Introduction
Ovarian cancer is the leading cause of mortality
among women with gynecological malignancies. It is frequently
diagnosed at an advanced, incurable stage and has a poor survival
rate, owing to its asymptomatic development (1–3).
High-grade serous ovarian cancer (HGSOC) is the most aggressive and
common subtype of ovarian cancer, accounting for two-thirds of
cancer-associated mortalities (1).
HGSOC is characterized by a mutation in tumor protein p53 (TP53), a
low rate of other mutations and extensive DNA copy number changes
(4,5).
Therefore, it is crucial to elucidate the etiology and
carcinogenesis of ovarian cancer, particularly in patients with
HGSOC. The traditional theory, which states that ovarian cancer
originates from ovarian surface epithelium (OSE), has been
challenged fundamentally in recent years (6,7).
Currently, a new paradigm for the pathogenesis of HGSOCs, with its
origins in the fallopian tube epithelium (FTE), has been proposed;
it is supported by numerous studies and has been termed ‘tubal
origin’ theory (8–10). It has been identified that paired box
gene 8 (PAX8), which is considered as the marker of the organs of
Müllerian origin including FTE, was highly expressed in HGSOC and
FTE, but not OSE (11). PAX8 serves
an important role as a distinguished factor in comparing the
immunophenotype of HGSOC, FTE and OSE. It indicates that the
immunophentype of HGSOC and FTE showed more similarity than that of
OSE, which supports ‘tubal origin’ theory (11).
Considering the traditional theory focuses on the
differentially expressed genes between HGSOC and OSE, it is of
clinical importance to establish the true origin of HGSOC and
identify the genes differentially expressed between HGSOC and FTE,
which may be useful in investigating ovarian carcinogenesis and
providing early detection in clinics, based on the understanding of
tubal origin theory. We hypothesize that HGSOC arises from FTE, and
that there should be certain differentially expressed genes between
them. Consequently, there were two steps in the present study.
Firstly, the immunophenotype and gene expression profiling among
HGSOC, OSE and FTE were compared and analyzed to find evidence
supporting tubal origin theory through immunohistochemistry and
microarray analysis. Secondly, the candidate genes of HGSOC that
were possibly associated with ovarian carcinogenesis were
identified through clustering analysis and expression
confirmation.
Materials and methods
Sample collection
All tissue samples were obtained from the Department
of Gynecology in The Affiliated Hospital of Qingdao University
between January, 2011 and May, 2012, following approval of the
institutional review board from the Affiliated Hospital of Qingdao
University (Qingdao, China). A total of 61 cases were used in the
present study, which were assigned to three groups: 21 cases of
HGSOC (range 33–69 years, median 46.6 years), 20 cases of OSE
(range 47–73 years, median 49.2 years) and 20 cases of FTE (range
47–73 years, median 49.2 years) were used. The tissues samples of
HGSOC were collected from patients who underwent surgical resection
of ovarian tumors with written consent, while samples of FTE and
OSE were obtained with written consent from patients who underwent
a hysterectomy and bilateral adnexectomy owing to benign uterine
disease.
Immunohistochemistry (IHC) and
scoring
Immunostaining was performed to evaluate the
expression of paired box gene 8 (PAX8) in the samples using
Image-Pro Plus v6.0 software (Media Cybernetics, Silver Spring, MD,
USA). Following fixation with 4% paraformaldehyde for 20 h at 23°C,
5 µm paraffin-embedded sections were deparaffinized at 67°C for 5
min, and rehydrated with double-distilled water. The antigens were
unmasked with the heat-mediated antigen retrieval method in citrate
buffer (pH 6.0) (11). Incubation
with the anti-PAX8 (cat. no. 10336-1-AP; Proteintech, Chicago, IL)
(dilution: 1:100) primary antibody was performed at 4°C overnight.
Specific signals were visualized by incubation with a
peroxidase-conjugated secondary antibody (cat. no. ab6721; Abcam,
Cambridge, MA, USA) for 60 min at 23°C followed by incubation with
3,3/-diaminobenzidine (DAB) as the chromogen at 23°C for 5 min,
creating a brown stain. Counterstaining with in hematoxylin for 5
min at 23°C was performed, and a coverslip was placed on the
samples. Cases were scored as positive if nuclear staining was
observed in >5% of cells using a light Leica DM6000 (Leica
Microsystems, GmbH, Wetzlar, Germany) at magnification, ×200, as
described previously (11). The
scoring process was supervised by two pathologists.
mRNA expression profiling
Four samples in each of the HGSOC, FTE and OSE
groups were randomly selected and prepared for microarray analysis.
Total RNA was extracted from all sample tissues using the RNeasy
kit (Qiagen, Inc., Valencia, CA, USA) according to the
manufacturer's protocol. Microarray studies were performed by
Capital Medical University Microarray Centre (Beijing, China) using
Illumina humanHT-12 v4 expression BeadChip (Illumina, Inc., San
Diego, CA, USA), based on the Illumina BeadStation500. Biotinylated
cRNA preparation, hybridization and scanning of microarrays were
performed according to the manufacturer's protocol. Triple
biological replicates were used to reduce errors. Illumina Gene
Expression BeadChip possesses internal control features to monitor
data quality. The GenomeStudio software (version 2009.2, Illumina,
Inc. San Diego, CA, USA) calculated and reported a detection
P-value, which determined whether a transcript on the array was
detected. In the present study, a detection value of P<0.01
indicated that a gene could be considered as expressed.
Differentially expressed genes between HGSOC and FTE were also
identified and analyzed. The output was filtered to include genes
whose expression was altered at least two-fold. Gene Ontology (GO)
analysis was performed to explore the cell function of
differentially expressed genes using The Database for Annotation,
Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/tools.jsp) and the data
was summarized in Results section. The microarray analysis dataset
was deposited in ArrayExpress (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3706/,
last access date on 2 July 2015). Through the literature review in
NCBI Pubmed (https://www.ncbi.nlm.nih.gov/pubmed, search term:
TACTSD, ovarian cancer), the differentially expressed genes were
evaluated for additional study.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to confirm differential gene
expression of candidate markers between HGSOC and FTE using the
BIO-RAD IQ5 Real-Time PCR System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The source of total RNA was obtained from the
tissues samples of HGSOC were collected from patients who underwent
surgical resection of ovarian tumors with written consent, while
samples of FTE were obtained with written consent from patients who
underwent a hysterectomy and bilateral adnexectomy owing to benign
uterine disease. The total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
cDNA was synthesized using 1 µg total RNA, oligo (dT) 18 primer
(Invitrogen; Thermo Fisher Scientific, Inc.), and Superscript™ III
Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). SYBR Green I was used as the fluorophore
(Invitrogen; Thermo Fisher Scientific, Inc.). Synthesis was
performed according to the manufacturer's protocol (42°C 5 min,
95°C for 10 sec, 58°C for 30 sec, for 40 cycles) and quantified
using the 2−ΔΔCt quantitative method (12). All the primers for TACSTD2 were
designed with Primer Express software 3.0.1 (forward
5′-GCTTCCCTGTTCTGATCCTATC-3′, and reverse
5′-TCTTATACTCTACCCGACCTGC-3′). β-actin was used as a reference
(forward 5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse
5′-GCTGTCACCTTCACCGTTCC-3′) (Applied Biosystems; Thermo Fisher
Scientific, Waltham, MA, USA). Predicted PCR product sequences were
verified using Basic local alignment search tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) for
recognition of target and non-target sequences.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
(SPSS, Inc., Chicago, IL, USA). An unpaired Student's t-test was
used to test for statistical significance. Data are presented as
the mean ± standard error of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
Immunophenotypic similarity between
HGSOC and FTE
Previously, the fallopian tube was considered to be
the organ of Müllerian origin, whereas OSE was associated with a
mesothelial origin (8). To compare
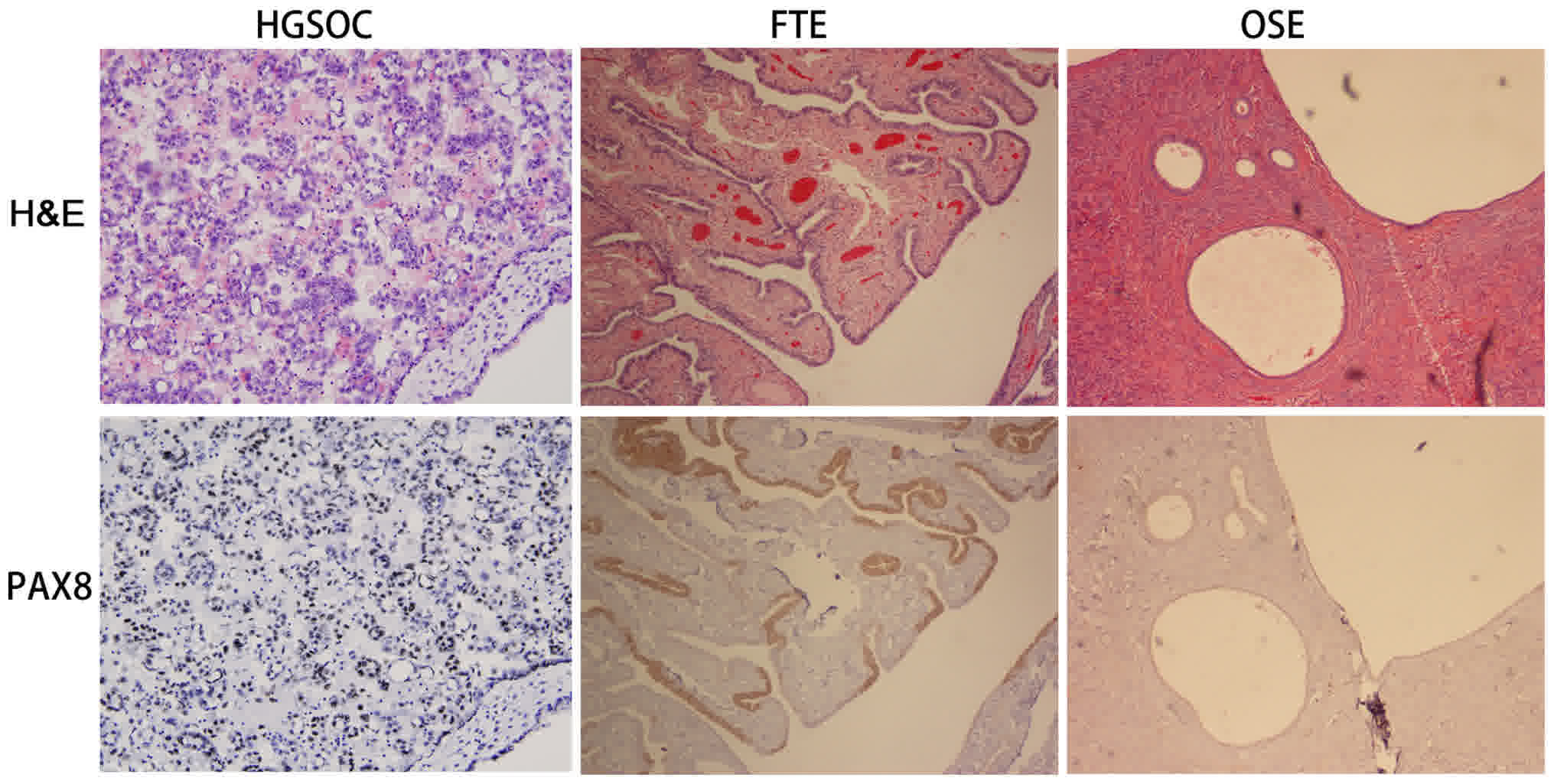
the immunophenotype of HGSOC, FTE and OSE, IHC staining was
performed to confirm the expression of PAX8 (Fig. 1), which is considered to be a
potential marker for organs of Müllerian origin (11). The results of the present study
demonstrated that PAX8 was highly expressed in HGSOC (19/21, 90.4%)
and FTE (20/20, 100%), but not in OSE (3/20, 14.3%). The
consistency of PAX8 expression in HGSOC and FTE indicated the
immunophenotypic similarity, thereby supporting a tubal origin
theory.
Similarities in the gene expression
profile of HGSOC and FTE
A dendrogram illustrated that HGSOC and FTE were
clustered to the same branch with closer distance, when compared
with OSE, indicating the presence of similarities in the gene
expression profiles of HGSOC and FTE (Fig. 2A).
 | Figure 2.Gene expression profiling of HGSOC,
FTE and OSE. (A) Dendrogram demonstrating that HGSOC and FTE were
clustered to the same branch, compared with OSE, indicating
similarities in the gene expression profiles of HGSOC and FTE.
S1-S4, HGSOC samples; F1-F4, FTE samples; T1-T4, OSE samples. (B)
Heatmap plot of scaled gene-expression levels through hierarchical
clustering. S1-S4, HGSOC samples; F1-F4, FTE samples; T1-T4, OSE
samples. (C) Gene Ontology analysis indicating that these genes,
including upregulated genes and downregulated genes, were primarily
involved in endoplasmic reticulum, extracellular region, cell
fraction and structural molecule activity. HGSOC, high-grade serous
ovarian carcinoma; FTE, fallopian tube epithelium; OSE, ovarian
surface epithelium; GO, gene ontology. |
Differentially expressed genes between
HGSOC and FTE
In total, 2,412 differentially expressed genes were
identified in the microarray (absolute fold-change >2) between
HGSOC and FTE, including 822 upregulated genes and 1,590
downregulated genes. The differentially expressed genes are
summarized in Fig. 2B and Table I. Furthermore, Gene Ontology (GO)
analysis revealed that these genes were primarily involved in the
endoplasmic reticulum, extracellular region, cell fraction and
structural molecule activity (Fig.
2C). The differentially expressed genes were also annotated in
several Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways,
including focal adhesion (hsa04510), pathways in cancer (hsa05200),
and the peroxisome proliferator-activated receptor (PPAR) signaling
pathway (hsa03320) (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3706/).
 | Table I.Representative differentially
expressed genes between high-grade serous ovarian carcinoma and
fallopian tube epithelium identified by gene expression
profiling. |
Table I.
Representative differentially
expressed genes between high-grade serous ovarian carcinoma and
fallopian tube epithelium identified by gene expression
profiling.
| Gene | Full gene name | Fold-change |
|---|
| Upregulated |
|
|
|
S100P | S100 calcium binding
protein P | 61.3 |
|
RASIP1 | Ras interacting
protein 1 | 50.6 |
|
WNT5A | Wingless-type MMTV
integration site family, member 5A | 28.3 |
|
TACSTD2 | Tumor-associated
calcium signal transducer 2 | 25.9 |
|
PTGES | Prostaglandin E
synthase | 23.3 |
|
TPD52L1 | Tumor protein
D52-like 1 | 19.7 |
|
MAP1LC3A |
Microtubule-associated protein 1 light
chain 3 alpha | 19.4 |
|
MGST1 | Microsomal
glutathione S-transferase 1 | 16.7 |
|
TMPRSS3 | Transmembrane
protease, serine 3 | 14.7 |
| Downregulated |
|
|
| DKK3 | Dickkopf WNT
signaling pathway inhibitor 3 | −117.2 |
|
LDOC1 | Leucine zipper,
down-regulated in cancer 1 | −28.3 |
|
TUSC3 | Tumor suppressor
candidate 3 | −25.9 |
| LXN | Latexin | −25.0 |
Confirmation of the differential
expression of candidate genes in HGSOC and FTE
A total of six genes differentially expressed
between HGSOC and FTE were selected as candidate genes, and were
confirmed via RT-qPCR using triplicate samples, including four
upregulated [(S100 calcium binding protein P (S100P),
Ras-interacting protein 1 (RASIP1), Wnt family member 5A (WNT5A),
and tumor-associated calcium signal transducer 2 (TACSTD2)] and two
downregulated [Dickkopf Wnt signaling pathway inhibitor 3 (DKK3 and
tumor suppressor candidate 3 (TUSC3)] genes in cancer. A
significant difference was detected between the expression of
candidate genes in HGSOC and FTE (P<0.05). The data are
presented in Fig. 3.
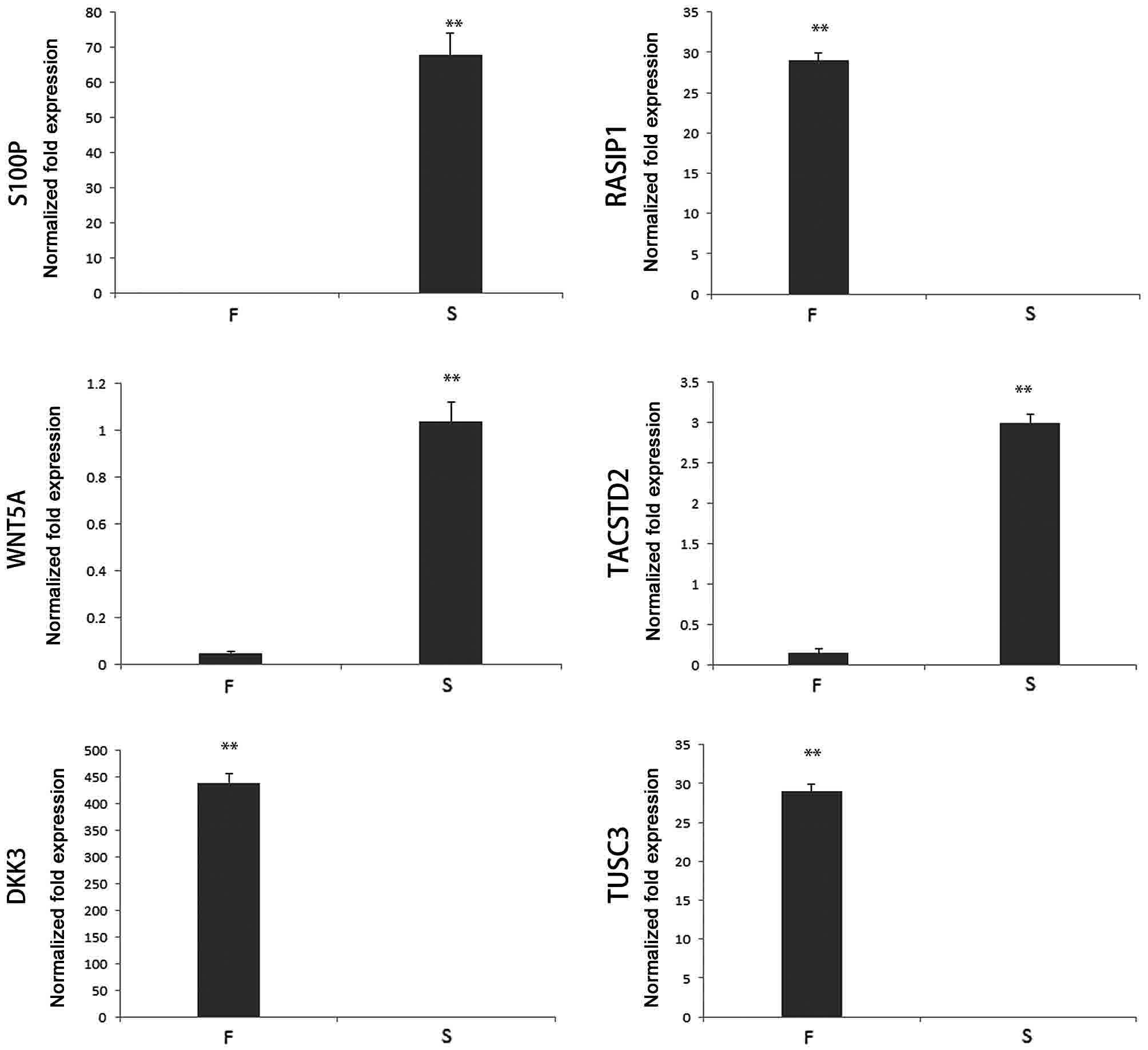 | Figure 3.Differential expression of candidate
genes in HGSOC and FTE (**P<0.01). The candidate genes included
genes upregulated in ovarian cancer (S100P, RASIP1, WNT5A,
TACSTD2), as well as genes downregulated in ovarian cancer DKK3 and
TUSC3. The differential expression of 6 genes in the FTE and HGSOC
was confirmed by reverse transcription-quantitative polymerase
chain reaction. HGSOC, high-grade serous ovarian carcinoma; FTE,
fallopian tube epithelium; OSE, ovarian surface epithelium; F, FTE;
S, HGSOC; S100P, S100 calcium-binding protein P; RASIP1,
Ras-interacting protein 1; WNT5A, Wnt family member 5A; TACSTD2,
tumor-associated calcium signal transducer 2; DKK5, Dickkopf Wnt
signaling pathway inhibitor 3; TUSC3, tumor suppressor candidate
3. |
Discussion
Ovarian cancer is the most lethal gynecological
cancer in women. An estimated 22,240 new cases of ovarian cancer
will be diagnosed and 14,070 cancer-associated mortalities will
occur in the United States in 2018, whereas rates are substantially
higher in China, where a total of 52,100 incidence of ovarian
cancer and 22,500 mortalities (2,3).
Epithelial cancer is considered to be the most common type of
ovarian cancer and is responsible for ~90% of cases. Ovarian
epithelial cancer consists of a heterogeneous group of histological
subtypes, including serous, endometrioid, clear cell, and mucinous
carcinoma. HGSOC represents the majority of cases of advanced-stage
ovarian cancer and is frequently associated with a poor prognosis
(7). Therefore, it is crucial to
elucidate the carcinogenesis of HGSOC and identify useful tumor
markers to improve treatment and prognosis.
The traditional theory that ovarian cancer arises
from OSE was initially proposed by Fathalla in 1971 (13). It was assumed that that the repeated
overuse and repair of ovarian epithelium culminated in
transformations, a concept that was further supported by
epidemiological evidence revealing an increased incidence of
ovarian cancer in women who had never been pregnant (14). However, the origin of ovarian cancer
has been subject to controversy, and the conventional theory has
been challenged: The process of transformation from OSE to ovarian
cancer has never been precisely identified and defined, and ovarian
cancer is more histologically similar to FTE rather than OSE
(15).
A model proposing that FTE may be the origin of
HGSOC has been developed. Initially, it was reported that tubal
carcinoma was detected in 5 of 13 cases in prophylactic
adnexectomies from women with breast cancer susceptibility protein
(BRCA) mutations (BRCA positive) following a protocol of sectioning
and extensively examining the fimbria (SEE-FIM); however, no
ovarian carcinomas were identified, indicating that the fimbria was
the most common location for early serous carcinoma in this series
of BRCA-positive women (16).
Subsequently, Callahan et al (17) reported that 7 consecutive cancer
cases, from 123 cases of BRCA-positive women undergoing surgery for
ovarian cancer risk reduction, originated in the fimbrial or
ampullary region of the tube, 6 of which had an early
(intraepithelial) component. This was corroborated by evidence
supporting the tubal origin of ovarian cancer (18,19).
Clonal alterations in TP53 in benign tubal epithelium, which are
referred to as p53 signatures, and generic secretory cell outgrowth
(SCOUT) in the FTE associated with altered PAX2 expression has
established a foundation for a serous cancer precursor in the
fimbria (10). Accordingly, the
Society of Gynecologic Oncology recommendations for the prevention
of ovarian cancer indicate the importance of the FTEs as a
potential source of HGSOC (20).
Furthermore, in parallel with the implementation of the new
International Federation of Gynecology and Obstetrics (FIGO)
staging classification (21), the
revised World Health Organization (WHO) classification eliminates
the previous focus on the mesothelial origin of ovarian cancer, and
features a discussion of tubal carcinogenesis of hereditary and
other types of high-grade serous carcinomas (6).
Evidence has been provided for the theoretical tubal
origin of HGSOC, and gynecologists recommend a preventative
bilateral salpingectomy for women with a high-risk of ovarian
cancer (20). However, at the time of
writing, the tubal origin of HGSOC has not been demonstrated
completely or become the established clinical guideline based on
FIGO and WHO files (6,20–22). The
present study aimed to assess the possibility and feasibility of
identifying the genes differentially expressed between HGSOC and
FTE, which may be useful in investigating ovarian carcinogenesis.
The candidate genes identified in the present study are relatively
novel, including 4 upregulated (S100P, RASIP1, WNT5A and TACSTD2),
and 2 downregulated (DKK3 and TUSC3) genes in cancer. In the
present study, PAX8 was used for immunostaining as a recognized
marker for FTE and HGSOC, hence its selection for the
immunophenotypic comparison of FTE, HGSOC and OSE. The similarity
establishment of FTE and HGSOC was the first step in the present
study. Gene expression profile similarity was then confirmed and
the differentially expressed genes were identified. There were a
total of 2,412 differentially expressed genes analyzed recognized
in the microarray of the present study. Factors, including
fold-change, association with tumor origin and development, and
current research status were taken into consideration when deciding
which genes to study further. Upregulated and downregulated genes
in cancer following the order of fold-change were listed and the
published articles of the top 100 genes in each list were reviewed
to estimate their research status and tumor association. Through
the literature review in NCBI Pubmed, S100P, RASIP1, WNT5A,
TACSTD2, DKK3 and TUSC3, which are closely associated with tumor
development, were chosen as the candidate markers in the present
study.
Accumulating evidence has demonstrated that the
aforementioned candidate genes are involved in the tumorigenesis
and progression of multiple cancer types, including ovarian cancer.
For example, high expression of S100P is associated with
unfavorable prognosis and tumor progression in patients with
epithelial ovarian cancer (23,24). Post
et al (25) reported that
RASIP1 mediates Rap1 regulation of Rho in endothelial barrier
function through ArhGAP29, which may serve an important role in the
development of tumors (25). Studies
have demonstrated that WNT5A exerts immunomodulatory activity and
influences viability, migration, adhesion, colony formation and the
expression of E- and N-cadherin in the human ovarian cancer SKOV-3
cell line (26,27). TACSTD2 is an intracellular calcium
signal transducer that is differentially expressed in a number of
cancer types (28). DKK3 may serve a
key role in the inhibition of cancer cell proliferation by
regulating Wnt signaling (29).
Expression of TUSC3 prevents the epithelial-to-mesenchymal
transition and inhibits tumor growth by modulating the endoplasmic
reticulum stress response in ovarian cancer cells (30). Further study of these candidate genes
is necessary to investigate the molecular mechanism of cancer
development.
To conclude, in the present study, a greater
similarity of immunophenotype and gene expression profile was
observed between HGSOC and FTE when compared with OSE, and a total
of 6 candidate genes that are potentially associated with the
carcinogenesis of HGSOC were identified. It is evident that the
present study is relatively superficial, and merely offers
supportive evidence for tubal origin theory and potential gene
markers for future study. However, it is possible that any novel
knowledge regarding the tubal origin of ovarian cancer may open
novel pathways in basic research and clinical studies.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Fund (grant nos. 81402157 and 81502310) and the Doctoral
Fund of The Affiliated Hospital of Qingdao University (grant no.
2075). The funders had no role in study design, data collection and
analysis, decision to publish, or preparation of the
manuscript.
Availability of data and materials
The microarray analysis dataset was deposited in
ArrayExpress (accession no. E-MTAB-3706, https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3706/).
Author contributions
XL conceived and designed the experiments, GR and ZJ
performed the experiments: GR and WZ analyzed the data, SF
collected the samples and cases and XL wrote the paper.
Ethics approval and consent to
participate
The study was approved by the Institutional Review
Board and Human Ethics Committee of Affiliated Hospital of Qingdao
University. Written informed consent for using the samples for
research purposes was obtained from all patients prior to surgical
resection.
Consent for publication
The authors declare that the patient, parent,
guardian or next of kin (in case of deceased patients) provided
written informed consent for the publication of any associated data
and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kujawa KA and Lisowska KM: Ovarian
cancer-from biology to clinic. Postepy Hig Med Dosw (Online).
69:1275–1290. 2015.(Article in Polish). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eng KH, Hanlon BM, Bradley WH and Szender
JB: Prognostic factors modifying the treatment-free interval in
recurrent ovarian cancer. Gynecol Oncol. 139:228–235. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ramalingam P: Morphologic,
immunophenotypic, and molecular features of epithelial ovarian
cancer. Oncology (Williston Park). 30:166–176. 2016.PubMed/NCBI
|
|
6
|
Meinhold-Heerlein I, Fotopoulou C, Harter
P, Kurzeder C, Mustea A, Wimberger P, Hauptmann S and Sehouli J:
The new WHO classification of ovarian, fallopian tube, and primary
peritoneal cancer and its clinical implications. Arch Gynecol
Obstet. 293:695–700. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kurman RJ: Origin and molecular
pathogenesis of ovarian high-grade serous carcinoma. Ann Oncol. 24
Suppl 10:S16–S21. 2013. View Article : Google Scholar
|
|
8
|
Perets R and Drapkin R: It's totally
Tubular…Riding the new wave of ovarian cancer research. Cancer Res.
76:10–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ning G, Bijron JG, Yamamoto Y, Wang X,
Howitt BE, Herfs M, Yang E, Hong Y, Cornille M, Wu L, et al: The
PAX2-null immunophenotype defines multiple lineages with common
expression signatures in benign and neoplastic oviductal
epithelium. J Pathol. 234:478–487. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mehra K, Mehrad M, Ning G, Drapkin R,
McKeon FD, Xian W and Crum CP: STICS, SCOUTs and p53 signatures; a
new language for pelvic serous carcinogenesis. Front Biosci (Elite
Ed). 3:625–634. 2011.PubMed/NCBI
|
|
11
|
Xiang L, Zheng W and Kong B: Detection of
PAX8 and p53 is beneficial in recognizing metastatic carcinomas in
pelvic washings, especially in cases with suspicious cytology.
Gynecol Oncol. 127:595–600. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Dalta Dalta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fathalla MF: Incessant ovulation-a factor
in ovarian neoplasia? Lancet. 2:1631971. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fraumeni JF Jr, Lloyd JW, Smith EM and
Wagoner JK: Cancer mortality among nuns: Role of marital status in
etiology of neoplastic disease in women. J Natl Cancer Inst.
42:455–468. 1969.PubMed/NCBI
|
|
15
|
Mhawech-Fauceglia P, Wang D, Samrao D,
Godoy H, Ough F, Liu S, Pejovic T and Lele S: Pair Box 8 (PAX8)
protein expression in high grade, late stage (stages III and IV)
ovarian serous carcinoma. Gynecol Oncol. 127:198–201. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Medeiros F, Muto MG, Lee Y, Elvin JA,
Callahan MJ, Feltmate C, Garber JE, Cramer DW and Crum CP: The
tubal fimbria is a preferred site for early adenocarcinoma in women
with familial ovarian cancer syndrome. Am J Surg Pathol.
30:230–236. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Callahan MJ, Crum CP, Medeiros F,
Kindelberger DW, Elvin JA, Garber JE, Feltmate CM, Berkowitz RS and
Muto MG: Primary fallopian tube malignancies in BRCA-positive women
undergoing surgery for ovarian cancer risk reduction. J Clin Oncol.
25:3985–3990. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seidman JD: Serous tubal intraepithelial
carcinoma localizes to the tubal-peritoneal junction: A pivotal
clue to the site of origin of extrauterine high-grade serous
carcinoma (ovarian cancer). Int J Gynecol Pathol. 34:112–120. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Erickson BK, Conner MG and Landen CN Jr:
The role of the fallopian tube in the origin of ovarian cancer. Am
J Obstet Gynecol. 209:409–414. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Walker JL, Powell CB, Chen LM, Carter J,
Jump Bae VL, Parker LP, Borowsky ME and Gibb RK: Society of
gynecologic oncology recommendations for the prevention of ovarian
cancer. Cancer. 121:2108–2120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duska LR and Kohn EC: The new
classifications of ovarian, fallopian tube, and primary peritoneal
cancer and their clinical implications. Ann Oncol. 28 Suppl 8:2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roche Long KC, Abu-Rustum NR, Nourmoussavi
M and Zivanovic O: Risk-reducing salpingectomy: Let us be
opportunistic. Cancer. 123:1714–1720. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Tian T, Li X, Zhao M, Lou Y, Qian
J, Liu Z, Chen H and Cui Z: High expression of S100P is associated
with unfavorable prognosis and tumor progression in patients with
epithelial ovarian cancer. Am J Cancer Res. 5:2409–2421.
2015.PubMed/NCBI
|
|
24
|
Surowiak P, Maciejczyk A, Materna V,
Drag-Zalesińska M, Wojnar A, Pudelko M, Kedzia W, Spaczyński M,
Dietel M, Zabel M and Lage H: Unfavourable prognostic significance
of S100P expression in ovarian cancers. Histopathology. 51:125–128.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Post A, Pannekoek WJ, Ross SH, Verlaan I,
Brouwer PM and Bos JL: Rasip1 mediates Rap1 regulation of Rho in
endothelial barrier function through ArhGAP29. Proc Natl Acad Sci
USA. 110:11427–11432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arabzadeh S, Hossein G and Zarnani AH:
Wnt5A exerts immunomodulatory activity in the human ovarian cancer
cell line SKOV-3. Cell Biol Int. 40:177–187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jannesari-Ladani F, Hossein G, Monhasery
N, Shahoei SH and Mood Izadi N: Wnt5a influences viability,
migration, adhesion, colony formation, E- and N-cadherin expression
of human ovarian cancer cell line SKOV-3. Folia Biol (Praha).
60:57–67. 2014.PubMed/NCBI
|
|
28
|
Shvartsur A and Bonavida B: Trop2 and its
overexpression in cancers: Regulation and clinical/therapeutic
implications. Genes Cancer. 6:84–105. 2015.PubMed/NCBI
|
|
29
|
Mohammadpour H, Pourfathollah AA, Zarif
Nikougoftar M and Khalili S: Key role of Dkk3 protein in inhibition
of cancer cell proliferation: An in silico identification. J Theor
Biol. 393:98–104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kratochvílová K, Horak P, Ešner M, Souček
K, Pils D, Anees M, Tomasich E, Dráfi F, Jurtíková V, Hampl A, et
al: Tumor suppressor candidate 3 (TUSC3) prevents the
epithelial-to-mesenchymal transition and inhibits tumor growth by
modulating the endoplasmic reticulum stress response in ovarian
cancer cells. Int J Cancer. 137:1330–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|