-mediated Bcl-2 silencing. Biochem Biophys Res
Commun. 390:642–647. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song B, Wang Y, Xi Y, Kudo K, Bruheim S,
Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al:
Mechanism of chemoresistance mediated by miR-140 in human
osteosarcoma and colon cancer cells. Oncogene. 28:4065–4074. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaplan RN, Riba RD, Zacharoulis S, Bramley
AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et
al: VEGFR1-positive haematopoietic bone marrow progenitors initiate
the pre-metastatic niche. Nature. 438:820–827. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kato Y, Ozawa S, Miyamoto C, Maehata Y,
Suzuki A, Maeda T and Baba Y: Acidic extracellular microenvironment
and cancer. Cancer Cell Int. 13:892013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McIntyre A and Harris AL: The role of ph
regulation in cancer progression. Recent Results Cancer Res.
207:93–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Trédan O, Galmarini CM, Patel K and
Tannock IF: Drug resistance and the solid tumor microenvironment. J
Natl Cancer Inst. 99:1441–1454. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Damaghi M, Wojtkowiak JW and Gillies RJ:
pH sensing and regulation in cancer. Front Physiol. 4:3702013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Avnet S, Lemma S, Cortini M, Pellegrini P,
Perut F, Zini N, Kusuzaki K, Chano T, Grisendi G, Dominici M, et
al: Altered pH gradient at the plasma membrane of osteosarcoma
cells is a key mechanism of drug resistance. Oncotarget.
7:63408–63423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luciani F, Spada M, De Milito A, Molinari
A, Rivoltini L, Montinaro A, Marra M, Lugini L, Logozzi M, Lozupone
F, et al: Effect of proton pump inhibitor pretreatment on
resistance of solid tumors to cytotoxic drugs. J Natl Cancer Inst.
96:1702–1713. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pérez-Sayáns M, Somoza-Martin JM,
Barros-Angueira F, Diz PG, Rey JM and Garcia-Garcia A: Multidrug
resistance in oral squamous cell carcinoma: The role of vacuolar
ATPases. Cancer Lett. 295:135–143. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nabeshima K, Iwasaki H, Koga K, Hojo H,
Suzumiya J and Kikuchi M: Emmprin (basigin/CD147): Matrix
metalloproteinase modulator and multifunctional cell recognition
molecule that plays a critical role in cancer progression. Pathol
Int. 56:359–367. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang JM, Xu Z, Wu H, Zhu H, Wu X and Hait
WN: Overexpression of extracellular matrix metalloproteinase
inducer in multidrug resistant cancer cells. Mol Cancer Res.
1:420–427. 2003.PubMed/NCBI
|
|
21
|
Marieb EA, Zoltan-Jones A, Li R, Misra S,
Ghatak S, Cao J, Zucker S and Toole BP: Emmprin promotes
anchorage-independent growth in human mammary carcinoma cells by
stimulating hyaluronan production. Cancer Res. 64:1229–1232. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou S, Liao L, Chen C, Zeng W, Liu S, Su
J, Zhao S, Chen M, Kuang Y, Chen X and Li J: CD147 mediates
chemoresistance in breast cancer via ABCG2 by affecting its
cellular localization and dimerization. Cancer Lett. 337:285–292.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qin Z, Dai L, Bratoeva M, Slomiany MG,
Toole BP and Parsons C: Cooperative roles for emmprin and LYVE-1 in
the regulation of chemoresistance for primary effusion lymphoma.
Leukemia. 25:1598–1609. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Slomiany MG, Grass GD, Robertson AD, Yang
XY, Maria BL, Beeson C and Toole BP: Hyaluronan, CD44, and emmprin
regulate lactate efflux and membrane localization of
monocarboxylate transporters in human breast carcinoma cells.
Cancer Res. 69:1293–1301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gallagher SM, Castorino JJ, Wang D and
Philp NJ: Monocarboxylate transporter 4 regulates maturation and
trafficking of CD147 to the plasma membrane in the metastatic
breast cancer cell line MDA-MB-231. Cancer Res. 67:4182–4189. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao S, Chen C, Liu S, Zeng W, Su J, Wu L,
Luo Z, Zhou S, Li Q, Zhang J, et al: CD147 promotes MTX resistance
by immune cells through up-regulating ABCG2 expression and
function. J Dermatol Sci. 70:182–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen X, Su J, Chang J, Kanekura T, Li J,
Kuang YH, Peng S, Yang F, Lu H and Zhang JL: Inhibition of CD147
gene expression via RNA interference reduces tumor cell
proliferation, activation, adhesion, and migration activity in the
human Jurkat T-lymphoma cell line. Cancer Invest. 26:689–697. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. 25:402–408. 2001.
|
|
29
|
Lebeau A, Kriegsmann M, Burandt E and Sinn
HP: Invasive breast cancer: The current WHO classification.
Pathologe. 35:7–17. 2014.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McCarty KS Jr, Miller LS, Cox EB, Konrath
J and McCarty KS Sr: Estrogen receptor analyses. Correlation of
biochemical and immunohistochemical methods using monoclonal
antireceptor antibodies. Arch Pathol Lab Med. 109:716–721.
1985.PubMed/NCBI
|
|
32
|
Chen M, Huang SL, Zhang XQ, Zhang B, Zhu
H, Yang VW and Zou XP: Reversal effects of pantoprazole on
multidrug resistance in human gastric adenocarcinoma cells by
down-regulating the V-ATPases/mTOR/HIF-1α/P-gp and MRP1 signaling
pathway in vitro and in vivo. J Cell Biochem. 113:2474–2487. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Franck P, Petitipain N, Cherlet M,
Dardennes M, Maachi F, Schutz B, Poisson L and Nabet P: Measurement
of intracellular pH in cultured cells by flow cytometry with
BCECF-AM. J Biotechnol. 46:187–195. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liao L, Song M, Li X, Tang L, Zhang T,
Zhang L, Pan Y, Chouchane L and Ma X: E3 ubiquitin ligase UBR5
drives the growth and metastasis of triple negative breast cancer.
Cancer Res. 77:2090–2101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vichai V and Kirtikara K: Sulforhodamine B
colorimetric assay for cytotoxicity screening. Nat Protoc.
1:1112–1116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cotter K, Capecci J, Sennoune S, Huss M,
Maier M, Martinez-Zaguilan R and Forgac M: Activity of plasma
membrane V-ATPases is critical for the invasion of MDA-MB231 breast
cancer cells. J Biol Chem. 290:3680–3692. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hrabeta J, Groh T, Khalil MA, Poljakova J,
Adam V, Kizek R, Uhlik J, Doktorova H, Cerna T, Frei E, et al:
Vacuolar-ATPase-mediated intracellular sequestration of ellipticine
contributes to drug resistance in neuroblastoma cells. Int J Oncol.
47:971–980. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
García-García A, Pérez-Sayáns García M,
Rodríguez MJ, Antúnez-López J, Barros-Angueira F, Somoza-Martín M,
Gándara-Rey JM and Aguirre-Urízar JM: Immunohistochemical
localization of C1 subunit of V-ATPase (ATPase C1) in oral squamous
cell cancer and normal oral mucosa. Biotech Histochem. 87:133–139.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Muramatsu T: Basigin (CD147), a
multifunctional transmembrane glycoprotein with various binding
partners. J Biochem. 159:481–490. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kasinrerk W, Tokrasinwit N and Phunpae P:
CD147 monoclonal antibodies induce homotypic cell aggregation of
monocytic cell line U937 via LFA-1/ICAM-1 pathway. Immunology.
96:184–192. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Curtin KD, Meinertzhagen IA and Wyman RJ:
Basigin (EMMPRIN/CD147) interacts with integrin to affect cellular
architecture. J Cell Sci. 118:2649–2660. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Takahashi M, Suzuki S and Ishikawa K:
Cyclophilin A-EMMPRIN interaction induces invasion of head and neck
squamous cell carcinoma. Oncol Rep. 27:198–203. 2012.PubMed/NCBI
|
|
44
|
Grass GD, Dai L, Qin Z, Parsons C and
Toole BP: CD147: Regulator of hyaluronan signaling in invasiveness
and chemoresistance. Adv Cancer Res. 123:351–373. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li QQ, Wang WJ, Xu JD, Cao XX, Chen Q,
Yang JM and Xu ZD: Involvement of CD147 in regulation of multidrug
resistance to P-gp substrate drugs and in vitro invasion in breast
cancer cells. Cancer Sci. 98:1064–1069. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jamali S, Klier M, Ames S, Barros LF,
McKenna R, Deitmer JW and Becker HM: Hypoxia-induced carbonic
anhydrase IX facilitates lactate flux in human breast cancer cells
by non-catalytic function. Sci Rep. 5:136052015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Toole BP and Slomiany MG: Hyaluronan, CD44
and Emmprin: Partners in cancer cell chemoresistance. Drug Resist
Updat. 11:110–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li X, Yu X, Dai D, Song X and Xu W: The
altered glucose metabolism in tumor and a tumor acidic
microenvironment associated with extracellular matrix
metalloproteinase inducer and monocarboxylate transporters.
Oncotarget. 7:23141–23155. 2016.PubMed/NCBI
|
|
49
|
Kendrick AA, Schafer J, Dzieciatkowska M,
Nemkov T, D'Alessandro A, Neelakantan D, Ford HL, Pearson CG,
Weekes CD, Hansen KC and Eisenmesser EZ: CD147: A small molecule
transporter ancillary protein at the crossroad of multiple
hallmarks of cancer and metabolic reprogramming. Oncotarget.
8:6742–6762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ellsworth RE, Decewicz DJ, Shriver CD and
Ellsworth DL: Breast cancer in the personal genomics era. Curr
Genomics. 11:146–161. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ferrari S, Perut F, Fagioli F, Del Prever
Brach A, Meazza C, Parafioriti A, Picci P, Gambarotti M, Avnet S,
Baldini N and Fais S: Proton pump inhibitor chemosensitization in
human osteosarcoma: From the bench to the patients' bed. J Transl
Med. 11:2682013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Spugnini EP, Buglioni S, Carocci F,
Francesco M, Vincenzi B, Fanciulli M and Fais S: High dose
lansoprazole combined with metronomic chemotherapy: A phase I/II
study in companion animals with spontaneously occurring tumors. J
Transl Med. 12:2252014. View Article : Google Scholar : PubMed/NCBI
|