Introduction
Circular RNA (circRNA) was first detected in RNA
viruses in the 1970s (1,2), in which the 3′ and 5′ ends are joined
together. Compared with the linear structure of coding RNAs, the
closed structure of circRNA means it is more conserved (3). CircRNA has been detected in several
species, including viruses (2),
plants (4), archaea (5)[Salzman, 2013 #198; Memczak, 2013 #291]
and animals (6). In eukaryotic cells,
circRNA has attracted attention due to its unique characteristics,
including high stability, specificity and evolutionary conservation
(7). The majority of circRNAs are
derived from exons of coding regions, 3′ UTRs, 5′ UTRs, introns,
intergenetic regions and antisense RNAs (8). CircRNAs, which have attracted attention
in recent years, can be produced by canonical and noncanonical
splicing as distinguished from the linear RNAs (9). Through high-throughput sequencing, three
types of circRNAs have been identified: Exonic circRNAs (7), circular intronic RNAs (ciRNAs) (9), and retained-intron circular RNAs or
exon-intron circRNAs (elciRNAs) (10). To date, evidence suggests that
circRNAs may regulate transcription and pathways by manipulating
microRNAs (11,12). circRNA_100290 acts as a competing
endogenous RNA to regulate the expression of cyclin-dependent
kinase (CDK)6 through regulating microRNA (miR)-29b family members
(13). circRNA-100269 and miR-630
comprise a novel pathway, which regulates the proliferation of
gastric cancer cells (14).
Circ-ABCB10 has provided novel insight into the pathogenesis of
breast cancer through regulating miR-1271 (15). Therefore, circRNAs are involved in the
development of diseases and have become novel biomarkers in cancer
(16,17).
CDR1as, as a circRNA with the most known miR-7
binding sites (>70), is known for its ‘circular RNA sponge for
miR-7’ (ciRS-7 and CDR1as), and there are >70 miR-7 binding
sites on this circRNA (3,18). CDR1as, as an endogenous RNA,
indirectly regulates microRNAs and is involved in the development
of multiple diseases through a variety of pathways (18,19). A
previous study showed that the CDR1as-miR-7 axis can regulate the
neocortical and hippocampal neurons in mouse brains (17,20). In
the embryonic zebrafish midbrain, the overexpression of CDR1as
induces developmental defects (6).
These findings indicate that CDR1as is important in different
mechanisms, acting as an miR-7 buffer (18,21). Other
functional investigations have shown that ciRS-7 inhibits the
activity of miR-7 and activates downstream genes of miR-7 to
perform a regulatory role in various types of cancer, including
colorectal cancer (17),
hepatocellular carcinoma (16) and
glioblastoma multiforme (22).
Melanoma, the most life-threatening form of skin
cancer, has changed markedly in the 21st century. Early detection,
genetic testing and substantial improvements in advanced melanoma
therapies are examples of such progress. However, the role of
circRNA in melanoma remains to be fully elucidated. miR-7 can
target melanoma through inhibiting the mitogen-activated protein
kinase (MAPK) and phosphoinositide 3-kinase (PI3K)/protein kinase B
(AKT) signaling pathways (23),
therefore, CDR1as serves as a novel and stable biomarker for the
diagnosis and progression of several types of cancer, including
melanoma, which is the most difficult form of skin cancer to treat
(24).
In the present study, the unknown roles of CDR1as in
a highly complex metabolic network were examined through novel
bioinformatics analysis as a novel field of investigation to
elucidate its full biologic functions in cellular regulation and in
human diseases.
Materials and methods
Bioinformatics databases and
software
Several databases and software packages were used in
the present study. The University of California, Santa Cruz (UCSC)
Genome Browser (http://genome.UCSC.Edu/), TargetScan (25) (http://www.Targetscan.Org/vert_61) predicts biological
targets of miRNAs by searching for the presence of conserved 8mer,
7mer and 6mer sites, which match the seed region of each miRNA
(26). In mammals, predictions are
ranked based on the predicted efficacy of targeting as calculated
using cumulative weighted context++ scores of the sites (27).
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.abcc.Nciferf.gov/) (28,29)
involves the Statistical Consultation Group, Computer &
Statistical Services Program, NCI-Frederick and NIH (30).
CircBase (http://www.circbase.org/) (31) contains data from all studies of
large-scale circRNA identification published to date. In addition
to the information made available by the investigators, the present
study annotated all circRNA transcripts and predicted their
putative spliced forms. Transcript reannotation was necessary to
standardize the content and level the quantity of information
contained across certain publications (10,32).
For CircNet (http://circnet.mbc.nctu.edu.tw/) (33), perfect complementarity was required of
sequences for a circRNA-miRNA target association to be identified.
To normalize the number of occurrences of these sites, the
following formula was used: Frequency of Nmer=(number of target
seeds × 1,000)/(N × length of circRNA). With this formula, four
frequency numbers can be acquired from each circRNA-miRNA pair. To
distinguish circRNA from linear isoforms, frequency values were
also calculated for linear mRNA and miRNA pairs. P-values for each
circRNA and miRNA association were acquired by one minus the
calculated cumulated distribution function of the frequency Z
score. For a circRNA-miRNA pair, P<0.001 indicates high
regulatory potential between the circRNA and miRNA (33).
CircInteractome (http://circinteractome.nia.nih.gov/) comprises
datasets, including the binding sites of 35 RBPs identified using
PAR-CLIP, HITS-CLIP, or iCLIP, retrieved from starBase v2.0
(http://starbase.sysu.edu.cn/), which
decodes miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction
networks from large-scale CLIP-Seq data (34).
Using Circ2Traits (http://gyanxet-beta.com/circdb/), a measure was
implemented for the likelihood of a circRNA to be associated with a
disease in terms of the significance of the interaction between the
circRNA and miRNAs associated with the disease of interest. In the
present study, P-values were calculated for the association between
each circRNA interacting with disease-associated miRNAs. The
p-value for the likelihood of the circRNA to be enriched for
interaction with miRNAs associated with that disease was calculated
as follows: P=∑i=mdmin (mc, Md) (Mdi) (MT-Mdmc-i) (MTmc). MT is the
total number of miRNAs in the human genome, mc is the number of
miRNAs interacting with the circRNA, Md is the total number of
miRNAs associated with the disease, md is the number of miRNAs
associated with the disease which interact with the circRNA.
Following Bonferroni correction, P<0.05 indicated the circRNA
was likely to be associated with the disease, and
pthreshold=0.05/mm indicated the total number of circRNAs
interacting with any miRNA associated with the disease (35).
MiRanda (http://.Mirorna.Org/microrna/getDownload) is an mirSVR
predicted target site scoring method: Comprehensive modeling of
microRNA targets predicts functional non-conserved and
non-canonical sites (mirSVRscore <-1) (36).
Methods of analysis
The location of CDR1as in human and mouse genomes
were supplied by the UCSC Genome Browser database. The roles of
CDR1as in cancer are summarized in the circ2Traits database. The
CircBase, circNet, miRanda, TargetScan and circInteractome
databases were used to detect the interaction and co-expression of
CDR1as, miRNAs and proteins. The functions and the Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathways of the associated
proteins were predicted using DAVID.
Results
Location of CDR1as
CDR1as is derived from an antisense transcript of
the CDR1 at chromosome X (NC_004065) in humans and chromosome X
(NC_000086.7) in mice, located in the ChrX:139865339-139866824 and
chX:58436422-58539349 (Fig. 1).
MicroRNA prediction
Correlations between circRNAs and microRNAs were
determined using CircInteractome and TargetScan. CDR1as contained
>70 conserved sites for miR-7, in addition to miR-671-5p,
miR-3134 and miR-139-3p, which are important target miRNAs
regulating metabolism. The predicted microRNAs in these two
databases comprised 15 combined microRNAs, as shown in the Table I (context score percentile
>90).
 | Table I.miRs interacting with CDR1as. |
Table I.
miRs interacting with CDR1as.
| Circular RNA | miR |
|---|
| CDR1as | miR-7-5p,
miR-671-5p, miR-3134, miR-139-3p, miR-6807-5p, miR-3617-5p,
miR-3617-3p, miR-4254, miR-3145-3p, miR-4291, miR-3201, miR-4306,
miR-471-5p, miR-3065-3p, miR-3132, |
Protein prediction
The predicted proteins were all targeted by the
microRNAs interacting with CDR1as. A total of 60 proteins were
identified using the cirNet database and miRanda database in total
(P<0.05, mirSVRscore <-1). Several of these proteins are
associated with cancer invasion and metastasis, including AKT3,
Secretory carrier-associated membrane protein 2 (SCAMP2), SLU7,
B-cell lymphoma 2 (BCL2) and CDK1. BCL2 is an important gene, which
inhibits the apoptotic death of certain cells, including
lymphocytes. CDK1 functions as a serine/threonine kinase, and is key in cell cycle regulation (Table II).
 | Table II.Target proteins of microRNAs
interacting with CDR1as. |
Table II.
Target proteins of microRNAs
interacting with CDR1as.
| Circular RNA | MicroRNA-interacted
protein |
|---|
| CDR1as | PAPD7, PTN3,
CDKIAP2, C11orf74, PPIF, TUBA1B, CDR1, ABHD14A-ACY1, GNA13, AKT3,
GJC1, SCAMP2, CNE3, MED16, WASF2, LRPPRC, URGCP-MRPS24, OCID2,
OCIAD2, SNRNP27, ZNF730, PARP2, TMEM170A, CHEK1, SMC4, MGAT4A,
TRAF3IP2, ZNF689, SCUBE3, FAM13A, MTRNRIL3, RAB10, Bcl2l2-PABPN1,
ARIH2, CYB5D1, ZBTB33, FEM1B, GNA13, CDK6, MTHFD2, IVNS1ABP, NUPL2,
ARPP19, C17orf49, PNRC1, RNF103-CHMP3, USF, MICA, SLU7, BTBD9,
MYL12B, CDKN2B, C1QTNF6, DCAF7, NBPF10, STX1B, ABCC5, SNX20,
VAS1V |
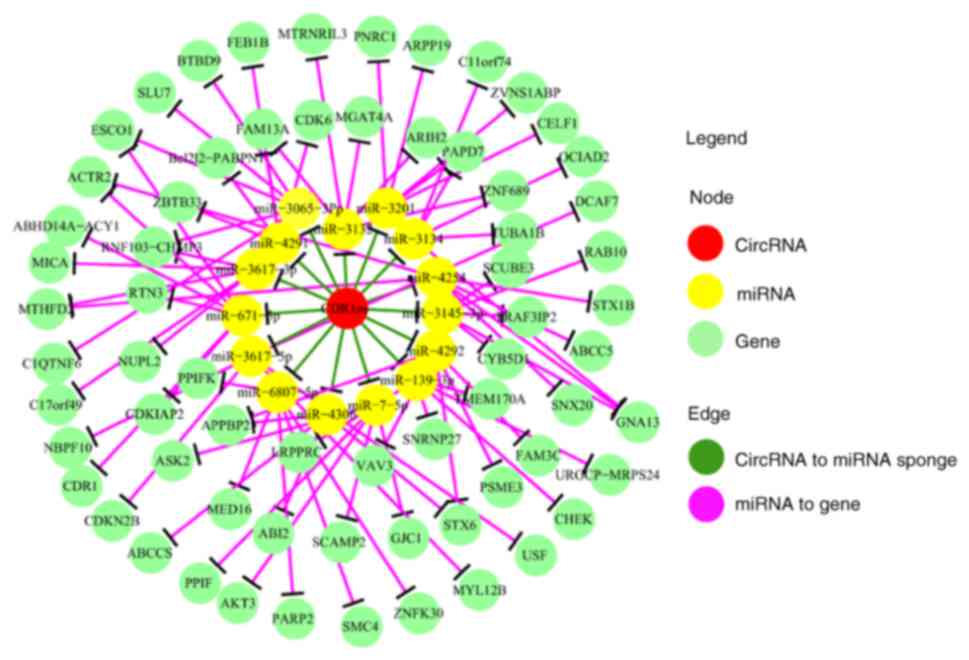
circRNA-microRNA-protein co-expression
network
The potential microRNA-circRNA-mRNA interaction
analysis, which was determined using the circNet database, was used
to elucidate the correlations between circRNAs and microRNA. In the
present study, the top 15 miRNAs that interacted with CDR1as were
selected (yellow in Fig. 2), and 66
mRNAs (light green in Fig. 2), were
identified as the targets of these miRNAs (P<0.001 and support
experiment >5).
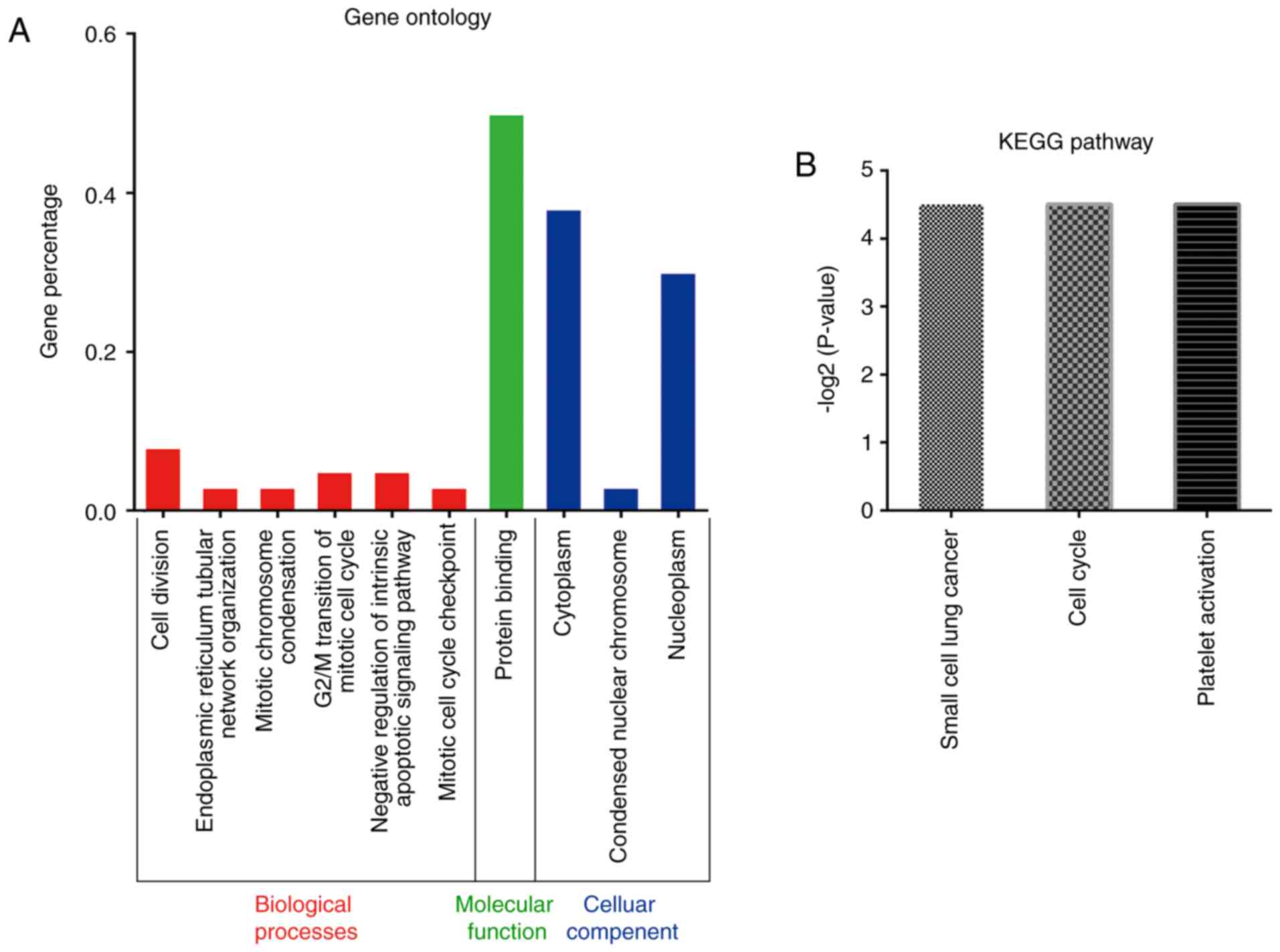
Gene Ontology (GO) and KEGG) pathway
analysis of target genes
The results of the GO analysis revealed target genes
(P<0.05, Bonferroni corr <0.05) involved in biological
processes, including cell division and G2/M transition, which can
negatively regulate the intrinsic apoptotic signaling pathway in
the mitotic cell cycle. The main molecular function of these target
genes was protein binding. The cellular components of the target
genes comprised nucleoplasms, cytoplasm and condensed nuclear
chromosomes (Fig. 3A). These
processes are all associated with human tumorigenesis and cancer
metastasis. The results of the KEGG analysis showed three pathways
(P<0.05) associated with the target genes; these pathways were
small cell lung cancer, cell cycle and platelet activation
(Fig. 3B).
CDR1as and cancer
CDR1as was found to be involved in several types of
cancer, including gastric cancer, hepatic carcinoma, lung cancer,
breast cancer, glioma, diabetes, Parkinson's disease, cervical
cancer and melanoma. These findings were predicted in the
Circ2Traits database, which also supplies the associated protein.
BCL-2 is regulated by the CDR1as/miR-7 axis, which is involved in
Parkinson's disease and lung cancer. Phosphatidylinositol 3-kinase
catalytic subunit δ is associated with hepatic carcinoma.
p21-activated kinase-1 (PAK1) and Kruppel-like factor 4 (KLF4) are
important in breast cancer. Insulin receptor substrate (IRS)-2,
IRS-1 and PAK are associated with glioma. IRS-2, epidermal growth
factor receptor (EGFR), IGE-1, CRAF and paired box 6 (PAX6) have
all been reported be vital in the melanoma through interacting with
miR-7, therefore, CDR1as may offer novel clues for the treatment of
melanoma. The predictions of CDR1as-miR-7-mRNAs in different types
of cancer are shown in. Fig. 4. These
mRNAs associated with cancer have been reported and are the targets
of miR-7, therefore, CDR1as may be important in regulating these
types of cancer.
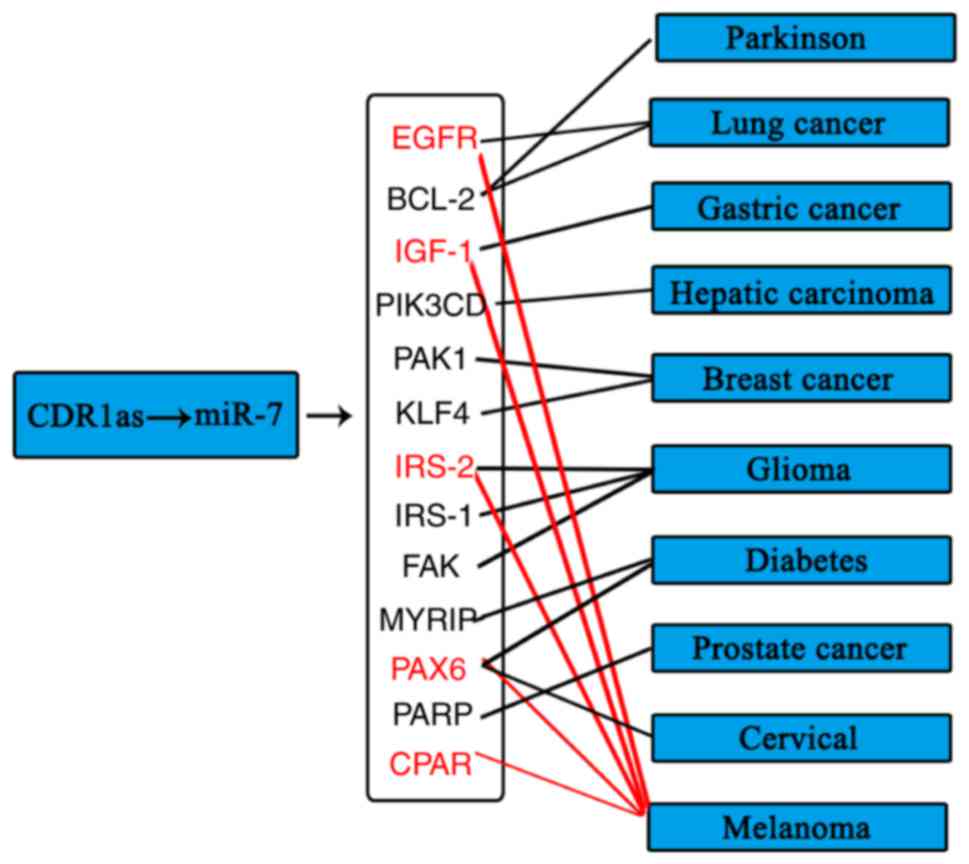 | Figure 4.CDR1as/miR-7 and cancer. CDR1as/miR-7
was found to regulate the expression of several cytokines and
oncogenes in different types of cancer. The red names and lines
indicate the genes and relationship with melanoma. The black names
and lines indicate the genes and relationship with other cancers.
miR, microRNA; CDR1as, circular RNA CDR1 antisense RNA; EGFR,
epidermal growth factor receptor; BCL-2, B cell lymphoma-2; IGF-1,
insulin-like growth factor-1; PIK3CD, phosphatidylinositol 3-kinase
catalytic subunit δ; PAK, p21-activated kinase; KLF4, Kruppel-like
factor 4; IRS, insulin receptor substrate; FAK, focal adhesion
kinase; MYRIP, myosin VIIA and Rab interacting protein; PARP, poly
(ADP-ribose) polymerase; CPAR, histone H3-like centromeric protein
cpar-1. |
Discussion
With the development of novel biochemical and
computational approaches in the RNA field, circRNAs have been an
active area of investigation (37).
CircRNAs, unlike linear RNAs, form a covalently closed continuous
loop (38); circRNAs are also
resistant to RNase R treatment (39).
CircRNAs interact with other molecules or microRNAs to regulate
gene expression in metabolism at the post-transcriptional or
transcriptional level (40). CircRNAs
are also crucial in various biological processes associated with
cancer cell development and proliferation, in addition to cancer
metastasis and invasion (3,19).
CDR1as, which is also considered a circRNA sponge
for miR-7 (ciRS-7), is derived from the antisense of cerebellar
degeneration-related protein 1 transcript (40), and has attracted attention in the RNA
field. CDR1as has numerous pathways and diseases of note, including
its function as a ceRNA of microRNA, particularly miR-7, in several
types of cancer. miR-7 can bind in the 3′ UTRs of mRNAs, which is
vital in these types of cancer, including breast cancer (41), glioma (42), melanoma (23), diabetes (43), Parkinson's disease (44) and cervical cancer (45). As CDR1as/miR-7 is likely to be
involved in various types of cancer, including breast cancer,
glioma, melanoma, diabetes, Parkinson's disease and cervical
cancer, detecting the function of CDR1as in the initiation and
progression of cancer is of interest for further investigations
(9). CDR1as is a common genetic
trigger in human diseases and a specific combination of various
markers for early diagnosis in cancer.
In the present study, GO enrichment analysis
revealed that the miR-7 target genes are associated with several
crucial biological processes and are involved in the cellular
response during cancer development. Among the pathways identified
in the present study, platelet activation has been shown to be a
main regulator in the tumor cell response (46). Negative regulation of the intrinsic
apoptotic signaling pathway can increase the repair space of DNA
double-strand breaks (47). miR-7 can
markedly decrease the expression levels of EGFR, IGF-1R and CRAF,
and suppress the activation of MAPK and the PI3K/AKT pathway in
VemR A375 melanoma cells (23). IRS-2
is a target of miR-7, and promotes migration and Akt signaling in
melanoma (24). Pax6 is associated
with the Wnt signaling pathway, which regulates the invasion and
migration of glioblastoma (48),
prostate cancer (49) and bladder
cancer (50). Pax6 is also important
in the growth of malignant melanoma (51). As Pax6 is the target of miR-7
(52), the CDR1as-miR-7-Pax6 axis may
be important in cancer invasion and migration.
Melanoma is a the main type of refractory skin
cancer due to its poor prognosis and high chronicity (53). The CDR1as/miR-7 axis is likely to be
involved in several types of cancer, including melanoma (54). Additionally, IRS-2 is a target of
miR-7-5p in melanoma cells and activates Akt, which promotes
melanoma cell migration. The inhibition of IRS-2 reduces the
activity of Akt, a key oncogenic effector molecule in melanoma, and
this finding suggests that the co-interaction of IRS-2 and CDR1as
is a therapeutic target in melanoma, with the potential function to
inhibit PI3K/Akt signaling, cell migration and tumor metastasis
(55).
The genome-wide identification of circRNAs depends
on efficient bioinformatics analysis. The prediction and functional
analysis of target genes are under strict conditions, which can
effectively reduce the false prediction rate. However, there can be
errors in the screening rates of the analysis, and a more accurate
method for subsequent analysis and further assessment is required.
Several results have been collected from several studies, including
the interaction of CDR1as and miR-7 (56), and the interaction of miR-7 and
proteins (23,57), however, there has been no systematic
investigation of the CDR1as-miRNA-protein axis in melanoma or
several other types of cancer. The present study aimed to offer
guidelines for further investigations of the CDR1as-miRNA-protein
axis in cancer.
In conclusion, decades have passed since circRNAs
were first detected. CircRNAs have attracted increasing attention
due to improvements in high-throughput sequencing technologies and
bioinformatics progression. In the present study, GO enrichment
analysis suggested that the protein associated with CDR1as is
mainly regulated in the cytoplasm, as the molecular protein
binding, the biological process of cell division, and the
endoplasmic reticulum tubular network may be involved in cancer.
The KEGG pathway enrichment cell cycle is closely associated with
the treatment of cancer. Co-expression network analysis revealed
the association between CDR1as, miR-7 and proteins; and CDR1as was
been found to be important in several types of cancer, including
breast cancer, glioma, diabetes, Parkinson's disease, and melanoma
in particular. These results are likely to assist in future
investigations of the function of CDR1as as a novel therapeutic
target in cancer.
Acknowledgements
The authors would like to thank Biomarker
Technologies, Inc. (Beijing, China) for technical support.
Funding
This study was supported by a grant from the Science
and Technology of Shanxi Agricultural University, Shanxi, China
(grant no. 2016ZZ07) and the National Natural Science Foundation of
China (grant no. 31402156).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on request.
Authors' contributions
LZ and YL performed most of the research and were
major contributors towards writing the manuscript. WL and HL
collected and analyzed the data. ZZ and LZ made substantial
contributions to the design of the work, drafted the manuscript and
revised it critically for important intellectual content. LZ and YL
gave final approval of the version to be published.
Ethics approval and consent to
participate
Not applicable
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kos A, Dijkema R, Arnberg AC, van der
Meide PH and Schellekens H: The hepatitis delta (delta) virus
possesses a circular RNA. Nature. 323:558–560. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H,
Zhao Q, Zhou C, Zhao Y, Lu D, et al: Transcriptome-wide
investigation of circular RNAs in rice. RNA. 21:2076–2087. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: circRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan Z, Hao L, Li W, Yu J, Li J, Shen Z, Ye
G, Qi X and Li G: CircRNA_100269 is downregulated in gastric cancer
and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017.PubMed/NCBI
|
|
15
|
Liang HF, Zhang XZ, Liu BG, Jia GT and Li
WL: Circular RNA circ-ABCB10 promotes breast cancer proliferation
and progression through sponging miR-1271. Am J Cancer Res.
7:1566–1576. 2017.PubMed/NCBI
|
|
16
|
Xu L, Ming Z, Zheng X, Yi P, Lan C and Xu
M: The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of
hepatic microvascular invasion in hepatocellular carcinoma. J
Cancer Res Clin Oncol. 143:17–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang W, Ji M, He G, Yang L, Niu Z, Jian M,
Wei Y, Ren L and Xu J: Silencing CDR1as inhibits colorectal cancer
progression through regulating microRNA-7. Onco Targets Ther.
10:2045–2056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu H, Guo S, Li W and Yu P: The circular
RNA Cdr1as, via miR-7 and its targets, regulates insulin
transcription and secretion in islet cells. Sci Rep. 5:124532015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barbagallo D, Condorelli A, Ragusa M,
Salito L, Sammito M, Banelli B, Caltabiano R, Barbagallo G, Zappalà
A, Battaglia R, et al: Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1
axis is involved in glioblastoma multiforme. Oncotarget.
7:4746–4759. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun X, Li J, Sun Y, Dong L, Shen C, Yang
L, Yang M, Li Y, Shen G, Tu Y and Tao J: miR-7 reverses the
resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and
inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget.
7:53558–53570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Giles KM, Brown RA, Epis MR, Kalinowski FC
and Leedman PJ: miRNA-7-5p inhibits melanoma cell migration and
invasion. Biochem Biophys Res Commun. 430:706–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jr PA, Hood N, Lyn S, Foote J and Bennett
J: MP-4.09: TargetScan®: A novel approach for outpatient
prostate biopsy with the potential for use as an aid to focal
prostate therapy. Urology. 72:S86–S87. 2008. View Article : Google Scholar
|
|
26
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
28
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Glažar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44:D209–D215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
John B, Sander C and Marks DS: Prediction
of human miRNA targets. Methods Mol Biol. 342:101–113.
2006.PubMed/NCBI
|
|
37
|
Ebbesen KK, Kjems J and Hansen TB:
Circular RNAs: Identification, biogenesis and function. Biochim
Biophys Acta. 1859:163–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen I, Chen CY and Chuang TJ: Biogenesis,
identification, and function of exonic circular RNAs. Wiley
Interdiscip Rev RNA. 6:563–579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu
S, Huang Y, Zhao X, Huang L, Wang Z, et al: Reciprocal changes of
circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute
myocardial infarction. Sci Rep. 6:223842016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Raychaudhuri M, Bronger H, Buchner T,
Kiechle M, Weichert W and Avril S: MicroRNAs miR-7 and miR-340
predict response to neoadjuvant chemotherapy in breast cancer.
Breast Cancer Res Treat. 162:511–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu LH, Zhang HY, Liu XZ, Zhang LD, Jiang
ZM and Hospital TH: Correlation between miR-7 and expression of
EGFR/PI3K signal pathway related protein in glioma. Shandong Med J.
2014.
|
|
43
|
Wan S, Wang J, Wang J, Wu J, Song J, Zhang
CY, Zhang C, Wang C and Wang JJ: Increased serum miR-7 is a
promising biomarker for type 2 diabetes mellitus and its
microvascular complications. Diabetes Res Clin Pract. 130:171–179.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li S, Lv X, Zhai K, Xu R, Zhang Y, Zhao S,
Qin X, Yin L and Lou J: MicroRNA-7 inhibits neuronal apoptosis in a
cellular Parkinson's disease model by targeting Bax and Sirt2. Am J
Transl Res. 8:993–1004. 2016.PubMed/NCBI
|
|
45
|
Liu S, Zhang P, Chen Z, Liu M, Li X and
Tang H: MicroRNA-7 downregulates XIAP expression to suppress cell
growth and promote apoptosis in cervical cancer cells. FEBS Lett.
587:2247–2253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Toulany M and Rodemann HP:
Phosphatidylinositol 3-kinase/Akt signaling as a key mediator of
tumor cell responsiveness to radiation. Semin Cancer Biol.
35:180–190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim Y, Kim KH, Lee J, Lee YA, Kim M, Lee
SJ, Park K, Yang H, Jin J, Joo KM, et al: Wnt activation is
implicated in glioblastoma radioresistance. Lab Invest. 92:466–473.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cojoc M, Peitzsch C, Kurth I, Trautmann F,
Kunz-Schughart LA, Telegeev GD, Stakhovsky EA, Walker JR, Simin K,
Lyle S, et al: Aldehyde dehydrogenase is regulated by β-Catenin/TCF
and promotes radioresistance in prostate cancer progenitor cells.
Cancer Res. 75:1482–1494. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pierre Renaud Q: MIR-7 targets pax6 and
modulates bladder cancer cell migration. Feb 2–2017.
|
|
51
|
Treszl A, Ladanyi A, Rakosy Z, Buczko Z,
Adany R and Balazs M: Molecular cytogenetic characterization of a
novel cell line established from a superficial spreading melanoma.
Front Biosci. 11:1844–1853. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
52
|
Needhamsen M, White RB, Giles KM, Dunlop
SA and Thomas MG: Regulation of Human PAX6 Expression by miR-7.
Evol Bioinform Online. 10:107–113. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bonazzi VF, Stark MS and Hayward NK:
MicroRNA regulation of melanoma progression. Melanoma Res.
22:101–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Geng HH, Li R, Su YM, Xiao J, Pan M, Cai
XX and Ji XP: The Circular RNA Cdr1as promotes myocardial
infarction by mediating the regulation of miR-7a on its target
genes expression. PLoS One. 11:e01517532016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma.
Hepatology. 55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shen Z, Qin X, Yan M, Li R, Chen G, Zhang
J and Chen W: Cancer-associated fibroblasts promote cancer cell
growth through a miR-7-RASSF2-PAR-4 axis in the tumor
microenvironment. Oncotarget. 8:1290–1303. 2017.PubMed/NCBI
|