Introduction
Lung cancer is a leading cause of cancer mortalities
worldwide, with 226,160 new cases and 160,340 mortalities in 2012
(1). As a type of lung cancer,
non-small cell lung carcinoma (NSCLC) consists of adenocarcinoma
(AC), squamous cell carcinoma and large cell carcinoma (2). Lung AC, a heterogeneous group of tumors
ranging in aggressiveness from noninvasive bronchioloalveolar
carcinoma (BAC) to microinvasive tumors, mixed-type tumors and pure
invasive AC, is the most common histological type of lung cancer
(3,4).
The first treatment for patients with early-stage lung AC is
surgical resection, but the 5-year overall survival rate remains at
~80% for stage IA disease (5).
Notably, patients with invasive lung AC have a lower mean 5-year
survival rate compared with that of patients with BAC (59 vs. 100%,
respectively) (6). Therefore, it is
important to better understand the molecular mechanisms of invasive
lung AC in order to develop effective preventive and curative
strategies.
Cancer invasion is a highly complicated process,
with the early events being the proteolytic degradation of
extracellular matrix (ECM) components to provide room for
infiltration, followed by migration into the adjacent tissues
(7,8).
Recent studies suggest that matrix metalloproteinases (MMPs),
members of the matrixin subfamily of zinc metalloproteinases, are
involved in the breakdown of the ECM (9,10).
Over-expression of MMPs has been detected in a number of tumor
types, including invasive lung AC (11). MMP2, MMP7, MMP9 and tissue inhibitor
of metalloproteinase 2 are upregulated in the lung tissue of
patients with primary spontaneous pneumothorax (PSP), and the
imbalance of their expression may be implicated in the pathogenesis
of PSP (12). The signal module and
sequence variant module appearing in lung AC, in which the
expression of MMP12 is upregulated but that of MMP11 is
downregulated, can promote the invasion of cancer cells (13). Chemotherapy drugs can markedly inhibit
the invasive ability of human lung AC cells via reducing the
expression of MMP2 and MMP9 (14,15).
Additionally, rosuvastatin and simvastatin can function in the
treatment of lung cancer by regulating the expression of MMP2,
MMP9, RAS and nuclear factor-κB-p65 (16). However, the current number of screened
MMPs is limited, and further studies are required to identify genes
with a similar function to that of MMPs.
In addition, it has been reported that the
transcription factor signal transducer and activator of
transcription 3 (STAT3) can regulate the transcription of MMP2 and
promote melanoma cell invasion (17).
Blocking STAT3 signaling significantly inhibits the invasion of
melanoma cells (17). Storz et
al (17) have demonstrated that
forkhead box O3 promotes invasion and progression in numerous human
solid tumors by inducing the expression of MMP9 and MMP13. However,
the regulatory mechanisms of MMPs are not well understood in lung
AC.
The present study aimed to identify differentially
expressed genes (DEGs), particularly MMPs-associated genes, using
gene expression profile data of invasive lung AC and adjacent
normal samples collected from a publicly available database.
Besides, the transcriptional factors regulating MMPs-associated
genes were also investigated.
Materials and methods
Ethical statement
The present study was approved by the Ethics
Committees of the Beijing Shijitan Hospital (Beijing, China) and
the Chest Hospital Affiliated to Shanghai Jiaotong University
(Shanghai, China), and it was performed in accordance with the
ethical standards (18). In addition,
written informed consent was obtained from all patients, prior to
enrollment in the present study.
Microarray data
The gene expression dataset GSE2514, which is based
on two platforms [GPL81 (MG_U74Av2) Affymetrix Murine Genome U74A
Version 2 Array and GPL8300 (HG_U95Av2) Affymetrix Human Genome U95
Version 2 Array], was downloaded from Gene Expression Omnibus (GEO;
https://www.ncbi.nlm.nih.gov/geo/)
database (19). Microarray data
obtained with the platform GPL8300 (HG_U95Av2; Affymetrix Human
Genome U95 Version 2 Array; Affymetrix, Inc., Santa Clara, CA, USA)
was downloaded from the GEO database (19) in our study. It contained 20 lung AC
samples and 19 adjacent lung samples (~1 cm away from the tumor
site), which were obtained from 5 male and 5 female patients
undergoing lobectomy (9) and wedge
resection (1). Of these patients, 9
had a history of tobacco smoking. The ages of the patients ranged
from 45 to 73 years. Their tumors were all invasive lung AC. The
majority of tumors were low-to-intermediate grade and low stage,
although 2 stage III tumors were included in the analysis. Probe
annotation files were also acquired.
Preprocessing and differential
analysis
The raw array data in the CEL file (Affymetrix,
Inc.) were transformed into recognizable gene expression data using
the robust multi-array average algorithm from the affy package in R
(20). Upon normalization, probes
were mapped to genes according to the annotation files. The levels
of probes corresponding to one gene were averaged and used as the
final gene expression value. DEGs were screened using the limma
package (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
(21) in R based on the cut-offs of
P<0.05 and |log fold-change| >1.
Cluster analysis
Cluster analysis of DEGs was performed with Cluster
3.0 (http://bonsai.ims.u-tokyo.ac.jp/mdehoon/software/cluster)
using the K-means clustering algorithm (22), which was conducted on data with K (the
number of clusters)=5.
Pearson correlation analysis
The method of Pearson correlation analysis (23) in the cor function in R (https://www.r-project.org/) was used to perform the
correlation analysis between the expression of DEGs and MMPs. A
correlation coefficient of >0.8 was used as the cut-off
criterion.
Western blot analysis
A total of 6 lung AC and matched adjacent lung
tissue biopsy samples were obtained in June 2014 from Shanghai Lung
Cancer Center, Chest Hospital Affiliated to Shanghai Jiaotong
University (Shanghai, China). Tissues were washed with ice-cold PBS
and lysed in ice-cold radioimmunoprecipitation assay lysis buffer
(Sangon Biotech Co., Ltd., Shanghai, China). Extracted proteins
were quantified using a BCA Protein Assay kit (Sangon Biotech Co.,
Ltd., Shanghai, China) and separated with SDS-PAGE with a 10%
separating gel and a 5% stacking gel. Subsequently, proteins were
transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA). Then, membranes were blocked in 5%
nonfat dry milk and probed with primary rabbit antibodies against
MMPs (anti-MMP1, cat. no. sc-58377; anti-MMP7, cat. no. sc-80205;
anti-MMP9, cat. no. sc-13520; and anti-MMP12, cat. no. sc-8839; all
diluted to 1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
overnight at 4°C. Horseradish peroxidase-conjugated anti-rabbit
immunoglobulin G (cat. no. 111-035-003; 1:5,000; Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA) served as a
secondary antibody and was incubated for 1 h at 4°C. The
anti-β-actin antibody (cat. no. 8227; 1:10,000; Abcam, Cambridge,
CA, USA) was used as the control. The position of protein bands was
developed with ECL chemoluminescence kit (Merck Millipore) and
visualized under a ChemiDoc MP imaging system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Functional and pathway enrichment
analysis
Gene Ontology (GO; http://www.geneontology.org/) (24) and Kyoto Encyclopedia of Genes and
Genomes (KEGG; http://www.genome.jp/kegg/) (25) pathway enrichment analyses were
performed for the MMPs-associated genes using the Database for
Annotation, Visualization and Integrated Discovery (DAVID) online
tool (26) to reveal altered
biological functions in lung AC. P<0.05 and a false discovery
rate (FDR) adjusted by the Benjamini and Hochberg method (27) of <0.01 were set as the
thresholds.
Selection of disease-associated
genes
Diseases associated with the DEGs were identified by
gene set enrichment analysis against the Genetic Association
Database Disease Class (http://geneticassociationdb.nih.gov/) using the
annotation server DAVID (P<0.05) (26).
Construction of protein-protein
interaction (PPI) network
PPI network analysis for the DEGs was carried out
with the Search Tool for the Retrieval of Interacting Genes
(STRING; http://string-db.org/), and the PPI
network involving MMPs-associated genes was selected. Interaction
associations with a combined score of ≥0.4 were retained and the
PPI network was visualized by Cytoscape software (version 2.8;
http://www.cytoscape.org) (28).
Transcriptional regulatory network
analysis
Transcriptional regulatory network analysis was
performed with DAVID (26) for the
group of DEGs including MMPs, and the transcriptional regulatory
network was visualized by Cytoscape software (version 2.8;
http://www.cytoscape.org) (28).
Results
DEGs analysis
Upon normalization of the raw data (Fig. 1), the DEGs were screened using the
limma package in R. As a result, 269 DEGs were identified between
lung AC and adjacent normal lung samples, including 78 upregulated
and 191 downregulated genes.
MMPs-associated genes screening
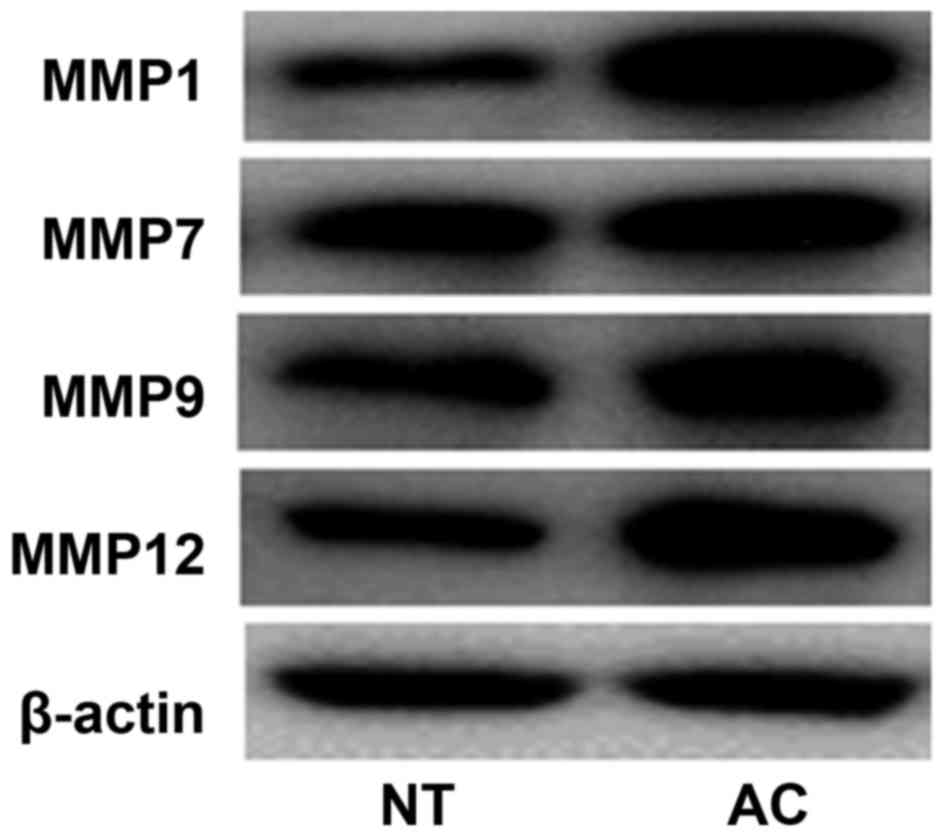
MMP1, MMP7, MMP9 and MMP12 were clustered into one
group with a number of other DEGs (Table
I), which were upregulated in lung AC compared with their
expression in normal lung tissues. These results were further
confirmed by Pearson correlation analysis and western blotting. The
present findings also demonstrated that MMP9 expression exhibited a
significant correlation with DSP (correlation
coefficient=0.8309295) and CLDN3 (correlation
coefficient=0.8058015) expression. Additionally, MMP12 expression
was significantly correlated with DSP expression (correlation
coefficient=0.8127249). The expression of MMP1, MMP7, MMP9 and
MMP12 was also observed to be upregulated in lung AC compared with
that in adjacent lung tissues (Fig.
2).
 | Table I.List of genes clustered with
MMPs. |
Table I.
List of genes clustered with
MMPs.
| ID | Log
(fold-change) | Mean
expression | t | P-value | False discovery
rate |
|---|
| RAMP1 | 1.803939312 | 6.214583105 | −10.555338710 |
4.33×10−13 |
3.50×10−11 |
| FAP | 1.561195027 | 6.500446054 | −9.026036586 |
3.63×10−11 |
1.48×10−9 |
| CLDN3 | 1.625063261 | 9.253029938 | −7.959600860 |
9.43×10−10 |
2.41×10−8 |
| CP | 1.533166764 | 7.200364641 | −7.654471654 |
2.45×10−9 |
5.46×10−8 |
| ZWINT | 1.534376695 | 6.188206312 | −7.484374926 |
4.18×10−9 |
8.53×10−8 |
| MDK | 1.627179122 | 9.344798091 | −7.324035903 |
6.94×10−9 |
1.32×10−7 |
| MMP9 | 1.708966359 | 7.597559232 | −6.771606537 |
4.04×10−8 |
5.82×10−7 |
| COL3A1 | 1.643158431 | 9.138801925 | −6.532240647 |
8.72×10−8 |
1.11×10−6 |
| COL11A1 | 2.387669841 | 5.767529739 | −6.412529728 |
1.28×10−7 |
1.54×10−6 |
| TOX3 | 1.770307511 | 4.159327973 | −6.212832065 |
2.44×10−7 |
2.68×10−6 |
| GABBR1 UBD | 1.619335627 | 7.242024866 | −6.016224500 |
4.60×10−7 |
4.64×10−6 |
| MMP12 | 1.644563956 | 5.138055675 | −5.757801490 |
1.06×10−6 |
9.61×10−6 |
| POSTN | 1.502940090 | 7.019690170 | −5.698113750 |
1.28×10−6 |
1.13×10−5 |
| SPINK1 | 2.517220534 | 7.945875667 | −5.613361864 |
1.69×10−6 |
1.44×10−5 |
| NQO1 | 1.527576974 | 7.820504924 | −5.374097053 |
3.64×10−6 |
2.77×10−5 |
| SLC7A5 | 1.517121297 | 7.685388227 | −5.301930776 |
4.59×10−6 |
3.37×10−5 |
| MMP7 | 2.087445737 | 6.987333841 | −5.300686125 |
4.61×10−6 |
3.38×10−5 |
| MMP1 | 1.553980336 | 4.289661258 | −5.235072252 |
5.68×10−6 |
4.05×10−5 |
| DSP | 1.669305242 | 7.306405073 | −5.162024033 |
7.18×10−6 |
4.94×10−5 |
| CEACAM5 | 2.090196480 | 7.403316921 | −4.560737891 |
4.80×10−5 |
2.43×10−4 |
| S100P | 1.513031206 | 7.826887416 | −3.965427875 |
2.97×10−4 |
1.12×10−3 |
Functional and pathway enrichment
analysis
Functional enrichment analysis was applied for the
above group of DEGs with the DAVID online tool. As shown in
Table II, collagen metabolism
(P=4.37×10−6) and multicellular organism macromolecule
metabolic processes (P=5.98×10−6) were enriched for
these DEGs.
 | Table II.GO terms over-represented for the
MMPs-associated genes. |
Table II.
GO terms over-represented for the
MMPs-associated genes.
|
|
| Gene |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Category | Term | N | % | P-value | Genes | False discovery
rate |
|---|
| GO_BP | GO:0032963 collagen
metabolic process | 4 | 20 |
4.37×10−6 | MMP9, COL3A1, MMP7
and MMP1 | 0.005547327 |
| GO_BP | GO:0044259
multicellular organism macromolecule metabolic process | 4 | 20 |
5.98×10−6 | MMP9, COL3A1, MMP7
and MMP1 | 0.007594962 |
| GO_CC | GO:0005578
proteinaceous ECM | 7 | 35 |
2.30×10−6 | MMP9, COL3A1, MMP7,
POSTN, COL11A1, MMP12 and MMP1 | 0.002198433 |
| GO_CC | GO:0031012 ECM | 7 | 35 |
3.56×10−6 | MMP9, COL3A1, MMP7,
POSTN, COL11A1, MMP12 and MMP1 | 0.003399869 |
Disease-relevant genes selection
Relevant diseases to the MMPs-associated DEGs were
retrieved with DAVID. Under the Genetic Association DB Disease
Class, MMP9, NAD(P)H quinone oxidoreductase 1 (NQO1), MMP12 and
MMP1 were linked with lung AC, while MMP9, MMP12 and MMP1 were
associated with lung function (Table
III).
 | Table III.Diseases associated with
MMPs-associated genes. |
Table III.
Diseases associated with
MMPs-associated genes.
| Term | P-value | Genes |
|---|
| Nasopharyngeal
cancer |
8.72×10−6 | MMP9, MMP7, NQO1,
MMP12 and MMP1 |
| Abdominal aortic
aneurysm |
1.18×10−4 | MMP9, COL3A1, MMP12
and MMP1 |
| Brain cancer |
6.15×10−4 | MMP9, MMP7, NQO1
and MMP1 |
| Gastric ulcer |
6.37×10−4 | MMP9, MMP7 and
MMP1 |
| Subarachnoid
hemorrhage |
1.68×10−3 | MMP9, MMP12 and
MMP1 |
| Lung function |
3.48×10−3 | MMP9, MMP12 and
MMP1 |
| Ovarian cancer |
3.78×10−3 | MMP9, MMP7, NQO1
and MMP1 |
| Bladder cancer |
5.85×10−3 | MMP9, NQO1, MMP12
and MMP1 |
| Colorectal
cancer |
6.11×10−3 | MMP9, MMP7, NQO1,
MDK and MMP1 |
| Coronary artery
luminal dimensions |
7.91×10−3 | MMP7 and MMP12 |
| Osseointegrated
implant failure |
7.91×10−3 | MMP9 and MMP1 |
| Alzheimer's disease
dementia, vascular |
1.18×10−2 | MMP9 and MMP1 |
| Chronic obstructive
pulmonary disease |
1.84×10−2 | MMP9, MMP12 and
MMP1 |
| Rheumatoid
arthritis |
1.85×10−2 | MMP9, MMP7, MMP12
and MMP1 |
| Uterine
leiomyoma |
1.97×10−2 | MMP9 and MMP1 |
| Adenomyosis
endometriosis |
1.97×10−2 | MMP9 and MMP7 |
| Breast cancer |
2.09×10−2 | MMP9, POSTN, NQO1,
MMP12 and MMP1 |
| Cervical artery
dissection, spontaneous |
2.36×10−2 | MMP9 and
COL3A1 |
| Aneurysm |
2.36×10−2 | MMP9 and MMP12 |
| Lung cancer |
2.51×10−2 | MMP9, NQO1, MMP12
and MMP1 |
| Coronary artery
disease |
3.07×10−2 | MMP9, COL3A1 and
MMP12 |
| H. pylori
infection stomach cancer |
3.52×10−2 | MMP9 and MMP7 |
| Left ventricular
remodeling |
3.90×10−2 | MMP9 and MMP1 |
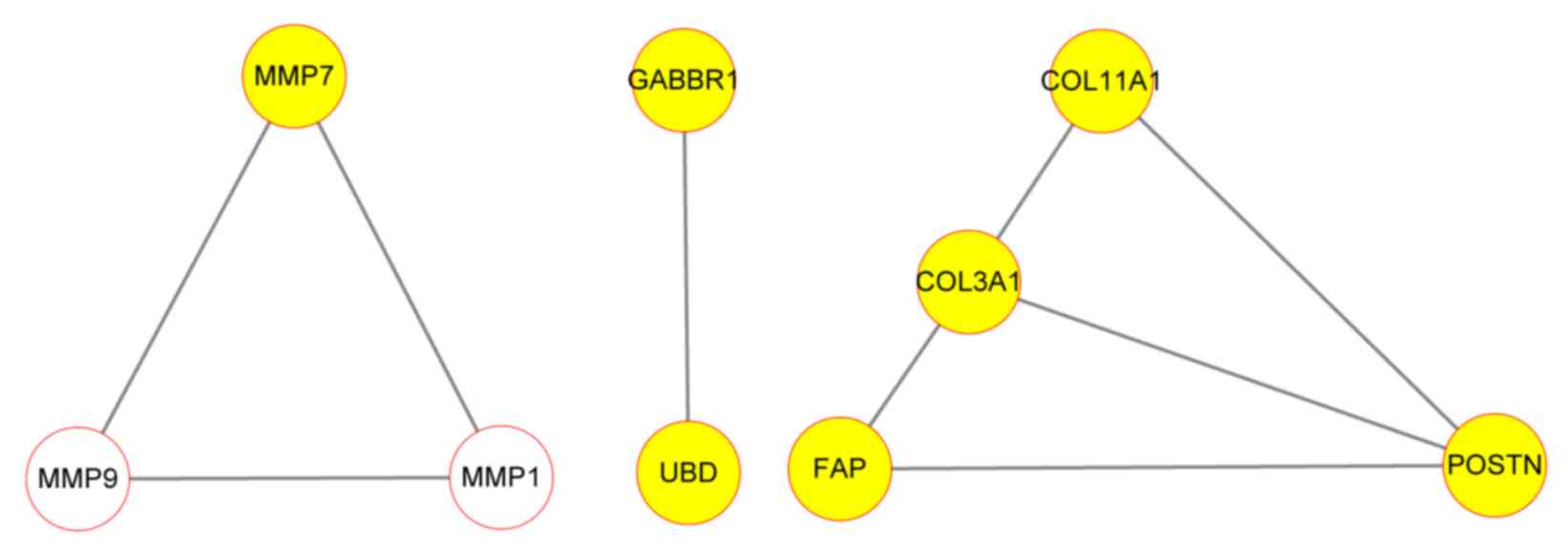
PPI network analysis
A PPI network was constructed for the
MMPs-associated DEGs using STRING (Fig.
3). The results revealed that interaction associations existed
among MMP9, NQO1 and MMP7. In addition, the other genes clustered
into the MMPs group also interacted with each other.
Transcriptional regulatory network
analysis
Transcriptional regulatory network analysis
indicated that 9 genes, including MMP1, MMP9 and NQO1, were
commonly regulated by the CCAAT/enhancer binding protein (C/EBP) α
(CEBPA) transcription factor (Fig.
4).
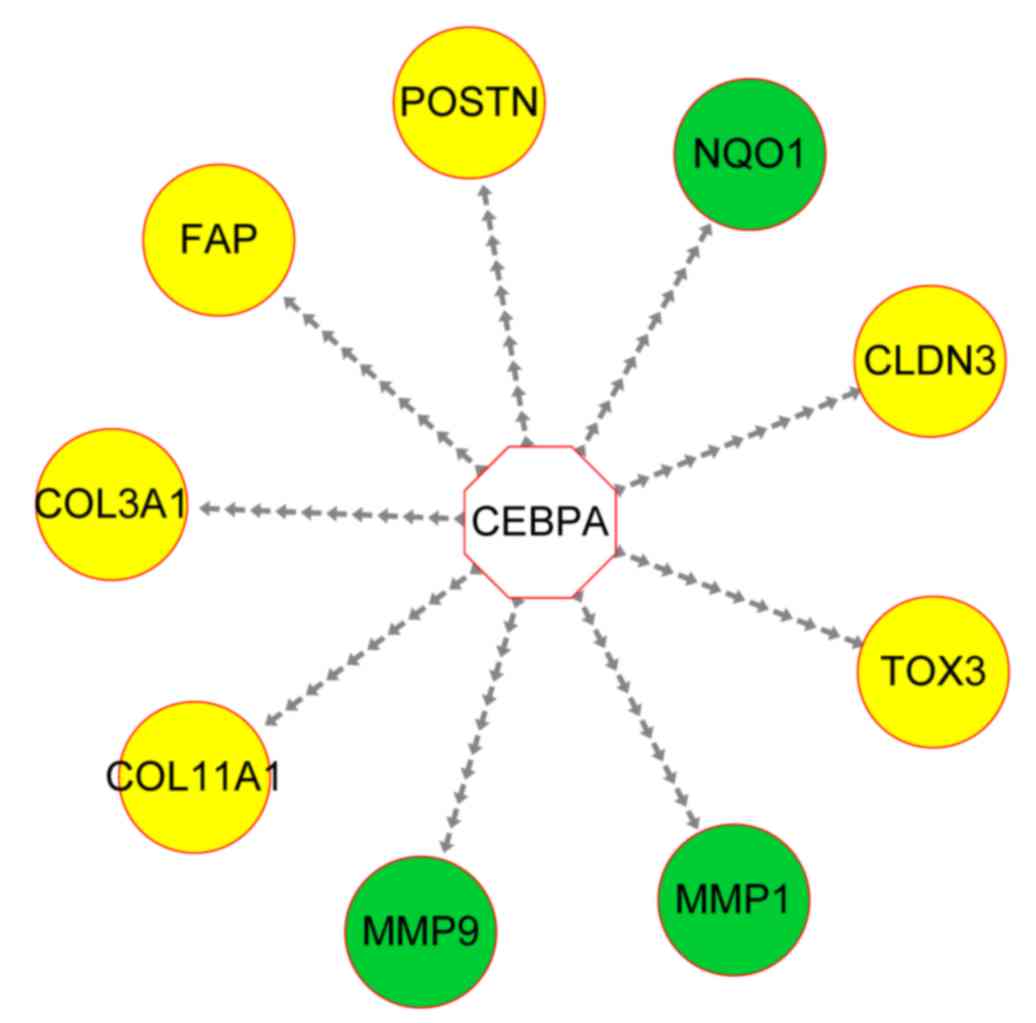 | Figure 4.Transcriptional regulatory network
containing MMPs. The transcription factor CEBPA is marked as white,
while lung cancer-associated genes are presented as green and other
genes as yellow. The dashed arrows indicate regulatory
associations. MMP, matrix metalloproteinase; FAP, familial
adenomatous polyposis; NQO1, NAD(P)H quinone oxidoreductase 1;
POSTN, periostin; COL11A1, collagen type XI α I; COL3A1, collagen
type III α 1; TOX3, TOX high mobility group box family member 3;
CLDN3, claudin 3; CEBPA, CCAAT/enhancer binding protein (C/EBP)
α. |
Discussion
In the present study, cluster analysis demonstrated
that 4 MMPs, including MMP1, MMP7, MMP9 and MMP12, were upregulated
in lung AC samples, and may be involved in ECM metabolic processes,
thus promoting cancer invasion. These results were validated by
western blotting and were in accordance with previous studies
(29–32). MMP1 is the most highly expressed
interstitial collagenase for degrading fibrillar collagens
(33). Overexpression of MMP1 has
been associated with tumor invasion and metastasis by modulating
the polarization of T helper (Th)1/Th2 inflammatory responses
(29). Downregulation of MMP-7
mediated by antisense oligonucleotide changes the ultrastructure of
lung AC A549 cells, leading to decreased microvilli, endoplasmic
reticulum dilation, swelling of mitochondria and formation of
apoptotic bodies, which eventually inhibits invasion in lung AC
A549 cells (30). Similarly, the
expression levels of MMP9 and MMP12 were observed to be higher in
NSCLC than in normal samples (31).
Upregulation of MMP12 and MMP9 may be one of the mechanisms to
promote lung cancer cell invasion (32).
In addition, the present study identified several
DEGs that were clustered into one group with MMPs, thus indicating
the same function in cancer cell invasion. A number of these genes
have been linked to lung cancer in the following studies. NQO1 is a
member of the NQO family, and altered expression of this protein
has been reported in numerous tumors (34). Mutations in this gene contribute to
susceptibility to various forms of cancer, including lung cancer
(35,36). S100 calcium-binding protein P (S100P)
is a member of the S100 family of proteins containing two EF-hand
calcium-binding motifs, which are involved in the regulation of
cell cycle progression and differentiation (37). Bartling et al (31) reported that S100P expression is mainly
increased in AC, and that S100P upregulation is detected in early
rather than in advanced tumor stages. Overexpression of S100P may
lead to cancer cell invasion by changing the expression levels of
several cytoskeletal proteins (38).
Serine peptidase inhibitor, Kazal type 1 (SPINK1) is a secreted
serine protease inhibitor (39).
Overexpression of SPINK1 is associated with aggressiveness of
prostate cancer (40). A previous
study by Soon et al (34)
revealed that SPINK1 is an invasion factor associated with
prognosis in breast cancer patients. However, its role in lung
cancer remains unknown. The present study suggested that SPINK1 may
also be involved in lung AC invasion. Ubiquitin D (UBD, also known
as F-associated transcript 10) is a member of the ubiquitin-like
modifier family, and appears to be upregulated in hepatocellular
carcinoma, as well as in gastrointestinal and gynecological cancers
(41). Increased cytoplasmic UBD is
significantly associated with depth of colon cancer invasion
(42). Few studies have investigated
the roles of UBD in lung AC invasion, including the present study.
Collagen type XI α I (COL11A1) has also been demonstrated to be
overexpressed in NSCLC, and has been identified as a potential
invasion-associated gene in cancer (43). However, its mechanism of function is
not understood yet. The present study revealed that COL11A1 could
interact with the osteoblast-specific factor periostin (POSTN),
although this requires further experimental validation. In
addition, the associations between MMPs and various genes
(including MMP9-CLDN3, MMP9-DSP and MMP12-DSP) were also further
confirmed by Pearson correlation analysis. CLDN3, a component of
tight junctions, has been reported to be upregulated in NSCLC, and
may be important in invasion (44).
Agarwal et al (39) reported
that CLDN3-mediated increased invasion may be accomplished through
the activation of MMP2. In the present study, CLDN3 was
significantly correlated with MMP9 via Pearson correlation
analysis, indicating a potential link between CLDN3 and MMP9 in
lung AC cell invasion. DSP acts as a tumor suppressor via
inhibiting the Wnt/β-catenin signaling pathway in NSCLC (45). Thus, the expression patterns of DSP
and MMPs are theoretically opposite, which has been demonstrated in
the human epithelial carcinoma cell line A431 during the
epithelial-mesenchymal transition, with upregulated MMPs and
downregulated DSP levels (46).
According to the present transcriptional regulatory
network analysis, MMPs-associated genes were regulated by CEBPA.
CEBPA is a basic leucine zipper domain transcription factor that
not only can bind to certain promoters and enhancers as a
homodimer, but can also form heterodimers with the associated
proteins CEBPB and CEBPG (47). It
has been reported that CEBPB is an important mediator in the
activation of MMP genes (including MMP1, MMP3 and MMP10) in A549
lung carcinoma cells stimulated with the inflammatory cytokine
interleukin-1β (48). Therefore, it
could be speculated that CEBPA may also serve an important role in
upregulating the expression of MMP1 and MPP9, thus affecting the
invasion of lung cancer. Although the present results indicated
that CEBPA could modulate the expression of fibroblast activation
protein α, CLDN3, collagen type III α 1, COL11A1, TOX high mobility
group box family member 3, NQO1 and POSTN, no experimental evidence
could be obtained.
In conclusion, the present study identified several
genes such as SPINK1 and UBD that may serve important roles in lung
AC invasion with MMPs (including MMP1, MMP7, MMP9 and MMP12).
Several MMPs-associated genes were observed to regulated by the
CEBPA transcription factor. These findings may provide various
underlying targets for prevention of lung AC invasion. However,
further experimental investigations or studies on other datasets
are required to validate the present observations.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lazar V, Suo C, Orear C, van den Oord J,
Balogh Z, Guegan J, Job B, Meurice G, Ripoche H, Calza S, et al:
Integrated molecular portrait of non-small cell lung cancers. BMC
Med Genomics. 6:532013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Devesa SS, Bray F, Vizcaino AP and Parkin
DM: International lung cancer trends by histologic type:
Male:female differences diminishing and adenocarcinoma rates
rising. Int J Cancer. 117:294–299. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Borczuk AC, Qian F, Kazeros A, Eleazar J,
Assaad A, Sonett JR, Ginsburg M, Gorenstein L and Powell CA:
Invasive size is an independent predictor of survival in pulmonary
adenocarcinoma. Am J Surg Pathol. 33:462–469. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Rens MT, de la Rivière AB, Elbers HR
and van Den Bosch JM: Prognostic assessment of 2,361 patients who
underwent pulmonary resection for non-small cell lung cancer, stage
I, II and IIIA. Chest. 117:374–379. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakurai H, Maeshima A, Watanabe S, Suzuki
K, Tsuchiya R, Maeshima AM, Matsuno Y and Asamura H: Grade of
stromal invasion in small adenocarcinoma of the lung:
Histopathological minimal invasion and prognosis. Am J Surg Pathol.
28:198–206. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Veiseh O, Kievit FM, Ellenbogen RG and
Zhang M: Cancer cell invasion: Treatment and monitoring
opportunities in nanomedicine. Adv Drug Deliv Rev. 63:582–596.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kam Y, Rejniak KA and Anderson AR:
Cellular modeling of cancer invasion: Integration of in silico and
in vitro approaches. J Cell Physiol. 227:431–438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freije JM, Balbin M, Pendás AM, Sánchez
LM, Puente XS and López-Otin C: Matrix metalloproteinases and tumor
progression. Adv Exp Med Biol. 532:91–107. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kleiner DE and Stetler-Stevenson WG:
Matrix metalloproteinases and metastasis. Cancer Chemother
Pharmacol. 43 Suppl:S42–S51. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumaki F, Matsui K, Kawai T, Ozeki Y, Yu
ZX, Ferrans VJ and Travis WD: Expression of matrix
metalloproteinases in invasive pulmonary adenocarcinoma with
bronchioloalveolar component and atypical adenomatous hyperplasia.
Am J Pathol. 159:2125–2135. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen CK, Chen PR, Huang HC, Lin YS and
Fang HY: Overexpression of matrix metalloproteinases in lung tissue
of patients with primary spontaneous pneumothorax. Respiration.
88:418–425. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Y, Wang L, Jiang M, Huang J, Liu Z and
Wolfl S: Secreted phosphoprotein 1 upstream invasive network
construction and analysis of lung adenocarcinoma compared with
human normal adjacent tissues by integrative biocomputation. Cell
Biochem Biophys. 56:59–71. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan KH, Lee LM, Yan SH, Huang HC, Li CC,
Lin HT and Chen PS: Tomatidine inhibits invasion of human lung
adenocarcinoma cell A549 by reducing matrix metalloproteinases
expression. Chem Biol Interact. 203:580–587. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kao SJ, Su JL, Chen CK, Yu MC, Bai KJ,
Chang JH, Bien MY, Yang SF and Chien MH: Osthole inhibits the
invasive ability of human lung adenocarcinoma cells via suppression
of NF-κB-mediated matrix metalloproteinase-9 expression. Toxicol
Appl Pharmacol. 261:105–115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Falcone D, Gallelli L, Di Virgilio A,
Tucci L, Scaramuzzino M, Terracciano R, Pelaia G and Savino R:
Effects of simvastatin and rosuvastatin on RAS protein, matrix
metalloproteinases and NF-κB in lung cancer and in normal pulmonary
tissues. Cell Prolif. 46:172–182. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Storz P, Döppler H, Copland JA, Simpson KJ
and Toker A: FOXO3a promotes tumor cell invasion through the
induction of matrix metalloproteinases. Mol Cell Biol.
29:4906–4917. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Assembly WMAG: World medical association
declaration of Helsinki: Ethical principles for medical research
involving human subjects. Chinese J Integrative Med. 310:2191–2194.
2001.
|
|
19
|
Stearman RS, Dwyer-Nield L, Zerbe L,
Blaine SA, Chan Z, Bunn PA Jr, Johnson GL, Hirsch FR, Merrick DT,
Franklin WA, et al: Analysis of orthologous gene expression between
human pulmonary adenocarcinoma and a carcinogen-induced murine
model. Am J Pathol. 167:1763–1775. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Na S, Xumin L and Yong G: Research on
k-means clustering algorithm: An improved k-means clustering
algorithmProceedings of the Third International Symposium on
Intelligent Information Technology and Security Informatics
(IITSI). IITSI; Jian: pp. 63S–67S. 2010, View Article : Google Scholar
|
|
23
|
Månsson R, Tsapogas P, Akerlund M,
Lagergren A, Gisler R and Sigvardsson M: Pearson correlation
analysis of microarray data allows for the identification of
genetic targets for early B-cell factor. J Biol Chem.
279:17905–17913. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sherlock G: Gene Ontology: Tool for the
unification of biology. Canadian Institute Food Sci Technol J.
22:4152009.
|
|
25
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nature Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
27
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statistical Society. 57:289–300.
1995.
|
|
28
|
Yeung N, Cline MS, Kuchinsky A, Smoot ME
and Bader GD: Exploring biological networks with Cytoscape
software. Curr Protoc Bioinformatics. 23Chapter 8. (8.13):
8.13.1–8.13.20. 2008. View Article : Google Scholar
|
|
29
|
Fanjul-Fernández M, Folgueras AR, Fueyo A,
Balbín M, Suárez MF, Fernández-García MS, Shapiro SD, Freije JM and
López-Otín C: Matrix metalloproteinase Mmp-1a is dispensable for
normal growth and fertility in mice and promotes lung cancer
progression by modulating inflammatory responses. J Biol Chem.
288:14647–14656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang B, Gao J, Rao Z, Zhang B, Ouyang W
and Yang C: Antisense oligonucleotide targeting matrix
metalloproteinase-7 (MMP-7) changes the ultrastructure of human
A549 lung adenocarcinoma cells. Ultrastruct Pathol. 35:256–259.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bartling B, Rehbein G, Schmitt WD, Hofmann
H-S, Silber R-E and Simm A: S100A2–S100P expression profile and
diagnosis of non-small cell lung carcinoma: impairment by advanced
tumour stages and neoadjuvant chemotherapy. Eur J Cancer.
43:1935–1943. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hung WC, Tseng WL, Shiea J and Chang HC:
Skp2 overexpression increases the expression of MMP-2 and MMP-9 and
invasion of lung cancer cells. Cancer Lett. 288:156–161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sauter W, Rosenberger A, Beckmann L, Kropp
S, Mittelstrass K, Timofeeva M, Wölke G, Steinwachs A, Scheiner D,
Meese E, et al: Matrix metalloproteinase 1 (MMP1) is associated
with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev.
17:1127–1135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Soon WW, Miller LD, Black MA, Dalmasso C,
Chan XB, Pang B, Ong CW, SaltoTellez M, Desai KV and Liu ET:
Combined genomic and phenotype screening reveals secretory factor
SPINK1 as an invasion and survival factor associated with patient
prognosis in breast cancer. EMBO Mol Med. 3:451–464. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sunaga N, Kohno T, Yanagitani N, Sugimura
H, Kunitoh H, Tamura T, Takei Y, Tsuchiya S, Saito R and Yokota J:
Contribution of the NQO1 and GSTT1 polymorphisms to lung
adenocarcinoma susceptibility. Cancer Epidemiol Biomarkers Prev.
11:730–738. 2002.PubMed/NCBI
|
|
36
|
Chao C, Zhang ZF, Berthiller J, Boffetta P
and Hashibe M: NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser
polymorphism and the risk of lung, bladder, and colorectal cancers:
A meta-analysis. Cancer Epidemiol Biomarkers Prev. 15:979–987.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Du M, Wang G, Barraclough R and Rudland P:
P0017 S100P regulates cytoskeleton dynamics to promote cell
migration and metastasis. Eur J Cancer. 51:e6–e7. 2015. View Article : Google Scholar
|
|
38
|
Whiteman HJ, Weeks ME, Dowen SE, Barry S,
Timms JF, Lemoine NR and Crnogorac-Jurcevic T: The role of S100P in
the invasion of pancreatic cancer cells is mediated through
cytoskeletal changes and regulation of cathepsin D. Cancer Res.
67:8633–8642. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Agarwal R, D'Souza T and Morin PJ:
Claudin-3 and claudin-4 expression in ovarian epithelial cells
enhances invasion and is associated with increased matrix
metalloproteinase-2 activity. Cancer Res. 65:7378–7385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ateeq B, Tomlins SA, Laxman B, Asangani
IA, Cao Q, Cao X, Li Y, Wang X, Feng FY, Pienta KJ, et al:
Therapeutic targeting of SPINK1-positive prostate cancer. Sci
Transl Med. 3:72ra172011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL,
Tan Yong S, Kan A, Nuchprayoon I, Jin R, Lee KH, et al: Expression
of the FAT10 gene is highly upregulated in hepatocellular carcinoma
and other gastrointestinal and gynecological cancers. Oncogene.
22:2592–2603. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yan DW, Li DW, Yang YX, Xia J, Wang XL,
Zhou CZ, Fan JW, Wen YG, Sun HC, Wang Q, et al: Ubiquitin D is
correlated with colon cancer progression and predicts recurrence
for stage II-III disease after curative surgery. Br J Cancer.
103:961–969. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim H, Watkinson J, Varadan V and
Anastassiou D: Multi-cancer computational analysis reveals
invasion-associated variant of desmoplastic reaction involving
INHBA, THBS2 and COL11A1. BMC Med Genomics. 3:512010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li J, Tu Y, Jiang L, Xu H and Zhang S:
Expression and significance of Snail and Claudin-3 in non-small
cell lung cancer. Zhongguo Fei Ai Za Zhi. 15:583–590. 2012.(In
Chinese). PubMed/NCBI
|
|
45
|
Yang L, Chen Y, Cui T, Knösel T, Zhang Q,
Albring KF, Huber O and Petersen I: Desmoplakin acts as a tumor
suppressor by inhibition of the Wnt/β-catenin signaling pathway in
human lung cancer. Carcinogenesis. 33:1863–1870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lin CY, Tsai PH, Kandaswami CC, Lee PP,
Huang CJ, Hwang JJ and Lee MT: Matrix metalloproteinase-9
cooperates with transcription factor Snail to induce
epithelial-mesenchymal transition. Cancer Sci. 102:815–827. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nerlov C: The C/EBP family of
transcription factors: A paradigm for interaction between gene
expression and proliferation control. Trends Cell Biol. 17:318–324.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Armstrong DA, Phelps LN and Vincenti MP:
CCAAT enhancer binding protein-beta regulates matrix
metalloproteinase-1 expression in interleukin-1beta-stimulated A549
lung carcinoma cells. Mol Cancer Res. 7:1517–1524. 2009. View Article : Google Scholar : PubMed/NCBI
|